Abstract
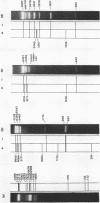
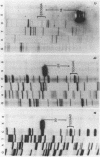
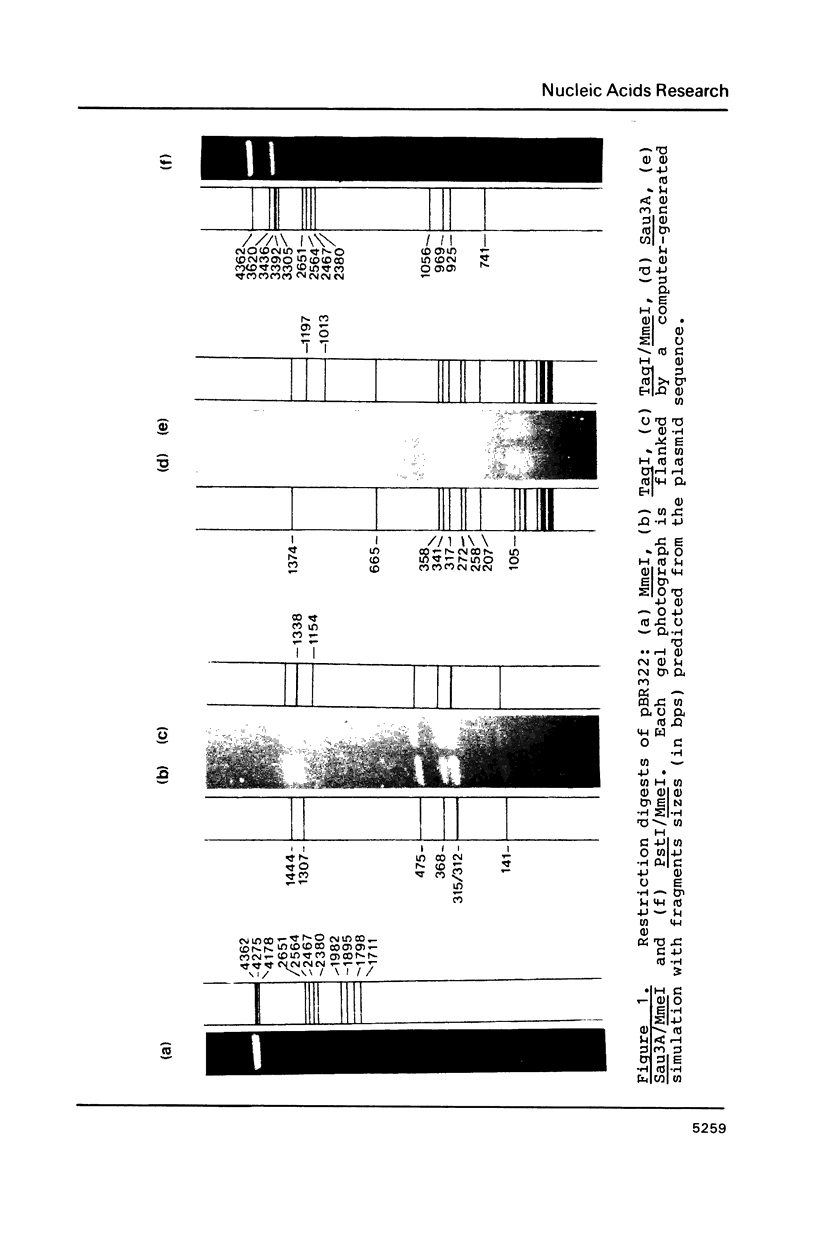
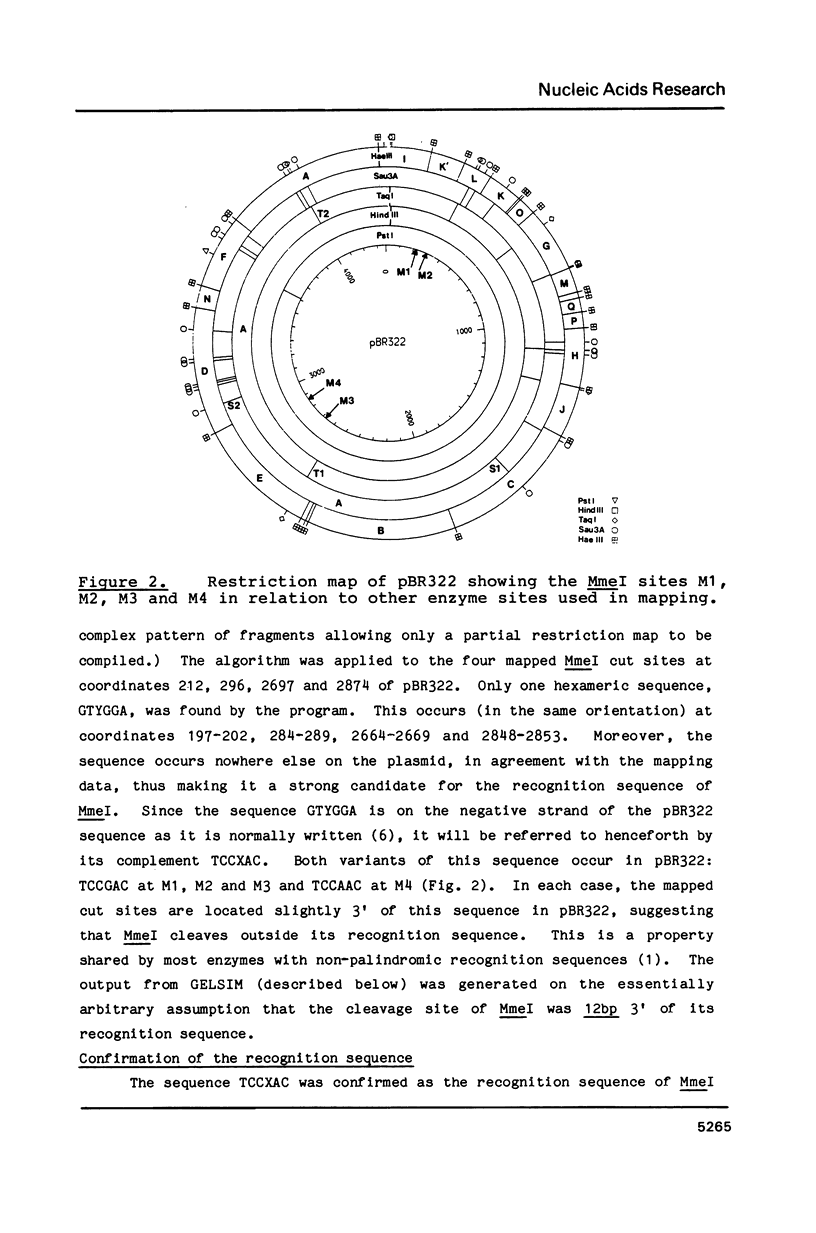
A Type II restriction endonuclease, MmeI, has been purified from the obligate methylotroph, Methylophilus methylotrophus. The enzyme was shown to have the non-palindromic recognition sequence 5'-T C C Pu A C (N)20-3', 3'-A G G Py T G (N)18-5' and to cleave (as indicated) on the 3' side, generating a two nucleotide 3' projection. Determination of the recognition sequence was achieved using two new computer programs; RECOG, which predicts recognition sequences from the pattern of restriction fragments obtained from DNAs of known sequence, and GELSIM, which generates graphical simulations of DNA band patterns obtained by gel electrophoresis of restriction digests of sequenced DNA molecules.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barker D., Hoff M., Oliphant A., White R. A second type II restriction endonuclease from Thermus aquaticus with an unusual sequence specificity. Nucleic Acids Res. 1984 Jul 25;12(14):5567–5581. doi: 10.1093/nar/12.14.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck E., Sommer R., Auerswald E. A., Kurz C., Zink B., Osterburg G., Schaller H., Sugimoto K., Sugisaki H., Okamoto T. Nucleotide sequence of bacteriophage fd DNA. Nucleic Acids Res. 1978 Dec;5(12):4495–4503. doi: 10.1093/nar/5.12.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck E., Zink B. Nucleotide sequence and genome organisation of filamentous bacteriophages fl and fd. Gene. 1981 Dec;16(1-3):35–58. doi: 10.1016/0378-1119(81)90059-7. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. L., Hutchison C. A., 3rd, Smith M. The specific non-symmetrical sequence recognized by restriction endonuclease MboII. J Mol Biol. 1980 Jun 15;140(1):143–148. doi: 10.1016/0022-2836(80)90360-5. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Haegemann G., Rogiers R., Van de Voorde A., Van Heuverswyn H., Van Herreweghe J., Volckaert G., Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978 May 11;273(5658):113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- Fuchs C., Rosenvold E. C., Honigman A., Szybalski W. A simple method for identifying the palindromic sequences recognized by restriction endonucleases: the nucleotide sequence of the AvaII site. Gene. 1978 Sep;4(1):1–23. doi: 10.1016/0378-1119(78)90011-2. [DOI] [PubMed] [Google Scholar]
- Geier G. E., Modrich P. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. J Biol Chem. 1979 Feb 25;254(4):1408–1413. [PubMed] [Google Scholar]
- Gingeras T. R., MIlazzo J. P., Roberts R. J. A computer assisted method for the determination of restriction enzyme recognifion sites. Nucleic Acids Res. 1978 Nov;5(11):4105–4127. doi: 10.1093/nar/5.11.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene P. J., Heyneker H. L., Bolivar F., Rodriguez R. L., Betlach M. C., Covarrubias A. A., Backman K., Russel D. J., Tait R., Boyer H. W. A general method for the purification of restriction enzymes. Nucleic Acids Res. 1978 Jul;5(7):2373–2380. doi: 10.1093/nar/5.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C., Corcoran M., Roberts R. J. Computer programs for handling nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):379–386. doi: 10.1093/nar/12.1part1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler C., Neumaier P. S., Wolf W. Recognition sequences of restriction endonucleases and methylases--a review. Gene. 1985;33(1):1–102. doi: 10.1016/0378-1119(85)90119-2. [DOI] [PubMed] [Google Scholar]
- Kleid D. G. Purification and properties of the HphI endonuclease. Methods Enzymol. 1980;65(1):163–166. doi: 10.1016/s0076-6879(80)65022-8. [DOI] [PubMed] [Google Scholar]
- Marinus M. G. Location of DNA methylation genes on the Escherichia coli K-12 genetic map. Mol Gen Genet. 1973 Dec 14;127(1):47–55. doi: 10.1007/BF00267782. [DOI] [PubMed] [Google Scholar]
- Matthes H. W., Zenke W. M., Grundström T., Staub A., Wintzerith M., Chambon P. Simultaneous rapid chemical synthesis of over one hundred oligonucleotides on a microscale. EMBO J. 1984 Apr;3(4):801–805. doi: 10.1002/j.1460-2075.1984.tb01888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden K. W. Revised sequence of the tetracycline-resistance gene of pBR322. Gene. 1983 May-Jun;22(2-3):277–280. doi: 10.1016/0378-1119(83)90112-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Schaffer H. E., Sederoff R. R. Improved estimation of DNA fragment lengths from Agarose gels. Anal Biochem. 1981 Jul 15;115(1):113–122. doi: 10.1016/0003-2697(81)90533-9. [DOI] [PubMed] [Google Scholar]
- Schroeder J. L., Blattner F. R. Least-squares method for restriction mapping. Gene. 1978 Oct;4(2):167–174. doi: 10.1016/0378-1119(78)90028-8. [DOI] [PubMed] [Google Scholar]
- Shinomiya T., Kobayashi M., Sato S. A second site specific endonuclease from Thermus thermophilus 111, Tth111II. Nucleic Acids Res. 1980 Aug 11;8(15):3275–3285. doi: 10.1093/nar/8.15.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Measurement of DNA length by gel electrophoresis. Anal Biochem. 1979 Dec;100(2):319–323. doi: 10.1016/0003-2697(79)90235-5. [DOI] [PubMed] [Google Scholar]
- Sproat B. S., Gait M. J. Chemical synthesis of a gene for somatomedin C. Nucleic Acids Res. 1985 Apr 25;13(8):2959–2977. doi: 10.1093/nar/13.8.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Tolstoshev C. M., Blakesley R. W. RSITE: a computer program to predict the recognition sequence of a restriction enzyme. Nucleic Acids Res. 1982 Jan 11;10(1):1–17. doi: 10.1093/nar/10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windass J. D., Worsey M. J., Pioli E. M., Pioli D., Barth P. T., Atherton K. T., Dart E. C., Byrom D., Powell K., Senior P. J. Improved conversion of methanol to single-cell protein by Methylophilus methylotrophus. Nature. 1980 Oct 2;287(5781):396–401. doi: 10.1038/287396a0. [DOI] [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- van Wezenbeek P. M., Hulsebos T. J., Schoenmakers J. G. Nucleotide sequence of the filamentous bacteriophage M13 DNA genome: comparison with phage fd. Gene. 1980 Oct;11(1-2):129–148. doi: 10.1016/0378-1119(80)90093-1. [DOI] [PubMed] [Google Scholar]