Abstract
Myosin-binding protein C (MyBP-C) is a ~130 kDa rod-shaped protein of the thick (myosin-containing) filaments of vertebrate striated muscle. It is composed of ten or eleven globular, 10-kDa domains from the immunoglobulin and fibronectin type III families, and an additional, MyBP-C-specific motif. The cardiac isoform, cMyBP-C, plays a key role in the phosphorylation-dependent enhancement of cardiac function that occurs upon β-adrenergic stimulation, and mutations in MyBP-C cause skeletal muscle and heart disease. In addition to binding to myosin, MyBP-C can also bind to actin, via its N-terminal end, potentially modulating contraction in a novel way via this thick-thin filament bridge. To understand the structural basis of actin binding, we have used negative stain electron microscopy and 3D reconstruction to study the structure of F-actin decorated with bacterially expressed N-terminal cMyBP-C fragments. Clear decoration was obtained under a variety of salt conditions varying from 25-180 mM KCl concentration. 3D helical reconstructions, carried out at the 180 mM KCl level to minimize non-specific binding, showed MyBP-C density over a broad portion of the periphery of subdomain 1 of actin and extending tangentially from its surface in the direction of actin's pointed end. Molecular fitting with an atomic structure of a MyBP-C Ig domain suggested that most of the N-terminal domains may be well ordered on actin. The location of binding was such that it could modulate tropomyosin position and would interfere with myosin head binding to actin.
Keywords: muscle structure, muscle regulation, cardiac contraction, cardiac regulation, cardiac muscle
Introduction
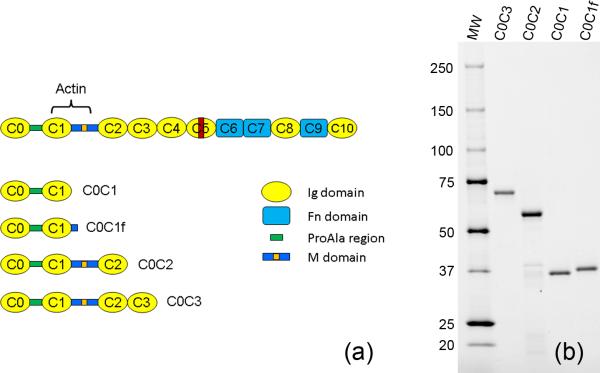
Myosin binding protein C (MyBP-C, C-protein)* is a ~130 kDa, 40 nm-long, rod-shaped accessory protein of the thick (myosin-containing) filaments of vertebrate striated muscle1-4 located in the C-zone of the A-band5-7. The skeletal isoform comprises ten tandemly arranged 10 kDa immunoglobulin (Ig) and fibronectin type III (Fn3) domains, numbered C1-C10 from the N-terminus, together with a MyBP-C-specific motif (also called the M-domain) between C1 and C2 and a Pro-Ala rich sequence at the N-terminus3, 8. The cardiac isoform (cMyBP-C) has an additional N-terminal Ig domain (C0), four phosphorylation sites in the M-domain, and a 28 residue insert in the C5 domain (Fig. 1a)3, 4, 9-11.
Figure 1. N-terminal fragments used for F-actin decoration.
(a) Layout of cMyBP-C molecule and the N-terminal fragments used. Orange, phosphorylation sites in M-domain; red, cardiac-specific insert in C5; actin, main actin-binding site. (b) SDS-PAGE of the four fragments used. MW = molecular weight markers shown in kDa (Bio-Rad Precision Plus Protein Dual Color Standards).
MyBP-C is essential for the normal functioning of striated muscle. The cardiac isoform plays a key role in the enhancement of cardiac contraction that occurs in response to β-adrenergic stimulation, and mutations in cMyBP-C are a major cause of inherited Hypertrophic Cardiomyopathy (HCM)3, 4, 11-14. In the case of skeletal MyBP-C, mutations can cause distal arthrogryposis, a disease characterized by joint contractures and abnormal muscle development15. Apart from its role in modulating cardiac contraction, MyBP-C has also been reported to function in the regulation of myosin filament assembly in sarcomerogenesis16, in stabilization of the thick filament through its phosphorylation17, and in the modulation of myosin head organization18-20.
MyBP-C attaches to the thick filament surface via its C-terminal domains (C8-C10; Fig. 1a)3. In addition, its N-terminus has been shown to bind reversibly to myosin subfragment 221, 22. Surprisingly, MyBP-C, first isolated as a myosin-binding protein, can also bind to the other major filament of the sarcomere, the thin filament. In vitro evidence for MyBP-C-actin binding was first reported on the basis of centrifugation measurements and electron microscopy23. Later studies demonstrated that binding also occurs to regulated thin filaments (containing troponin and tropomyosin) in the presence24-26 and also in the absence of Ca2+ 25, 26. Experiments with expressed MyBP-C fragments show that binding occurs via the N-terminus, primarily the C1 and M domains25, 27, 28, at a saturating 1:1 molar ratio with actin25, although additional binding through the C-terminal half has also been reported29. Electron tomography of muscle sections has directly demonstrated that MyBP-C links thick and thin filaments in intact muscle, demonstrating the physiological relevance of these in vitro studies30. In the simple model suggested by this work, the thick filament binding domains C8-C10 run longitudinally along the surface of the filament, while the rest of the molecule extends out from the thick filament, binding to neighboring thin filaments by its N-terminus20, 30, 31.
These structural studies all suggest the possibility that MyBP-C might play a key role in muscle contraction, by modulating actin-myosin filament sliding through this physical link between filaments. Support for this comes from in vitro motility observations demonstrating that actin filament sliding over myosin is slowed by MyBP-C binding to actin27, 32-36. Interestingly, this effect is weakened when the M-domain is phosphorylated25, 36. Despite its potential importance in contraction, however, the structural mechanism by which MyBP-C binding to actin might modulate filament sliding is not understood. For example, it is not known whether MyBP-C might simply act as a tether, or play a more active role, possibly by physically interfering with attachment of myosin heads, or affecting tropomyosin positioning or movement upon Ca2+-activation. Structural studies are needed to answer this question. Based on neutron scattering of F-actin decorated with the N-terminal fragment C0C2*, a structure has been proposed in which the C0 and C1 domains bind specifically to actin near the DNase I binding loop and subdomain 1, resulting in a highly regular structure37. However, this work was carried out at relatively low ionic strength, and the proposed structure depends on model-building of relatively low resolution data, leading to uncertainty in interpretation. Direct imaging by electron microscopy of negatively stained F-actin decorated with C0C2 has shown an increase in filament diameter and alterations in the Fourier transform of decorated filaments, together demonstrating regular binding of the fragment25, 38. However, the location and mode of binding of the fragment on actin was not revealed. Here we have used negative staining EM to observe F-actin decorated with N-terminal fragments, and have computed three dimensional reconstructions of the filaments that reveal the mode and location of fragment binding. Comparison of structures decorated with three different fragments has helped in interpreting the location and organization of the different MyBP-C domains. We find that the N-terminus of cMyBP-C directly interacts with F-actin in a position that would compete with the binding of myosin heads and possibly also with tropomyosin.
Results
Characterization of N-terminal MyBP-C fragments
Four N-terminal fragments of mouse cMyBP-C were expressed in E. coli (see Materials and Methods). These corresponded to domains C0C1, C0C1f, C0C2, and C0C3 (Fig. 1a). All fragments contain C0 and the Pro-Ala rich region, and C0C2 and C0C3 also contain the M-domain. C0C1f was the same as C0C1, but contained, in addition, the first fifteen amino acids of the M domain39 which have been shown to be important in binding to actin34, 36. These fragments ran with SDS-PAGE mobilities of 31, 32, 54 and 66 kDa (Fig. 1b), approximately consistent with their theoretical molecular weights of 28, 30, 50 and 61 kDa respectively; they were 85-90% pure based on gel densitometry.
Electron microscopy of F-actin decorated with N-terminal cMyBP-C fragments
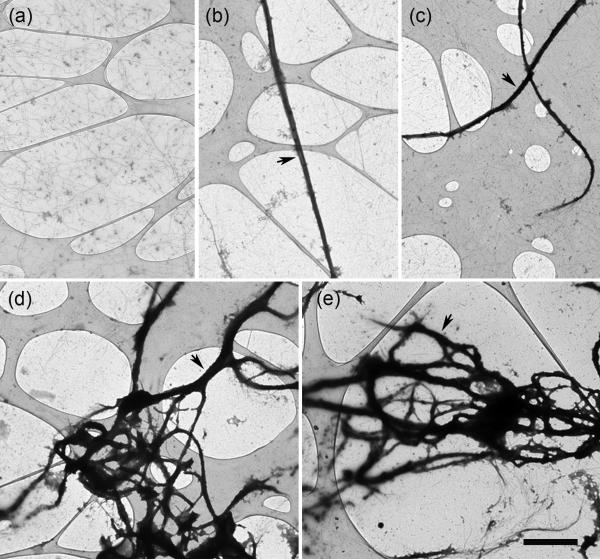
Fragments were mixed with F-actin at F-actin:cMyBP-C molar ratios of 1:1, 1:3 and 1:6 in solutions with KCl (or NaCl) concentrations varying from 25-180 mM (see Materials and Methods). The most obvious result of decoration was the bundling of actin filaments (Fig. 2b-e) compared to an F-actin control (Fig. 2a), as found previously 23, 25, 26. Bundling was present under all three ionic conditions, but was more prevalent at the lower salt levels. The amount of bundling was independent of the F-actin:cMyBP-C ratio, but significant differences were observed with the different fragments. The number and size of bundles observed with C0C1 (Fig. 2b) and C0C1f (Fig. 2c) was less than with C0C2 (Fig. 2d) and C0C3 (Fig. 2e).
Figure 2. Bundling of F-actin caused by binding of N-terminal cMyBP-C fragments.
F-actin alone (a), and decorated with C0C1 (b), C0C1f (c), C0C2 (d), and C0C3 (e), then negatively stained with 1% uranyl acetate. Decoration with C0C3 and C0C2 caused thicker bundles compared to C0C1 and C0C1f. Images are taken on holey carbon films with thin carbon over the holes (pale regions; see Materials and Methods). Single actin filaments are seen over the holes in (a). In (b)-(d), large, densely stained bundles of actin filaments are the most prominent feature (arrows), with single filaments in the background. Scale bar = 2 μm .
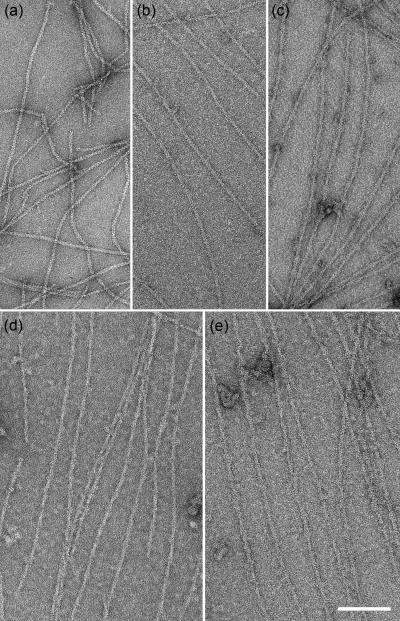
In addition to the bundles, numerous individual decorated filaments were also seen with each of the cMyBP-C fragments and in each salt condition (Fig. 3). The most obvious binding was seen with the larger fragments (C0C2 and C0C3), probably due largely to their greater size, but possibly also reflecting a greater amount of fragment bound. Binding was apparent to the eye as an increase in filament diameter (13-22 nm, depending on the fragment) compared with F-actin control filaments (10 nm; Table 1) and an enhancement of the visibility of the 36 nm crossover repeat of the F-actin double helix (Fig. 3). Decorated filaments showed lower contrast and higher background than the undecorated F-actin control due to the presence of excess unbound fragment. Details of F-actin substructure were also less apparent, presumably due to obscuring of the actin core by attachment of the fragment to the filament surface. This was most obvious with the larger fragments. Binding was usually clearer and better defined in the higher salt solutions, possibly due to reduction of non-specific binding.
Figure 3. Binding of cMyBP-C fragments to F-actin.
F-actin alone (a), F-actin decorated with C0C1 (b), C0C1f (c), C0C2 (d), and C0C3 (e). C0C1 filaments showed relatively small amounts of decoration. Scale bar = 100 nm.
Table 1. Filament diameters (nm) of decorated F-actin and F-actin control.
Values are mean ± SD (n = 20). For calculation of diameter, images were opened in ImageJ, straight regions selected, and lines drawn parallel to the filament touching the outermost edges. The distance between the lines was measured. The diameters for F-actin and C0C2-decorated filaments are very similar to published values38. Differences in filament diameter are statistically significant (p <0.05) for all pairs except A-C0C2/A-C0C3.
| Filament | Actin | A-C0C1 | A-C0C1f | A-C0C2 | A-C0C3 |
|---|---|---|---|---|---|
| Diameter | 10.3 ± 0.9 | 12.8 ± 1.0 | 16.1 ± 1.1 | 21.1 ± 1.4 | 22.2 ± 1.8 |
Three-dimensional reconstruction of F-actin decorated with N-terminal cMyBP-C fragments
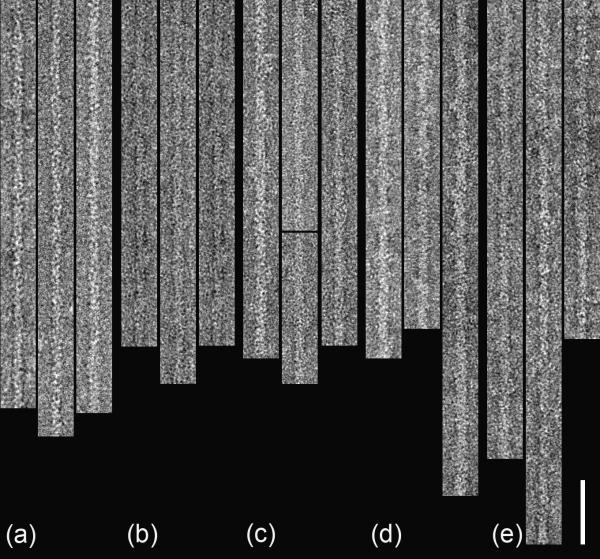
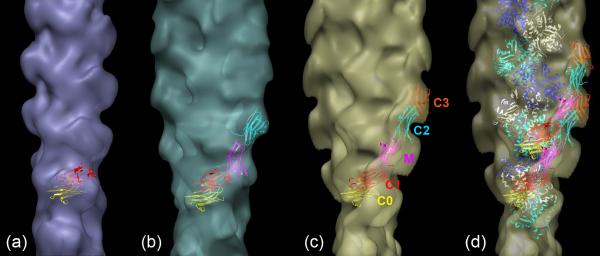
Three-dimensional helical reconstructions were carried out on selected, straightened filaments of F-actin alone and F-actin decorated with C0C3, C0C2 and C0C1f, all under the 180 mM salt conditions (used to minimize non-specific interactions; Fig. 4). Comparison of these reconstructions facilitated interpretation of the observed densities in terms of the different cMyBP-C domains. C0C1 filaments were not reconstructed as this fragment differs from C0C1f by only 1.5 kDa, and, lacking the first part of the M-domain present in C0C1f (thought to be a critical actin-binding peptide), decorated actin relatively poorly. Reconstruction of F-actin showed the well-known arrangement of four subdomains (SD) in each actin subunit, with SD1 and SD2 on the outside of the filament, and SD3 and SD4 near its center (Fig. 5a). Filaments decorated with the shortest fragment (C0C1f), revealed a compact shelf of density on actin SD1 (Fig. 5b). In the case of C0C2, similar binding on actin SD1 was observed, but with additional density projecting over SD2 (Fig. 5c). When filaments were decorated with the largest fragment, C0C3, density was again seen over the periphery of actin SD1, but now extended tangentially above the filament surface in the direction of actin's pointed end (Fig. 5d). Thus, the binding region on actin SD1 was similar for all fragments, while the additional domains present in C0C2 and C0C3 extended above actin in the direction of the pointed end.
Figure 4. Selected and straightened F-actin and decorated F-actin filaments.
(a) F-actin; (b)-(e) decorated with C0C1, C0C1f, C0C2, and C0C3, respectively. Scale bar = 50 nm.
Figure 5. Three-dimensional reconstructions of F-actin and F-actin decorated with cMyBP-C N-terminal fragments.
(a) F-actin; (b)-(d) decorated with C0-C1f, C0-C2, and C0-C3, respectively. (e) Enlargement of C0C3-decorated filament (d), fitted with ribbon depiction of F-actin atomic model40 (individual actin monomers are colored white, blue or cyan). Numbers on (a) indicate actin subdomains. Arrows point to additional density in decorated filaments. Filaments oriented with pointed end at top.
To better understand the molecular arrangement in these structures, we fitted an atomic model of F-actin40 into both the F-actin (Supplementary Fig. 1) and C0C3-decorated reconstructions (Fig. 5e). The excellent fit to F-actin clearly illustrates the organization of actin's four subdomains and validates this procedure for the decorated filaments. Fitting to the reconstruction of the C0C3-decorated filament confirmed that the major region of binding to actin was on SD1, centered approximately over actin's N-terminus. Insight into the organization of the cMyBP-C domains was gained by determining how the crystal structure of an Ig domain (C1 of cMyBP-C; PDB 3cx241) fitted into the cMyBP-C density in the three reconstructions. While the resolution of the reconstructions is limited and there may be some flexibility in the MyBP-C fragments, this approximate fitting provides an indication of the amount of MyBP-C fragment visualized and the possible orientation and location of the different domains. The different fragments (C0C1f, C0C2 and C0C3) consist of two, three and four Ig domains (C0, C1, C2, and C3) respectively, as well as the Pro-Ala-rich region between C0 and C1, and, in the case of C0C2 and C0C3, the M-domain as well. Although the structure of the M-domain is not yet known, it is similar in molecular weight to an Ig domain and, while extensible42, is thought to have a compact structure in solution similar to an Ig domain43. As a first approximation, we therefore modeled C0C1f, C0C2 and C0C3 as strings of two, four or five Ig domains respectively, and determined how these structures fitted into the cMyBP-C density in the different reconstructions.
Figure 6 shows that there is sufficient cMyBP-C density volume to accommodate all of the fitted domains in each case, suggesting that most of each fragment is present in the reconstructed volume and thus is relatively well ordered on actin (see also Supplementary Movie 1). The cMyBP-C density is best accounted for with C0 and C1 binding to SD1, the putative M-domain bridging over SD2 and possibly connecting SD1s of adjacent actins (cf. 44), and the C2 and C3 domains lying above the actin surface at higher radius, extending in the direction of the pointed end of the filament.
Figure 6. Fitting of Ig domains to MyBP-C fragment density on decorated actin reconstructions.
(a) Two domains fitted to C0C1f; (b) four domains fitted to C0C2; (c) five domains fitted to C0C3, with different domains identified; (d) fitting of actin atomic model (white, blue and cyan monomers) into (c); fitting of two C0C3 fragments in this case shows how the total fragment volume associated with a single actin monomer can be accounted for by overlapping MyBP-C molecules (e.g. C3 of lower molecule next to M of upper molecule).
Discussion
The concept that MyBP-C might bridge between thick and thin filaments and thus modulate muscle contraction has existed ever since it was first shown that this myosin-binding protein could also bind to F-actin23. This possibility has been strengthened by subsequent in vitro studies showing binding to regulated thin filaments24, 26, and the location of the interaction site has been narrowed to the N-terminal region, primarily the C1 and M domains25, 27, 28 (see also29). In vitro motility studies demonstrate that cMyBP-C slows actin-myosin sliding, an effect that is reduced by cMyBP-C phosphorylation27, 33-36. A simple interpretation of these results is that cMyBP-C acts as a phosphorylation-dependent tether between the two filaments, modulating their sliding. Direct observation of MyBP-C links between thick and thin filaments in intact muscle suggests that actin interaction is not an in vitro artifact, but occurs in vivo30. Our study of the binding of N-terminal fragments of MyBP-C to F-actin was designed to provide new insights into the nature of the actin end of this putatively crucial interaction.
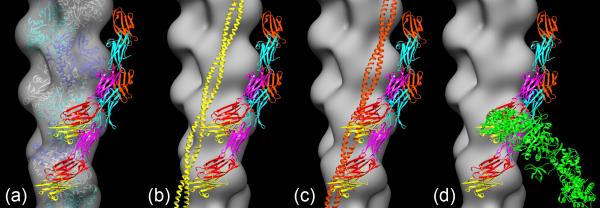
Our EM images and reconstructions directly support the view that it is the N-terminal domains of cMyBP-C that bind to actin25, 27, 28. First, even the shortest, most N-terminal fragments, C0C1 and C0C1f, appear to bind to actin. Second, we find that the diameter of the decorated filaments increases with fragment length (Table 1). All the fragments have C0 and C1 in common, and elongate in the C-terminal direction when additional domains are added (Fig. 1a). Assuming that the attachment site to actin is similar in all cases, the increase in diameter is consistent with the addition of C2 and C3 distal to the binding site and above the filament surface, rather than being directly attached to actin. The reconstructions support this view. C0C1f-decorated filaments show a small protrusion on SD1 of actin. With the addition of C2 and then C3, the attached density increases in length and extends further above the surface of the actin filament core in the direction of the pointed end. However, the actin attachment site itself appears similar in each case, covering a region of SD1 centered over the N-terminus of actin. While there is substantial coincidence between the distal, C-terminal regions of the C0C2 and C0C3 fragments in the two reconstructions, there appeared to be some variation in their precise positions. This may reflect some flexibility in the molecule (e.g. due to flexible linkers between the globular domains). In addition, as each domain is added, the fragments could adopt different conformations in their C-termini made possible by new potential interactions between the added domains. An example is shown with C0C3, where the C-terminal domains of one molecule come close to the more N-terminal domains of the next MyBP-C up (Fig. 7a, Supplementary Movie 2). While interaction between domains of neighboring MyBP-C molecules may be possible in this synthetic structure, it would not occur in intact muscle, being precluded by the low MyBP-C:myosin stoichiometry and large (43 nm) axial distance between neighboring MyBP-Cs on the thick filament (see later).
Figure 7. Best-fit position of two C0C3 fragments in C0C3 reconstruction (Fig. 6d), superimposed on F-actin reconstruction and shown in relation to tropomyosin and myosin head positions.
(a) F-actin reconstruction fitted with F-actin atomic model40. (b) Fitting of tropomyosin (yellow coiled-coil) in closed (high Ca2+) position45 to F-actin reconstruction with best-fit C0C3: there is little or no clash between C0C3 and tropomyosin. (c) Fitting of tropomyosin (orange coiled-coil) in blocked (low Ca2+) position45 to F-actin reconstruction with best-fit C0C3: there appears to be a clash between C0 and C1 domains and tropomyosin. (d) Fitting of myosin head (S1) in rigor position (green) to F-actin reconstruction with best-fit C0C3, showing clear steric clash of S1 with C0C3.
While most studies suggest that binding of MyBP-C's N-terminal region to actin is specific25, 27, 32, 36, this view is not universal, and it has been suggested that specific binding to actin might actually involve the C-terminal half of the molecule29. Our observation that N-terminal fragments bind in a regular way to the actin helix, as demonstrated by Fourier transforms of decorated filaments (not shown) supports the specificity of the interaction. The same conclusion was reached based on the appearance of filtered images and of actin layer lines in Fourier transforms of filaments decorated with C0C2, also under these salt conditions38. Specific binding is also indicated by our 3D reconstructions (also implied by44). Our use of helical reconstruction methods ensures that only features having the same helical symmetry as F-actin appear in the final density map; the presence in the reconstructions of substantial mass attached to actin with the actin helical symmetry implies a regular and specific interaction. Finally, while our experiments were carried out at three salt levels, all producing similar looking decoration, the highest was 180 mM which would minimize non-specific electrostatic binding; it was this level that was used for our reconstructions. We conclude that there is substantial regularity and hence specificity to the interaction with actin.
To understand whether and how MyBP-C might modulate filament interaction and contraction in muscle, we have compared its location on actin to that of tropomyosin and myosin heads. Fitting atomic models of thin filaments in high and low Ca2+ states45 to the reconstruction of C0C3-decorated filaments suggests that MyBP-C would not significantly interfere with tropomyosin in the high Ca2+ position (the “closed” state), but that there may be some interference in the low Ca2+ position (the “blocked” state; Fig. 7b, c, Supplementary Movies 3, 4). (As tropomyosin is located even further away from SD1 in the myosin-induced (“open”) state45, 46, any clash with cMyBP-C is even less likely than in the closed state; Supplementary Fig. 2.) However, there is a clear steric interference between the binding sites of MyBP-C and strongly bound myosin heads (rigor or end-of-power-stroke heads) on actin SD1, and this would prevent both from binding simultaneously (Fig. 7d, Supplementary Movie 5).
These structural results are in good agreement with biochemical and other observations. Several studies have shown that MyBP-C (and N-terminal fragments) can bind to thin filaments at high Ca2+ 24-26 and to actin-tropomyosin without troponin23: in both cases tropomyosin is in the closed position, which does not appear to clash with cMyBP-C in our reconstruction. In the low Ca2+ state (blocked position) binding is also found, although it may be weaker25, 26, consistent with the apparent partial overlap of tropomyosin and cMyBP-C binding positions seen in the reconstruction (Fig. 7b, c, Supplementary Movies 3, 4; 37). Electron tomography of muscle sections in the relaxed (low Ca2+) state shows that MyBP-C extends out from thick filaments and binds to thin filaments in the intact filament lattice30, suggesting that any steric interference in relaxed muscle in vivo is minimal. Such interactions may serve in vivo to organize and stabilize the filament lattice in the relaxed state. The strong binding of S1 to actin that occurs in the absence of ATP has been shown to greatly reduce MyBP-C binding23, 32, consistent with our observation of overlapping binding sites. Indeed, S1's high affinity for actin in the ATP-free state is sufficient to displace MyBP-C23, 32. The extension of MyBP-C axially over two actin subunits deduced from our fitting readily explains why S1 bound to actin at only half saturation is able to compete off essentially all MyBP-C attached to actin filaments23. In the presence of ATP, when myosin heads are actively cycling and bind more weakly to actin, both whole MyBPC and N-terminal fragments inhibit actomyosin ATPase activity and S1 binding23, 27, 32, again consistent with the location of cMyBP-C's N-terminal binding site on actin SD1.
In vitro motility assays of the effect of C0C2 on the movement of regulated thin filaments (containing tropomyosin and troponin) show that thin filaments at high Ca2+ (and also F-actin alone) are slowed by the presence of C0C2, whereas at low Ca2+ their velocity is increased in the presence of C0C227. Our reconstruction suggests a possible explanation in terms of the binding sites of cMyBP-C and tropomyosin-troponin. At high Ca2+ (tropomyosin in the closed position45) cMyBP-C and tropomyosin binding sites do not overlap, which would allow uninhibited binding of cMyBP-C to its site on actin SD1. Its binding could thus exert a significant drag on thin filament sliding. In contrast, the reconstruction suggests that cMyBP-C and tropomyosin compete for part of the same SD1 surface of actin when tropomyosin is in the low Ca2+ (blocking) position. If so, cMyBP-C could destabilize tropomyosin in its blocking position, tending to activate the thin filament, thus enhancing its motility.
If comparable effects occur in vivo, this would suggest that cMyBP-C may contribute (along with troponin) to a delicate balance between the blocked and closed positions of tropomyosin, poising the thin filament between its switched-off and switched-on regulatory states. In the case of cardiac MyBP-C, phosphorylation weakens MyBP-C actin binding, which could contribute to cMyBP-C's role in modulating cardiac activity. Other factors also come into play in intact muscle. The MyBP-C:actin molar ratio in the myofibril is approximately 1:30, and thus thin filaments could never be highly decorated in muscle as they are in our in vitro experiments. In addition, MyBP-C is confined to the middle third of each half sarcomere and here occurs only every 43 nm (similar to the actin helical repeat). Thus only a limited region of the thin filament can interact with MyBP-C at any particular sarcomere length, and here the interactions must be relatively sparse. Nevertheless, in light of the substantial stiffness of tropomyosin47, 48 and the consequent ability of structural changes to be transmitted considerable distances along the thin filament46, even these limited interactions could have significant effects in vivo. Such interactions, of course, must necessarily be relatively weak and in rapid equilibrium to enable the sliding of filaments that is essential for muscle shortening.
The arrangement of the C0C3 fragment on F-actin is such that the C0, C1 and M domains bind on or near SD1 while C2 and C3 extend to higher radius, lying above the next actin subunit in the direction of the pointed end (Fig. 7, Supplementary Movies 2-5). This domain arrangement is quite similar to that proposed by Whitten et al.37 based on modeling of neutron scattering data from C0C2-decorated F-actin, except that the M-domain in their model is well above, and does not bind to, the actin surface. Our fitting is consistent with the conclusion from comparative binding studies of different N-terminal fragments that the M-domain is critical for actin binding, and may interact with actin through electrostatic attraction between its high concentration of positively charged residues and negative charge on actin SD125, 27, 36. The stronger and more obvious binding that we see with C0C1f compared with C0C1 (which lacks the first part of the M-domain), supports a role of the M-domain in direct binding to actin. As suggested by Shaffer et al.25, the higher pH (8.0) of the neutron scattering experiments may cause the M-domain to dissociate from actin, accounting for its different positioning compared with our reconstruction (pH 7.4). The potential clash between the C0 and C1 domains and tropomyosin at low but not high Ca2+ that we observe is also supported by the model of Whitten et al.37 based on neutron scattering.
We conclude that cMyBP-C binds by its N-terminal domains to SD1 of actin at a site that can potentially modulate tropomyosin and myosin head binding in different physiological states. These findings, together with recent information showing that MyBP-C can bind to thin filaments in intact muscle30 and its known ability to bind to myosin subfragment 2 in a phosphorylation-dependent way21, 22, add support for and insight into the mounting evidence that MyBP-C plays a key role in muscle contraction.
Materials and Methods
Proteins
F-actin was purified from chicken pectoralis muscle according to Pardee et al.49. cMyBP-C fragments were bacterially expressed from mouse cardiac cDNA using the pET expression system (Novagen, Madison, WI). Four expressed N-terminal domains, C0C1 (amino acids 1-254), C0C1f (1-269), C0C2 (1-448), and C0C3 (1-539) were studied (Fig. 1a). The cDNA fragments were generated by PCR, incorporating compatible restriction enzyme sites for cloning into the pET28a vector, with 5’ Nco1 and a 3’ Xho1 sites. The Xho1 site was made so that the fragments were cloned in frame with a C-terminal 6-His tag. Protein expression was in BL21 (DE3) cells, grown for 4 hours after induction with IPTG. Cells were collected by centrifugation and lysed using the xTractor Buffer supplied in the HisTalon Buffer set (Clontech). Protein was purified according to the HisTalon protocol, using HisPur Cobalt Resin (Thermo). Protein extract from three liters of cells was passed over two 10 ml cobalt columns. The protein was eluted, the OD determined and the samples with the highest protein concentrations analyzed on an acrylamide gel. The samples of greater purity were pooled, dialyzed overnight against PBS, aliquoted and flash frozen in liquid nitrogen with storage at -80° C.
Electron microscopy
5 μM F-actin and 5-30 μM expressed fragment were mixed together under three different buffer conditions, each of which has been used in previous thin filament structural studies. The major difference between them was the KCl/NaCl concentration and hence the ionic strength. (1) 25 mM KCl, 4 mM MgCl2, 1 mM EGTA, 10 mM DTT, 1 mM ATP, 25 mM Imidazole, pH 7.434; (2) 100 mM NaCl, 3 mM MgCl2, 0.2 mM EGTA, 1 mM NaN3, 5 mM sodium phosphate, 5 mM Pipes, pH 7.148; (3) 180 mM KCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, 1 mM ATP, 20 mM imidazole, pH 7.425. Samples were incubated at room temperature for 30 min. 5 μl aliquots were then applied to EM grids coated with thin carbon supported by a holey carbon film and negatively stained with 1% uranyl acetate50. Dried grids were observed in a Philips CM120 electron microscope (FEI, Hillsboro, OR) at 80 KV under low dose conditions. Images of filaments were acquired at a pixel size of 0.35 nm, using a 2K × 2K CCD camera (F224HD, TVIPS GmbH, Gauting, Germany).
3D reconstruction
Helical reconstruction of filaments and averaging of reconstructions was carried out according to standard Fourier-based methods, using the Brandeis Helical Package51, 52. The resolutions of all the reconstructions were 2.5-3.0 nm51. Iterative helical real space reconstruction (IHRSR), also carried out (using SPIDER53), confirmed the results of helical reconstruction of the smallest (C0C1f) fragment, but was not as useful in preliminary reconstructions of filaments heavily decorated with the larger fragments, where projection matching was imprecise. UCSF Chimera54 was used for visualization, analysis, and atomic fitting of 3D volumes.
Supplementary Material
Acknowledgments
We thank Samantha Beck-Previs for making the F-actin. This work was supported by NIH grants AR034711 to RC, HL036153 to WL, and Program Project Grants P01 HL059408 to David Warshaw (RC, PI on Project 1, JR on Project 3) and P01 HL069779 to JR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: MyBP-C, myosin-binding protein C; cMyBP-C, cardiac myosin-binding protein C; 3D, three-dimensional; SD, subdomain
In the naming convention used, the fragment starts with the first domain listed, ends with the last domain, and includes all intervening domains in the structure. Thus C0C2 contains C0, the ProAla-rich domain, C1, the M-domain and C2.
References
- 1.Offer G, Moos C, Starr R. A new protein of the thick filaments of vertebrate skeletal myofibrils. Extraction, purification and characterization. J. Mol. Biol. 1973;74:653–676. doi: 10.1016/0022-2836(73)90055-7. [DOI] [PubMed] [Google Scholar]
- 2.Bennett PM, Furst DO, Gautel M. The C-protein (myosin binding protein C) family: regulators of contraction and sarcomere formation? Rev. Physiol Biochem. Pharmacol. 1999;138:203–234. doi: 10.1007/BFb0119628. [DOI] [PubMed] [Google Scholar]
- 3.Flashman E, Redwood C, Moolman-Smook J, Watkins H. Cardiac myosin binding protein C: its role in physiology and disease. Circ. Res. 2004;94:1279–1289. doi: 10.1161/01.RES.0000127175.21818.C2. [DOI] [PubMed] [Google Scholar]
- 4.Barefield D, Sadayappan S. Phosphorylation and function of cardiac myosin binding protein-C in health and disease. J. Mol. Cell Cardiol. 2010;48:866–875. doi: 10.1016/j.yjmcc.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craig R, Offer G. The location of C-protein in rabbit skeletal muscle. Proc. R. Soc. Lond B Biol. Sci. 1976;192:451–461. doi: 10.1098/rspb.1976.0023. [DOI] [PubMed] [Google Scholar]
- 6.Luther PK, Bennett PM, Knupp C, Craig R, Padron R, Harris SP, Patel J, Moss RL. Understanding the organisation and role of myosin binding protein C in normal striated muscle by comparison with MyBP-C knockout cardiac muscle. J. Mol. Biol. 2008;384:60–72. doi: 10.1016/j.jmb.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett P, Craig R, Starr R, Offer G. The ultrastructural location of C-protein, X-protein and H-protein in rabbit muscle. J. Muscle Res. Cell Motil. 1986;7:550–567. doi: 10.1007/BF01753571. [DOI] [PubMed] [Google Scholar]
- 8.Einheber S, Fischman DA. Isolation and characterization of a cDNA clone encoding avian skeletal muscle C-protein: an intracellular member of the immunoglobulin superfamily. Proc. Natl. Acad. Sci. U. S. A. 1990;87:2157–2161. doi: 10.1073/pnas.87.6.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia W, Shaffer JF, Harris SP, Leary JA. Identification of novel protein kinase A phosphorylation sites in the M-domain of human and murine cardiac myosin binding protein-C using mass spectrometry analysis. J. Proteome. Res. 2010;9:1843–1853. doi: 10.1021/pr901006h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yasuda M, Koshida S, Sato N, Obinata T. Complete primary structure of chicken cardiac C-protein (MyBP-C) and its expression in developing striated muscles. J. Mol. Cell Cardiol. 1995;27:2275–2286. doi: 10.1016/s0022-2828(95)91731-4. [DOI] [PubMed] [Google Scholar]
- 11.Gautel M, Zuffardi O, Freiburg A, Labeit S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: a modulator of cardiac contraction? EMBO J. 1995;14:1952–1960. doi: 10.1002/j.1460-2075.1995.tb07187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watkins H, Conner D, Thierfelder L, Jarcho JA, MacRae C, McKenna WJ, Maron BJ, Seidman JG, Seidman CE. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat. Genet. 1995;11:434–437. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- 13.Vikstrom KL, Leinwand LA. Contractile protein mutations and heart disease. Curr. Opin. Cell Biol. 1996;8:97–105. doi: 10.1016/s0955-0674(96)80053-6. [DOI] [PubMed] [Google Scholar]
- 14.Dhandapany PS, Sadayappan S, Xue Y, Powell GT, Rani DS, Nallari P, Rai TS, Khullar M, Soares P, Bahl A, Tharkan JM, Vaideeswar P, Rathinavel A, Narasimhan C, Ayapati DR, Ayub Q, Mehdi SQ, Oppenheimer S, Richards MB, Price AL, Patterson N, Reich D, Singh L, Tyler-Smith C, Thangaraj K. A common MYBPC3 (cardiac myosin binding protein C) variant associated with cardiomyopathies in South Asia. Nat. Genet. 2009;41:187–191. doi: 10.1038/ng.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurnett CA, Desruisseau DM, McCall K, Choi R, Meyer ZI, Talerico M, Miller SE, Ju JS, Pestronk A, Connolly AM, Druley TE, Weihl CC, Dobbs MB. Myosin binding protein C1: a novel gene for autosomal dominant distal arthrogryposis type 1. Hum. Mol. Genet. 2010;19:1165–1173. doi: 10.1093/hmg/ddp587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Z, Lu MH, Schultheiss T, Choi J, Holtzer S, DiLullo C, Fischman DA, Holtzer H. Sequential appearance of muscle-specific proteins in myoblasts as a function of time after cell division: evidence for a conserved myoblast differentiation program in skeletal muscle. Cell Motil. Cytoskeleton. 1994;29:1–19. doi: 10.1002/cm.970290102. [DOI] [PubMed] [Google Scholar]
- 17.Sadayappan S, Osinska H, Klevitsky R, Lorenz JN, Sargent M, Molkentin JD, Seidman CE, Seidman JG, Robbins J. Cardiac myosin binding protein C phosphorylation is cardioprotective. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16918–16923. doi: 10.1073/pnas.0607069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weisberg A, Winegrad S. Alteration of myosin cross bridges by phosphorylation of myosin-binding protein C in cardiac muscle. Proc. Natl. Acad. Sci. U. S. A. 1996;93:8999–9003. doi: 10.1073/pnas.93.17.8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine R, Weisberg A, Kulikovskaya I, McClellan G, Winegrad S. Multiple structures of thick filaments in resting cardiac muscle and their influence on cross-bridge interactions. Biophys. J. 2001;81:1070–1082. doi: 10.1016/S0006-3495(01)75764-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Starr R, Offer G. The interaction of C-protein with heavy meromyosin and subfragment-2. Biochem. J. 1978;171:813–816. doi: 10.1042/bj1710813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruen M, Prinz H, Gautel M. cAPK-phosphorylation controls the interaction of the regulatory domain of cardiac myosin binding protein C with myosin-S2 in an on-off fashion. FEBS Lett. 1999;453:254–259. doi: 10.1016/s0014-5793(99)00727-9. [DOI] [PubMed] [Google Scholar]
- 23.Moos C, Mason CM, Besterman JM, Feng IN, Dubin JH. The binding of skeletal muscle C-protein to F-actin, and its relation to the interaction of actin with myosin subfragment-1. J. Mol. Biol. 1978;124:571–586. doi: 10.1016/0022-2836(78)90172-9. [DOI] [PubMed] [Google Scholar]
- 24.Moos C. Fluorescence microscope study of the binding of added C protein to skeletal muscle myofibrils. J. Cell Biol. 1981;90:25–31. doi: 10.1083/jcb.90.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaffer JF, Kensler RW, Harris SP. The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner. J. Biol. Chem. 2009;284:12318–12327. doi: 10.1074/jbc.M808850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto K. The binding of skeletal muscle C-protein to regulated actin. FEBS Lett. 1986;208:123–127. doi: 10.1016/0014-5793(86)81545-9. [DOI] [PubMed] [Google Scholar]
- 27.Razumova MV, Shaffer JF, Tu AY, Flint GV, Regnier M, Harris SP. Effects of the N-terminal domains of myosin binding protein-C in an in vitro motility assay: Evidence for long-lived cross-bridges. J. Biol. Chem. 2006;281:35846–35854. doi: 10.1074/jbc.M606949200. [DOI] [PubMed] [Google Scholar]
- 28.Kulikovskaya I, McClellan G, Flavigny J, Carrier L, Winegrad S. Effect of MyBP-C binding to actin on contractility in heart muscle. J. Gen. Physiol. 2003;122:761–774. doi: 10.1085/jgp.200308941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rybakova IN, Greaser ML, Moss RL. Myosin binding protein C interaction with actin: characterization and mapping of the binding site. J. Biol. Chem. 2011;286:2008–2016. doi: 10.1074/jbc.M110.170605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luther P, Winkler H, Taylor K, Zoghbi ME, Craig R, Padrón R, Squire J, Liu J. Myosin binding protein C bridges myosin and actin filaments in intact muscle. Proc. Natl. Acad. Sci. U. S. A. 2011 doi: 10.1073/pnas.1103216108. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Squire JM, Luther PK, Knupp C. Structural evidence for the interaction of C-protein (MyBP-C) with actin and sequence identification of a possible actin-binding domain. J. Mol. Biol. 2003;331:713–724. doi: 10.1016/s0022-2836(03)00781-2. [DOI] [PubMed] [Google Scholar]
- 32.Saber W, Begin KJ, Warshaw DM, VanBuren P. Cardiac myosin binding protein-C modulates actomyosin binding and kinetics in the in vitro motility assay. J. Mol. Cell Cardiol. 2008;44:1053–1061. doi: 10.1016/j.yjmcc.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Previs MJ, Previs SB, Robbins J, Warshaw DM. Cardiac myosin binding protein-C (MyBP-C) impedes actin filament motility on native mouse ventricular thick filaments only within the C-zone . Biophys. J. 2011;100:370a. [Google Scholar]
- 34.Weith A, Sadayappan S, VanBuren P, Robbins J, Warshaw DM. N-terminal fragments of cardiac myosin binding protein-C inhibit actomyosin motility by tethering actin. Biophys. J. 2010;98:756a. [Google Scholar]
- 35.Weith A, Gulick J, VanBuren P, Robbins J, Warshaw DM. N-terminal fragment of cardiac myosin binding protein-C (cMyBP-C) reduces actomyosin power output in the laser trap assay. Biophys. J. 2011;100:454a. [Google Scholar]
- 36.Weith A, Sadayappan S, VanBuren P, Robbins J, Warshaw D. Cardiac myosin binding protein-C inhibits actomyosin motility by tethering actin. 2011. In preparation. [DOI] [PMC free article] [PubMed]
- 37.Whitten AE, Jeffries CM, Harris SP, Trewhella J. Cardiac myosin-binding protein C decorates F-actin: Implications for cardiac function. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18360–18365. doi: 10.1073/pnas.0808903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kensler RW, Shaffer JF, Harris SP. Binding of the N-terminal fragment C0-C2 of cardiac MyBP-C to cardiac F-actin. J. Struct. Biol. 2011;174:44–51. doi: 10.1016/j.jsb.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sadayappan S, Greis KD, Robbins J. Phosphorylation-dependent proteolysis and pathogenesis of cardiac myosin binding protein-C. J. Mol. Cell. Cardiol. 2008;44:S44. [Google Scholar]
- 40.Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 41.Fisher SJ, Helliwell JR, Khurshid S, Govada L, Redwood C, Squire JM, Chayen NE. An investigation into the protonation states of the C1 domain of cardiac myosin-binding protein C. Acta Crystallogr. D. Biol. Crystallogr. 2008;64:658–664. doi: 10.1107/S0907444908008792. [DOI] [PubMed] [Google Scholar]
- 42.Karsai A, Harris SP, Kellermayer M. The motif of myosin binding protein-C is mechanically weak and extensible. Biophys. J. 2011;100:453a–454a. [Google Scholar]
- 43.Jeffries CM, Whitten AE, Harris SP, Trewhella J. Small-angle X-ray scattering reveals the N-terminal domain organization of cardiac myosin binding protein C. J. Mol. Biol. 2008;377:1186–1199. doi: 10.1016/j.jmb.2008.01.080. [DOI] [PubMed] [Google Scholar]
- 44.Orlova A, Galkin VE, Jeffries CM, Trewhella J, Egelman EH. Binding of N-terminus fragments of cardiac myosin-binding C-protein to actin. Biophys. J. 2010;98:157a. [Google Scholar]
- 45.Poole KJ, Lorenz M, Evans G, Rosenbaum G, Pirani A, Craig R, Tobacman LS, Lehman W, Holmes KC. A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle X-ray fibre diagrams from non-overlap muscle. J. Struct. Biol. 2006;155:273–284. doi: 10.1016/j.jsb.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 46.Vibert P, Craig R, Lehman W. Steric-model for activation of muscle thin filaments. J. Mol. Biol. 1997;266:8–14. doi: 10.1006/jmbi.1996.0800. [DOI] [PubMed] [Google Scholar]
- 47.Sousa D, Cammarato A, Jang K, Graceffa P, Tobacman LS, Li XE, Lehman W. Electron microscopy and persistence length analysis of semi-rigid smooth muscle tropomyosin strands. Biophys. J. 2010;99:862–868. doi: 10.1016/j.bpj.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li XE, Tobacman LS, Mun JY, Craig R, Fischer S, Lehman W. Tropomyosin position on F-actin revealed by EM reconstruction and computational chemistry. Biophys. J. 2011;100:1005–1013. doi: 10.1016/j.bpj.2010.12.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85 Pt B:164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 50.Craig R, Lehman W. Crossbridge and tropomyosin positions observed in native, interacting thick and thin filaments. J. Mol. Biol. 2001;311:1027–1036. doi: 10.1006/jmbi.2001.4897. [DOI] [PubMed] [Google Scholar]
- 51.Lehman W, Hatch V, Korman V, Rosol M, Thomas L, Maytum R, Geeves MA, Van Eyk JE, Tobacman LS, Craig R. Tropomyosin and actin isoforms modulate the localization of tropomyosin strands on actin filaments. J. Mol. Biol. 2000;302:593–606. doi: 10.1006/jmbi.2000.4080. [DOI] [PubMed] [Google Scholar]
- 52.Owen CH, Morgan DG, DeRosier DJ. Image analysis of helical objects: the Brandeis Helical Package. J. Struct. Biol. 1996;116:167–175. doi: 10.1006/jsbi.1996.0027. [DOI] [PubMed] [Google Scholar]
- 53.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 54.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.