Abstract
HIV-1 integrase (IN) is one of three essential enzymes for viral replication, and a focus of ardent antiretroviral drug discovery and development efforts. Diligent research has led to the development of the strand transfer specific chemical class of IN inhibitors, with two compounds from this group, raltegravir and elvitegravir, advancing the farthest in the US FDA approval process for any IN inhibitor discovered thus far. Raltegravir, developed by Merck & Co., has been US FDA approved for HIV-1 therapy, whereas elvitegravir, developed by Gilead Sciences and Japan Tobacco, has reached Phase III clinical trials. Although this is an undoubted success for the HIV-1 IN drug discovery field, the development of HIV-1 IN strand transfer specific drug resistant viral strains upon clinical use of the compounds is expected in the patient. Furthermore the problem of strand transfer specific IN drug resistance will be exacerbated by the development of cross-resistant viral strains due to an overlapping binding orientation at the IN active site and an equivalent inhibitory mechanism for the two compounds. This inevitability will result in no available IN-targeted therapeutic options for HIV-1 treatment experienced patients. The development of allosterically targeted IN inhibitors presents an extremely advantageous approach for the discovery of compounds effective against IN strand transfer drug resistant viral strains, and would likely show synergy with all available FDA approved antiretroviral HIV-1 therapeutics, including the IN strand transfer specific compounds. Here we review the concept of allosteric IN inhibition, and the small molecules that have been investigated to bind non-active site regions to inhibit IN function.
Keywords: HIV-1 integrase, small molecule, inhibitors, allosteric, drug design
Introduction
HIV-1 integrase (IN) is one of three viral enzymes essential for viral replication and infectivity. Initially IN catalyzes the cleavage of a dinucleotide from each 3'-termini of the reverse transcribed viral DNA in the cytosol; a reaction termed 3'-processing which results in two 3'-recessed hydroxyl groups. This is followed by nuclear translcocation of IN, the processed viral DNA, and numerous cellular and viral proteins in a large nucleo-protein complex termed the preintegration complex. In the nuclear environment IN inserts the processed viral DNA into the host cell genome in a hydroxyl-mediated nucleophilic reaction termed strand transfer (Figure 1A).[1] Following viral DNA insertion into the cellular genome, late stage viral life cycle events, such as viral DNA transcription and translation by cellular machinery, virion assembly and budding commence to facilitate the generation of infectious virions and further viral propagation.
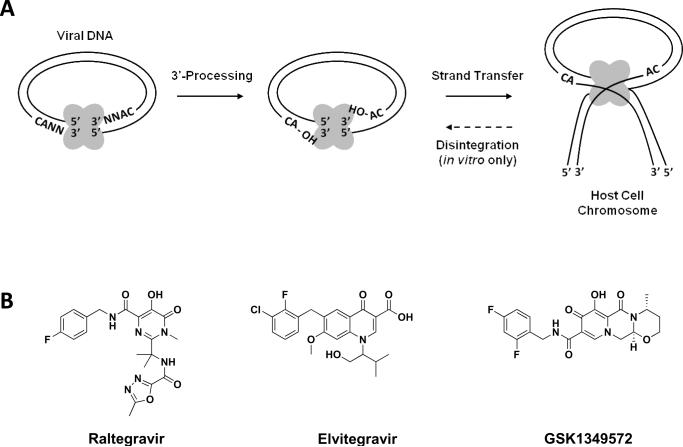
Figure 1.
(A) IN enzymatic reactions 3'-processing and strand transfer occur in vivo and can be mimicked in vitro with an oligonucleotide substrate, purified enzyme, and metal cofactor. IN is additionally capable of the reverse strand transfer reaction termed disintegration in vitro only. Oligomeric IN enzyme shown in grey. (B) Chemical structures of the clinically progressed strand transfer specific IN inhibitors raltegravir, elvitegravir, and the second generation strand transfer specific IN inhibitor GSK1349572.
IN drug design and discovery has mainly focused on the direct inhibition of enzyme catalytic activities due to the early advent of a convenient in vitro functional assay using recombinant IN, a DNA oligonucleotide substrate with ends corresponding to the U3 or U5 viral DNA termini, and Mg2+ or Mn2+ as a cofactor.[2] The purified IN enzyme can also catalyze the reverse of the strand transfer reaction, or disintegration (see Figure 1A), using the oligonucleotide substrate in this in vitro system. The disintegration reaction highlights the polynucleotidyl transfer activity of IN, but does not take place in the cellular context. These drug design efforts have resulted in the discovery of the strand transfer specific IN inhibitor chemical classes, which have experienced the most clinical success to date[3, 4]. Two strand transfer specific IN inhibitors have progressed the farthest in the US FDA drug approval process as compared to any previously discovered IN inhibitor. The pyrimidinone MK-0518 (raltegravir) was US FDA approved in 2007 for HIV/AIDS treatment, and displays excellent efficacy and safety profiles in both therapy-naïve and experienced patients.[5–8] The quinolone carboxylic acid GS-9137 (elvitegravir) has also exhibited favorable clinical results in phase II studies and preliminary phase III evaluations.[9, 10] A third strand transfer specific IN inhibitor (GSK1349572), a compound licensed from Shionogi and developed by GlaxoSmithKline, has also displayed favorable clinical results in late stages of phase II. The compound was designed as a second generation strand transfer specific IN inhibitor to be potent against viral species displaying resistance profiles to raltegravir and/or elvitegravir. GSK1349572 displays potent antiviral activity in treatment naïve and experienced patients (Shionogi-ViiV Healthcare LLC). The compound also displays activity in raltegravir-experienced patients suggesting limited cross-resistance between GSK1349572 and the first generation strand transfer specific IN inhibitors.[11] The chemical structures of raltegravir, elvitegravir, and GSK1349572 are depicted in Figure 1B.
Although the strand transfer specific IN inhibitors represent a great achievement in the IN drug design and discovery field, and the successful design of second generation strand transfer IN inhibitors is attainable, a continued pharmaceutical research effort targeting IN is founded. HIV-1 is characterized by a high turnover and mutational rate – which leads to drug resistant viral strains when the pathogen is forced to replicate under therapeutic pressure. The emergence of drug resistant viral strains to currently used highly potent antiretroviral drugs targeting the HIV-1 enzymes protease and reverse transcriptase (RT) forecast a similar fate for the strand transfer specific IN inhibitors upon clinical use. Furthermore, the strand transfer specific inhibitors each display a similar mechanism of action and binding mode at the enzyme active site, and multiple studies have already shown a significant degree of cross-resistance among first-generation inhibitors of this class.[12–15] HIV-1 infected patients undergoing raltegravir treatment regimens have already displayed the selection of raltegravir-resistant viral species.[16, 17] The design and use of second generation strand transfer specific IN inhibitors targeting the same enzymatic region will result in a temporary abatement of drug resistance at the active site. However, the inevitability of drug resistance prompts the need to discover altogether new classes of IN inhibitors targeting regions distant from the enzyme active site that are effective at disrupting IN function in vivo. Non-active site binding IN inhibitors would also display synergy with currently used strand transfer specific IN inhibitors and other antiretroviral agents in clinical use.
Allosteric Enzyme Inhibition and HIV-1
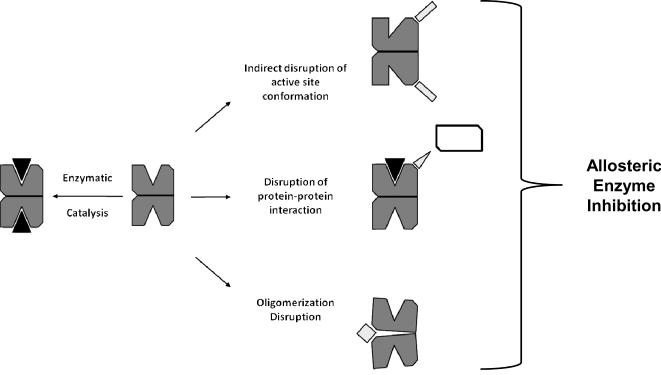
An allosteric mode of enzyme inhibition, whose term incorporates the Greek prefix allos- meaning `other' and suffix steros- meaning `place', refers to a mechanism of action where the inhibitor binds at a region distinct from the substrate binding active site to inhibit protein function. The binding of an allosteric drug ligand can indirectly modulate the enzyme's active site geometry creating an unfavorable substrate binding pocket, can block the protein-protein interaction(s) that may be required for enzyme function, and/or can disrupt the formation of necessary oligomeric complexes for enzymes that function as higher ordered protein structures (Figure 2). In the HIV-1 drug design and discovery field, the development of the non-nucleoside reverse transcriptase inhibitors (NNRTIs) has been the greatest accomplishment for allosteric antiretroviral drug design to date. Early pharmaceutical research efforts targeting RT were largely focused on the development of nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs) which mimicked natural nucleotides required for viral DNA synthesis. The NRTIs effectively compete with the natural nucleotide substrates for RT binding, and halt viral DNA synthesis following RT mediated viral DNA incorporation. In 1991 a series of tetrahydroimidazo-[4,5,1-jk] [1,4]-benzodiazepin-2(1H)-one and –thione (TIBO) derivatives were reported as highly selective RT inhibitors that did not compete with RT binding of polynucleotides.[18] Experimental evidence indicated the TIBO compounds bound a unique non-active site region of RT and displayed an alternative mechanism of action as compared to previously studied NRTIs. Within a few years a number x-ray crystallographic structures depicting RT in complex with diverse examples of NNRTIs were reported in the literature highlighting a common NNRTI allosteric binding site topographically removed from the polymerase dNTP binding site of the enzyme.[19–21] The binding of NNRTIs have been shown to indirectly alter RT active site geometry resulting in a decreased nucleotide incorporation rate.[19, 22] NNRTIs also act highly synergistically with NRTIs by inhibiting ATP-mediated NRTI removal from the growing viral DNA chain and phosphorylation by the enzyme.[23–25] By 1996, the first NNRTI, nevirapine, developed by Boehringer Ingelheim Pharmaceuticals, Inc., was US FDA approved for HIV/AIDS treatment.
Figure 2.
Allosteric inhibitory approaches for oligomeric functioning enzymes. Allosteric inhibitory agents bind to regions removed from the active site and can indirectly distort the enzyme active site, disrupt obligatory protein-protein interactions, or can disrupt oligomerization. The allosterically targeted enzyme is shown in dark grey, the natural enzymatic ligand in black, the different classes of allosteric acting inhibitory agents in light grey, whereas the protein binding partner required for enzyme function is in white.
HIV-1 IN as a Target for Allosteric Antiretroviral Drug Design
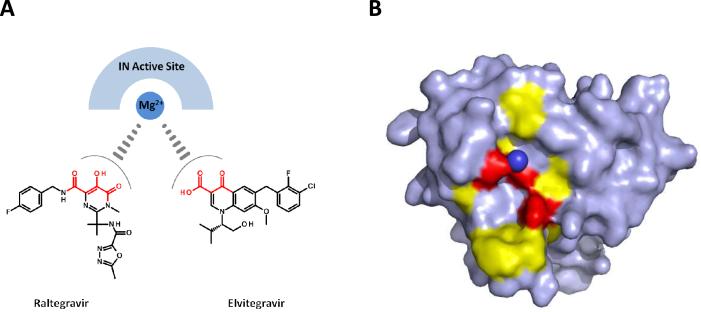
The requirement for developing allosteric binding IN inhibitors for clinical evaluation is based on the unavoidable emergence of HIV-1 strains harboring IN strand transfer specific drug resistant mutations upon the clinical use of this chemical class. Strand transfer specific inhibitors bind the IN active site and are thought to influence the IN coordination sphere of the Mg2+ cations required for DNA catalysis.[26] Chemical backbone similarities of the clinically progressed strand transfer specific inhibitors raltegravir and elvitegravir provide further evidence of their parallel inhibitory mechanism of action. Both these compounds, in addition to many other less clinically successful strand transfer specific inhibitors, contain a combination of carbonyl and a hydroxyl groups capable of chelating Mg2+ (Figure 3A). A recent crystal structure of the full-length prototype foamy virus IN in complex with its cognate DNA substrate was solved with each of raltegravir and elvitegravir separately, and has provided further insight into the similar binding orientations of the two inhibitors at the enzyme active site.[27] The metal chelating carbonyl and hydroxyl groups of each compound were observed to be directly orientated towards the metal cations in the active site, whereas the halobenzyl groups of each compound displaced the reactive 3' hydroxyl viral DNA adenosine (the product of 3'-processing). Binding of raltegravir or elvitegravir was observed to displace the reactive 3' hydroxyl group of the viral DNA end by more than 6 Å as compared to the non-drug bound IN active site.[27] Multiple studies have also identified the IN substitutions that emerge when the virus is passaged under the selective pressure of raltegravir or elvitegravir which emphasize each compound's overlapping binding at the IN active site as similar substitutions arise separately under the selective pressure of each inhibitor.[15, 28–30] The IN residues that confer resistance to each strand transfer specific inhibitor cluster around the aspartic and glutamic acid residues that coordinate the Mg2+ cations required for catalysis at the enzyme active site. Substitutions that confer resistance to raltegravir and elvitegravir surround the enzyme active site as depicted in Figure 3B using the IN crystal structure PDB 1BL3.[31] The high degree of similarity in binding and resistance profiles within the IN strand transfer specific inhibitory chemical class will result in the emergence of cross-resistant HIV-1 strains following clinical use. This clinical certainty prompts the need to identify and develop IN inhibitors with distinctive binding modes and inhibitory mechanisms to assure an IN inhibitor treatment option for patients experienced with the strand transfer specific inhibitory class. Further allosteric drug design efforts would provide chemical agents with unique binding and inhibitory modes that would be effective at targeting IN strand transfer resistant viral strains.
Figure 3.
Clinically progressed strand transfer specific IN inhibitors raltegravir and elvitegravir display equivalent mechanisms of action and bind the IN active site in an overlapping manner. (A) Chemical structures of raltegravir and elvitegravir highlighting in red the metal chelating motif thought to influence Mg2+ coordination in the IN active site. (B) IN catalytic core domain structure (PDB 1BL3) shown in surface representation with Mg2+ cation (only single cation resolved – dark blue sphere) coordination by the IN active site residues D64, D116, and E152 (red). IN positions that confer raltegravir and elvitegravir resistance upon substitution (yellow) surround the active site of the enzyme. Strand transfer specific inhibitor resistant IN positions are T66, E92, F121, G140, Y143, Q148, S153, and N155.
The IN enzyme is a highly amenable target for allosteric drug development as numerous allosteric inhibitory approaches are possible. First, IN is composed of three functional domains; the N-terminal, catalytic core, and C-terminal. The central catalytic core domain is well conserved, contains the active site acidic residues, and this domain alone can catalyze disintegration, or polynucleotidyl transfer in vitro, highlighting its importance for IN catalysis.[32] However, both the N- and C-terminal domains play critical roles for IN function through the promotion of DNA substrate binding and multimeric subunit formation. These domains present alternative protein regions for the design of allosteric IN inhibitors. Second, IN catalysis requires a multimeric state of the protein associated with substrate DNA – providing the opportunity to design inhibitors effective at disrupting the formation of oligomeric nucleoprotein IN complexes. Multiple studies have established that a dimeric species of IN is sufficient for 3-processing, whereas a tetrameric IN oligomer, stabilized by DNA contacts, is the oligomeric state required for strand transfer catalysis.[33–37] Additional evidence also indicates that IN-DNA complex formation for integration is a highly dynamic process that proceeds through a transient intermediary nucleoprotein assembly termed the synaptic complex (SC). The SC is formed by the alignment of two IN dimers on separate long terminal repeat (LTR) viral DNA ends, which is then followed by a juxtaposed rearrangement to form an active tetramer (one dimer contributed from each LTR end) for strand transfer catalysis.[38] Compounds efficient at disrupting any of the IN-IN and IN-DNA contacts critical for the nucleoprotein assembly process would be effective allosteric inhibitors of viral integration. A third effective allosteric inhibitory approach is targeting the disruption of IN-cellular cofactor interactions. The integration process in vivo depends on multiple interactions with different host cell cofactors.[39] Disrupting the protein-protein interactions between IN and cellular cofactors which are essential for viral replication represents a relatively unexploited allosteric approach for the design and development of HIV/AIDS antiretroviral agents.[40]
Targeting the N-Terminal Domain to Inhibit IN Function
The N-terminal domain spans IN residues 1–50, and contains a pair of histidine (H12 and H16) and cysteine (C40 and C43) residues, termed the HHCC motif, that is highly conserved across retroviral IN proteins.[41] The HHCC motif is analogous to zinc binding motifs found in many DNA binding proteins, and has been shown to bind zinc in a 1:1 ratio.[42, 43] The binding of zinc has been shown to enhance the formation of IN oligomers.[43, 44] The protein domain folds into a helix-turn-helix motif similar to known DNA binding domains,[45, 46] and recent progress uncovered that the HHCC motif mediates specific IN-DNA binding in the presence of the cognate viral DNA sequence and the Mg2+ cations required for catalysis.[47] Numerous viral mutagenic studies have determined that residues comprising the HHCC motif are essential for viral replication as substitutions to Ala, Asn, Leu, Ser, or Tyr have all rendered the virus non-infectious.[48–54] HHCC disruption blocks HIV-1 replication at two early stage events in the viral life cycle. Mutant viruses harboring HHCC substitutions are not only integration defective, but have also been shown to be incapable of initiating reverse transcription of viral DNA.[52, 54] IN makes direct contacts with RT, and is a known component of the reverse transcription complex.[54] Mutant IN H12Y proteins were also shown to disrupt the interaction with a known IN cellular cofactor potentially required for viral DNA synthesis and viral particle production, integrase interactor 1 (INI1), and mutant IN H12Y viruses were non-responsive to the antiretroviral effects of an INI1 derived peptide.[55]
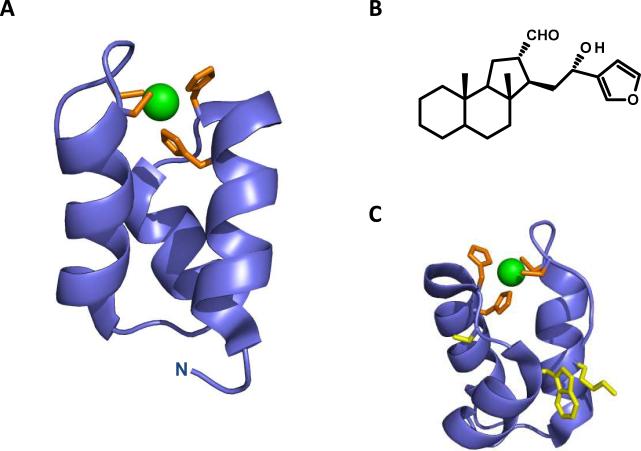
Allosteric inhibitors targeting the IN N-terminal domain would be effective at disrupting IN function through a range of inhibitory mechanisms. Inhibitors targeting zinc coordination by the HHCC motif would have a detrimental effect on IN-DNA binding, multimeric IN formation, reverse transcription, and/or integration. Additionally, the HHCC motif has a favorable three-dimensional geometry for drug design, with inter-atom distances between nitrogen and sulfydryl groups ranging from ~3.5–4 Å; dimensions that correspond appropriately with many drug-like small molecules. Shown in Figure 4A is the IN N-terminal domain highlighting the HHCC zinc coordination motif (modified from PDB 1K6Y[56]).
Figure 4.
Targeting the IN N-terminal domain for allosteric inhibition. (A) The IN N-terminal domain (modified from PDB 1K6Y) shown in blue, with the HHCC motif shown in orange stick representation, and zinc in green sphere representation. (B) Chemical structure of the natural product IN inhibitor hyrtiosal. (C) The IN N-terminal domain with the hyrtiosal binding residues S17, W19, and K34 shown in yellow stick representation.
A natural product, hyrtiosal, derived from a marine sponge, has been shown to inhibit IN through direct binding to the N-terminal domain. The compound inhibited IN-viral DNA binding at an IC50 value of 9.6 ± 0.86 μM. Hyrtiosal was shown to interact with IN only when residues 1–50 were present in the protein, indicating allosteric binding of the molecule to the N-terminal domain.[57] Molecular docking strategies were employed to discern the binding orientation of hyrtiosal in the N-terminal domain. The most energetically favorable conformation of the molecule is within a small pocket adjacent to the HHCC motif. Hyrtiosal interacted with IN residues S17, W19, K34, V37, A38, and S39.[57] To validate the molecular docking results, alanine substituted IN mutants at positions S17, W19, and K34 were constructed and investigated for binding capacity to the small molecule. Hyrtiosal-IN binding was significantly reduced in the S17A and W19A mutants, whereas it was completely abolished in the K34A mutant IN protein.[57] The chemical structure of hyrtiosal and the validated IN interacting residues are depicted in Figure 4B and C. The identified binding site for this natural product represents a useful platform for further target based allosteric drug design, whereas the compound hyrtiosal can be further used for the design of more potent analogues targeting the IN N-terminal domain.
Targeting the C-Terminal Domain to Inhibit IN Function
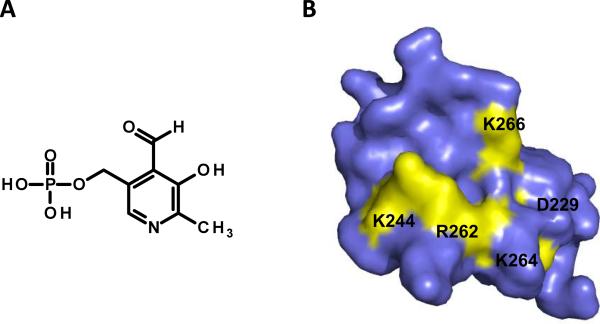
The IN C-terminal domain spans residues 220–288 and is much less evolutionarily conserved as compared to the N-terminal and core domains. Studies have shown this domain to possess non-specific DNA binding activity,[58–62] suggesting a potential cellular genomic DNA-binding role during integration in vivo. A nucleotide – based inhibitor binding site has been identified on the C-terminal domain of IN. The inhibitor, pyridoxal 5'-phosphate (PLP), was first covalently linked to the binding site, trypsin digested, and the resulting fragments were subjected to mass spectrometric (MS) analysis to identify the binding region.[63] PLP inhibited IN catalytic activities at an IC50 value of 15 μM. The inhibitor also disrupted IN-DNA binding at an IC50 value of 12 μM. Following incubation of IN with PLP, covalent linkage, and trypsin digestion, MS experiments revealed that one molecule of PLP bound to the IN peptide 241–258 region.[63] The peptide includes two lysine residues, K244 and K258, which contain the required primary amine functional group needed for covalent PLP-IN modification by means of Schiff – base chemistry employed by the study. Further trypsin digestion uncovered K244 as resistant to hydrolysis indicating this was the lysine residue covalently linked to the inhibitor PLP. IN protein K244 – PLP binding was also confirmed through MS/MS analysis.[63] Follow-up experiments were then conducted to determine if substitutions at IN K244 had any significant impact on HIV-1 replication. Mutant viruses containing an IN K244E substitution were impaired for replication due to a specific defect in viral DNA integration. The K244E IN protein was also deficient for both 3'-processing and strand transfer catalytic activities in vitro. Molecular modeling and dynamic simulations were utilized to gain more insight into the binding mode of PLP at the C-terminal domain of IN. In addition to interacting with K244, the inhibitor was observed to make energetically favorable hydrogen bond contacts with R262, K264, K266, and D229.[63] The chemical structure of PLP and the binding site identified (modified from PDB 1EX4[64]) are depicted in Figure 5A and B. The identified binding site represents a useful platform for further target based allosteric drug design at the C-terminal domain of IN. PLP can also be used for the design of more potent nucleotide-mimetic small molecule analogues to disrupt IN C-terminal DNA binding and catalytic activities of the enzyme.
Figure 5.
Targeting the IN C-terminal domain for allosteric inhibition. (A) Chemical structure of the nucleotide-based IN inhibitor PLP. (B) The IN C-terminal domain (modified from PDB 1EX4) shown in surface representation, with residues implicated in PLP binding highlighted yellow.
Disrupting IN Multimerization
Blocking the IN oligomerization process represents a very attainable allosteric inhibitory approach for the design of an altogether new class of IN inhibitors. Many different groups have used peptides derived from the IN interfacial region to show effective oligomerization disruption.[65–67] For small molecule inhibitor development, multiple independent studies have identified an inhibitor binding region at the IN dimeric interface of the catalytic core domain using chemically diverse compounds. Additionally, x-ray structure determinations of the IN allosteric region have been solved using two separate ligands. The identified inhibitor binding region at the dimeric interface is highly useful as a rational drug design platform to develop allosteric IN multimer disrupting agents with improved target affinity and inhibitory potency.
3,4-Dihydroxyphenyltriphenylarsonium (DHph-3ph-As)
As a means to identify the binding site of potential IN inhibitors, numerous compounds containing heavy atoms, which display useful x-ray absorption properties, were singled out by researchers to utilize for IN co-crystallization and x-ray diffraction studies.[68] This process led to the discovery of DHph-3ph-As (Figure 6A), which displayed modest IN inhibitory activity at an IC50 value of 150 μM. The compound was also found to block disintegration activity, providing evidence the binding site was located on the catalytic core domain. The replacement of the arsenic center with a phosphorous atom resulted in an enhanced inhibitory activity, with the phosphorous compound inhibiting both IN processes at an IC50 value of 13.5 μM. DHph-3ph-As was co-crystallized with IN and the structure was solved to a resolution of 2.3 Å.[68] Although the positioning of the catechol hydroxyls could not be resolved, the structure revealed the As center atom interacted with IN residue Q168, and both tryptophan residues at IN positions 131 and 132 made π electron stacking interactions with one of the phenyl rings.[68] The authors note that the binding of DHph-3ph-As to the IN dimeric interface could be a crystallographic artifact. However numerous studies detailed below using IN inhibitors with low micromolar IC50 values have also identified the same region at the IN dimeric interface which strengthens the idea of this protein region as amenable for further allosteric inhibitor development.
Figure 6.
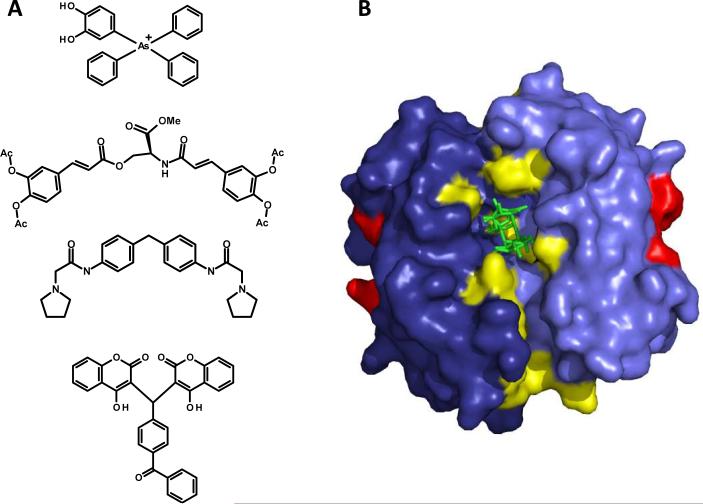
The identified IN allosteric inhibitor binding region located at the dimeric interface of the catalytic core domain. (A) Chemical structures of DHph-3ph-As, the acetylated bis-caffeoyl IN inhibitor, 1-pyrrolidineacetamide, and the coumarin-containing IN inhibitor each experimentally determined to bind the allosteric inhibitor binding region at the IN dimeric interface. (B) Structure of the IN catalytic core domain dimer (light and dark blue) with sucrose ligand (green) occupying the central allosteric binding cavity (PDB 3L3V). The active site D64, D116, and E152 residues are highlighted in red, whereas IN residues comprising the allosteric region at the dimeric interface are shown in yellow. IN allosteric inhibitor binding residues are E96, K103, C130, W131, W132, Q168, K173, T174, and M178.
Acetylated Bis-Caffeoyl IN Inhibitor
A novel affinity labeling approach using aryl-di-O-acetyl chemical groups, which specifically acetylate cysteine, lysine, and tyrosine functional groups, was used to identify the binding site of an acetylated bis-caffeoyl IN inhibitor (Figure 6A). The acetylated bis-caffeoyl compound inhibited IN catalytic activities at an IC50 value of 3 μM.[69] Full length IN protein was first treated with the acetylated IN inhibitor in solution, then subjected to in-gel proteolysis and MS analysis. Examination of the MS spectra revealed a single new peak as compared to a both a bis-caffeoyl (non-acetylated) IN treated control sample, and a non-inhibitor IN treated control sample. The molecular mass of the new peak was 2,220.13 Da, which directly corresponded to the IN tryptic peptide 167DQAEHLKTAVQMAVFIHNK185 (2,178.12 Da) plus one acetyl group (42.03 Da).[69] The identified IN 167–185 peptide contained two lysine residues, K173 and K185, susceptible to acetylation by the inhibitor. Further trypsin hydrolysis of the peptide revealed the K173 residue was resistant to digestion, and follow-up MS/MS analysis confirmed this residue as the site of acetyl modification. Subsequent molecular modeling of the IN-acetyl bis-caffeoyl inhibitor complex uncovered T174 and M178 as additional IN residues involved in binding the inhibitor.[69]
A follow-up study provided a detailed understanding of the mechanism of action of the acetyl bis-caffeoyl compound at the IN dimeric interface.[70] Using an IN subunit exchange assay, with histidine-tagged and un-tagged monomeric IN subunits, it was shown the acetyl bis-caffeoyl inhibitor directly disrupted the dynamic exchange of IN monomeric forms between IN multimers. The compound preferentially bound and stabilizes a multimeric IN complex, thereby disrupting the dynamics of free IN subunit exchanges that is required for IN-viral DNA nucleoprotein complex formation.[70]
1-Pyrrolidineacetamide
A symmetrical compound, 1-pyrrolidineacetamide (Figure 6A), was also shown to bind the IN dimeric interface, and specifically interact with residues K173, and T174.[71] A surface plasmon resonance (SPR) – based competitive assay determined the compound inhibited IN-viral DNA binding with an IC50 value of 7.29 ± 0.68 μM/L. 1-Pyrrolidineacetamide also showed moderate antiretroviral activity with an EC50 value of 17.05 μg/mL. Molecular modeling analysis suggested that the IN residue K173 interacted with the inhibitor through cation- π interactions, whereas IN residue T174 made hydrogen-bond interactions. Alanine substitutions at both positions K173 and T174 practically abolished binding of 1-pyrrolidineacetamide to IN.[71]
Coumarin-Containing IN Inhibitor
Our group identified the IN dimeric interface region as the binding site of a coumarin-containing inhibitor using a benzophenone photo-activatable affinity labeling approach.[72] The dimeric coumarin compound (Figure 6A) inhibited IN mediated 3'-processing and strand transfer catalytic activities at IC50 values of 27 ± 3 and 18 ± 3, respectively. A larger tetrameric coumarin compound inhibited the disintegration activity of IN, indicating this class of coumarins bound specifically to the catalytic core domain of IN. Specific photo-induced crosslinking of the inhibitor to the core domain followed by MS analysis revealed the coumarin bound the core domain in a 1:1 stoichiometric ratio. Tryptic hydrolysis of the inhibitor-IN complex followed by electrospray ionization MS experiments exposed the IN peptide 128AACWWAGIK136 as the site of inhibitor binding.[72] To further validate this region for coumarin binding we made conservative and non-conservative substitutions at IN positions C130, W131, W132, and K136 in the full length protein. We found that a Ser substitution at C130, and non-conservative substitutions (Ala, Gly, and Arg) at W132 each conferred major IN resistance (from two-fold to five-fold) to coumarin inhibition as compared to the wild type (WT) protein, providing evidence these IN residues were critical for binding the inhibitor. Molecular docking analysis revealed the coumarin compound bound a pocket at the IN dimeric interface suggesting it may disrupt the IN multimerization process. To test this inhibitory mechanism of action, we determined the inhibitory capacity of the coumarin compound on Ca2+ induced pre-assembled IN-DNA complexes. Ca2+ can facilitate the assembly of IN-DNA complex formation without enzymatic catalysis.[73, 74] IN mediated DNA cleavage can then be initiated by the addition of Mg2+ (or Mn2+) to the reaction conditions. The coumarin inhibitor was much less effective at inhibiting pre-assembled IN-DNA complexes, with IC50 values increasing by four-fold and two-fold for 3'-processing and strand transfer, respectively. The drop in coumarin inhibitory potency against IN-DNA pre-assembled complexes supports an inhibitory mechanism of action where the compound binds the allosteric site at the dimeric interface and disrupts IN multimerization.[72]
The fact that only non-conservative substitutions at IN residue W132 disrupted coumarin binding to the allosteric binding site at the IN dimeric interface prompted us to further explore the contributions of adjacent aromatic residues at the interface. In a follow-up study we determined that residues M178, F181, and F185 from one IN monomer contributed to an essential π electron orbital interaction with W132 from the adjacent IN monomer across the dimeric interface.[75] Disruption of the electron orbital interaction abolished IN strand transfer activity while leaving 3-processing activity unaffected. The uncoupling of IN enzymatic reactions by disrupting the electron orbital interaction bridging the dimeric interface indicates these residues contribute to a critical contact point for IN multimerization. Viruses containing non-conservative IN substitutions at this region were also defective at early stages of viral replication and non-infectious further underscoring this allosteric region at the dimeric interface as a valuable platform for inhibitor development.[75]
Allosteric Binding Region at IN Dimeric Interface at 2 Å Resolution
Recently, a report detailing the IN allosteric inhibitor binding region at the dimeric interface at a high resolution was disclosed in the literature.[76] The structure contained a molecule of sucrose bound to the central allosteric pocket as a result of crystal soaking in a sucrose containing cryoprotectant solution. The sucrose ligand made hydrogen-bond contacts with IN residues E96, K103, and K173, in addition to numerous water molecules present in the dimeric cavity after crystal packing.[76] Even though sucrose does not exhibit any IN inhibitory activity, the structure elucidation of the allosteric site it occupies represents a remarkably useful platform for the rational design of multimeric disrupting compounds with improved affinity and inhibitory potential. Depicted in Figure 6B is the 2 Å structure of the IN catalytic core domain with bound sucrose (PDB 3L3V[76]). Highlighted in the structure are the IN residues responsible for binding DHph-3ph-As, the acetylated bis-caffeoyl compound, 1-pyrrolidineacetamide, and the dimeric coumarin inhibitor, which together comprise the overlapping allosteric binding region at the IN dimeric interface.
Targeting IN – Cellular Cofactor Interactions
It is becoming increasingly clear that IN function in vivo requires numerous interactions will host cellular proteins. The targeted disruption of direct cellular IN cofactors required for viral replication has become a very sought-after allosteric inhibitory approach for the development of novel first in-class IN inhibitors for clinical development, and the IN cofactors identified thus far have been reviewed previously.[39, 40, 77] Although numerous IN cofactors have been identified, progress in developing IN-cofactor specific disrupting compounds is hampered by the difficulty in unambiguously establishing that the cellular protein is indeed absolutely required for viral replication. Although this is a frustrating impediment towards the development of new IN-cellular cofactor disrupting compounds, one cofactor, lens epithelium-derived growth factor (LEDGF)/p75, has been the subject of significant progress in the discovery and design of protein-protein disrupting compounds with a resulting allosteric inhibitory effect on IN.
The IN Cellular Cofactor LEDGF/p75
The association between IN and the cellular cofactor LEDGF/p75 is currently the most promising IN interaction for the design of protein-protein disrupting therapeutics. LEDGF/p75 is a pro-survival protein that activates the transcription of stress related/anti-apoptotic proteins, thus promoting cell growth and counteracting stress-induced cell death.[78] The cellular protein has been shown to be essential for the viral life cycle,[79, 80] acting as a chromatin tethering factor to facilitate the interaction between IN and nuclear chromatin.[81, 82] The LEDGF/p75 binding region on IN includes residues A128, A129, W131, W132, and I161-K173, which is located near the dimeric interface of the catalytic core domain.[83–85] An in-depth analysis of the biological role LEDGF/p75 plays in HIV-1 replication can be found in numerous reports.[86–89] Here we provide the ongoing progress in developing small molecule allosteric inhibitors effective at disrupting the IN – LEDGF/p75 interaction.
Benzoic Acid Derivative – D77
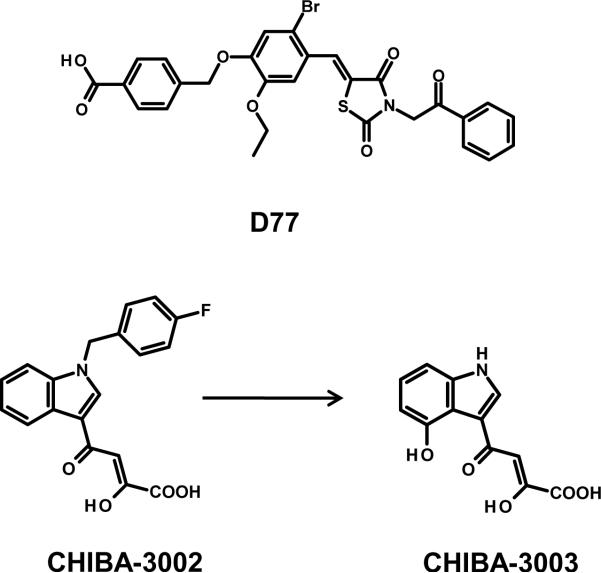
D77 (Figure 7), a benzoic acid derivative, was discovered from a library of ~300 compounds to have activity at disrupting the interaction between IN and the LEDGF/p75 integrase binding domain (IBD), residues 347–429, in a yeast and mammalian two-hybrid screening system.[90] A follow-up competitive SPR assay was then employed to determine whether D77 could directly disrupt the interaction between the IN catalytic core domain and the LEDGF/p75 IBD. The compound was observed to decrease IN – LEDGF/p75 IBD binding in a dose-dependent manner, although no specific concentration was disclosed as to indicate the compound's potency for disrupting the protein-protein interaction. At a concentration of 5 μM, added five hours post-transfection of fluorescently-tagged IN in 293T cells, the compound disrupted the nuclear accumulation of IN suggesting that the disruption of the IN-LEDGF/p75 interaction also disturbed IN nuclear translocation. D77 did show antiretroviral activity with an EC50 value of 23.8 μg/mL, and a cytotoxic concentration greater than three-folds higher (CC50 = 76.82 μg/mL) in MT-4 cells. The compound was determined to bind full length IN and the catalytic core domain with a dissociation constant (KD) of 5.81 and 6.83 μM, respectively.[90] Molecular docking analysis using the IN catalytic core domain revealed D77 made significant contacts with T174 from monomer A, and Q95, T125, and W131 from monomer B. An alanine substitution at position T125 significantly reduced the D77 – IN interaction, whereas the Q95A, W131A, and T174A substitutions practically abolished binding of the inhibitor to IN.[90]
Figure 7.
Small molecule IN inhibitors useful for analogue-based drug design targeting the IN-LEDGF/p75 protein-protein interaction. Chemical structures of the benzoic acid derivative D77, CHIBA-3002, and CHIBA-3003.
CHIBA-3003
A study using a structure-based pharmacophoric drug discovery platform, based on the co-crystal structure of the IN catalytic core domain and the LEDGF/p75 IBD (PDB 2B4J[91]), led to the discovery of a 2-hydroxy-4-(4-hydroxy-1H-indol-3-yl)-4-oxobut-2-enoic acid compound (CHIBA-3003, Figure 7) with micromolar activity against the IN-LEDGF/p75 protein-protein interaction.[92] The receptor-based pharmacophore model was largely modeled on two critical LEDGF/p75 IBD residues, I365 and D366, that have been shown to disrupt the IN interaction when substituted,[91] and were contained in a previous report disclosing a larger LEDGF/p75-derived peptide (residues 355–377) which displayed IN-LEDGF/p75 disrupting activity at an IC50 value of 25 μM.[93] The pharmcophore model was then used as a query in an in-house chemical database of 3,055 small molecules. After virtual screening and fitness refinement, one compound, termed CHIBA-3002 (Figure 7), exhibited the best fitness value. Testing revealed this compound to have modest activity against the IN-LEDGF/p75 interaction with 46% inhibition at a concentration of 100 μM.[92] Follow-up pharmacophoric model fitting of the compound revealed the molecule lacked a key hydrogen bonding functionality. Additionally, the halobenzyl group of CHIBA-3002 was found to be located outside the designated pharmacophore ligand space of the model. These observations prompted the research group to design chemical analogues aimed at incorporating a hydrogen-bonding chemical feature, and also the removal of the halogen-substituted phenyl ring. Rational chemical design of analogues resulted in the synthesis of CHIBA-3003 which incorporated a hydroxyl group and had the fluorine-substituted phenyl ring removed (structural comparison shown in Figure 7). CHIBA-3003 displayed an increased potency at disrupting the IN-LEDGF/p75 interaction, with an IC50 value of 35 μM.[92]
2-(quinolin-3-yl) Acetic Acids: “LEDGINs”
The rational design of a series of 2-(quinolin-3-yl) acetic acid based molecules termed “LEDGINs” marks the discovery of the first authentic small molecule allosteric inhibitor chemical class to display antiretroviral activity due to a specific disruption of the IN-LEDGF/p75 interaction.[94] A 2-(quinolin-3-yl) acetic acid was selected from a set of twenty-five molecules that remained after two in silico chemical filtering and screening processes. Initially a set of 200,000 commercially available compounds were filtered based on chemo-informatic parameters that incorporated known small molecule protein-protein interaction inhibitor chemical properties. After this initial filtering process, the remaining 160,000 compounds were screened by a pharmacophoric model using existing IN crystal structures,[31, 68] and importantly, the co-crystal structure of the IN-LEDGF/p75 binding domains.[91] The first hit LEDGIN molecule displayed modest activity against the IN-LEDGF/p75 interaction, with 36% inhibition at 100 μM.[94] Close structural analogues of the hit LEDGIN were either purchased commercially or chemically synthesized, and multiple rounds of structure-activity analysis were conducted to refine the 2-(quinolin-3-yl) acetic acid chemical class to identify a lead molecule with increased potency. The multiple rounds of structure-activity refinement resulted in a highly potent 2-(quinolin-3-yl) acetic acid derivative (structure shown in Figure 8). The lead LEDGIN was ten-fold more active (IC50 = 1.37 ± 0.36 μM) against the IN-LEDGF/p75 interaction in vitro, and exhibited a twenty-fold increase in antiretroviral activity (EC50 = 2.35 ± 0.28 μM) over compounds that were initially discovered.[94]
Figure 8.
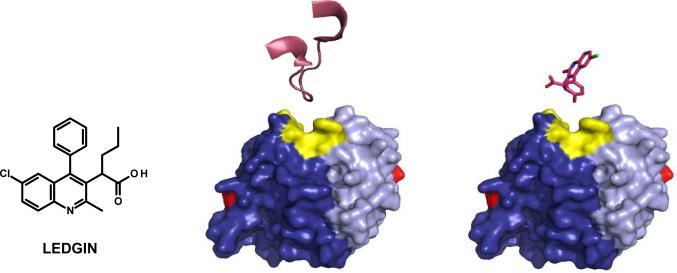
Successful design of the antiretroviral 2-(quinolin-3-yl) acetic acid compounds which display a potent and specific disruption of the IN-LEDGF/p75 interaction via mimicking the IN-LEDGF/p75 amino acid contact points. The chemical structure of the lead antiretroviral 2-(quinolin-3-yl) acetic acid LEDGIN compound, with a comparison of the binding orientations between the LEDGF/p75 IN interacting peptide (PDB 2B4J; residues 361–372) and the lead LEDGIN small molecule (PDB 3LPU), both depicted in magenta, at the IN catalytic core domain produced by overlapping the mimicked pharmacophoric groups of the LEDGIN compound with IN interacting LEDGF/p75 amino acids. The IN monomers are depicted in dark and light blue, with IN active site residues D64, D116, and E152 in red, and the IN LEDGF/p75 contact residues (Q168, E170, W131, and W132; 128AA129 not resolved) in yellow.
The compound was highly specific for the IN-LEDGF/p75 interaction, as it did not show any activity for disrupting LEDGF/p75 and other binding partners of the protein. The lead LEDGIN also retained antiretroviral activity in primary cells against both CXCR4 and CCR5 tropic viral strains. Importantly, the allosteric inhibitor was active against multiple clinically relevant drug resistant viral strains, including viral strains that were resistant to a NRTI (AZT), a NNRTI (efavirenz), a CXCR4 chemokine receptor agonist (AMD3100), and most significantly, the IN strand transfer specific inhibitor raltegravir, which targets the IN active site.[94] The potent inhibitory activity of the lead 2-(quinolin-3-yl) acetic acid derivative against a raltegravir resistant viral strain directly underscores the molecule's allosteric inhibitory mechanism of action, and the clinical importance of having viable allosteric IN inhibitors available to treat viral strains resistant to the strand transfer specific active site binding IN inhibitors currently used in the clinic. Providing added support that the LEDGIN inhibited viral replication through a direct disruption of the IN-LEDGF/IN interaction, the compound lost inhibitory activity against a mutant viral strain that contained IN substitutions (A128T/E170G) previously selected to be resistant to the transdominant inhibition of overexpressing the LEDGF/p75 IBD.[79] Virus passaged under the selective pressure of the LEDGIN also selected the A128T IN substitution located at the protein-protein interface clearly indicating the successful rational design of the 2-(quinolin-3-yl) acetic acids as effective allosteric LEDGF/p75-IN disrupting inhibitors.[94]
Also disclosed in the report were x-ray crystal structure determinations of the IN catalytic core domain in complex with two representative 2-(quinolin-3-yl) acetic acids, including the lead LEDGIN molecule, to further validate the pharmacophore models used in the inhibitor design process. Both LEDGIN molecules adopted conformations within the IN binding pocket that matched what was predicted in silico. The LEDGIN carboxyl moiety formed hydrogen bonds with both main chain nitrogen atoms of IN E170 and H171, mimicking the IN protein contacts of the LEDGF/p75 residue D366. The IN residue A128 occupied a space adjacent to the chlorine atom between the phenyl and conjugated ring system of the 2-(quinolin-3-yl) acetic acid derivative.[94] Shown in Figure 8 is the catalytic core IN-LEDGF/p75 IBD co-crystal structure (PDB 2B4J) highlighting the LEDGF/p75 IN interacting peptide (residues 361–372) in comparison with the LEDGIN small molecule orientation (PDB 3LPU). The similar orientation and IN contact region of both the LEDGF/p75 IN interacting peptide and the lead LEDGIN compound highlights that the 2-(quinolin-3-yl) acetic acid derivative successfully mimics the LEDGF/p75 contribution to the protein-protein interface with IN, and is a potent allosterically binding IN inhibitor effective at disrupting this critical protein-protein interaction. A series of 2-(quinolin-3-yl) acetic acid derivatives has also been disclosed in an international patent application by Boehringer Ingelheim Pharmaceuticals, Inc.[95] These compounds likely bind the same allosteric region of IN and disrupt the IN-LEDGF/p75 protein-protein interaction as their inhibitory mechanism of HIV-1 replication.
Outlook
Progress in developing clinically viable allosteric binding IN inhibitors targeting protein regions removed from the active site would provide a great benefit for HIV/AIDS patients. At present there is only one IN targeted treatment option to complement antiretroviral regimens – the strand transfer specific IN inhibitor raltegravir. A second strand transfer specific IN inhibitor, elvitegravir, may become the second IN targeted antiretroviral on the market pending positive results of Phase III clinical trials. However, both these inhibitors bind the IN active site in an overlapping orientation and display the same inhibitory mechanism. This commonality will undoubtedly cause difficulty in providing an effective IN targeted therapeutic option once the patient has experienced routine raltegravir and/or elvitegravir treatment. It is well known that HIV-1 is a pathogen that is characterized by a high turnover and mutation rate. The emergence of drug resistant viral strains once patients are routinely administered antiretroviral agents is an expected outcome. The challenge for HIV-1 drug discovery is therefore to stay one step ahead of drug resistant viral strains to ensure effective therapeutics for treatment experienced patients. Currently, there exists no IN targeted treatment options effective against IN strand transfer specific resistant viral strains that are FDA approved, although progress has been achieved in developing second generation strand transfer specific IN inhibitors like GSK1349572. However, clinically viable allosteric binding IN inhibitors would afford an altogether separate effective treatment option for raltegravir and/or elvitegravir experienced patients.
The IN protein is a very amenable target for allosteric inhibitor development as there exists numerous approaches to block the enzyme's function in vivo. Each allosteric IN inhibitory approach maps to regions removed from the enzyme active site (Figure 9). Progress has already been made in identifying compounds that bind both the N- and C-terminal domains of IN, which can be utilized for analogue based drug design approaches targeting these protein domains. Additionally, there have been significant advances detailing a common binding region at the IN dimeric interface using both affinity labeling approaches and x-ray structure determinations for a diverse set of allosteric acting small molecule inhibitors. This IN protein region is an extremely useful rational drug design platform for developing multimer disrupting and/or stabilizing agents with higher affinity and potency. Lastly, research aimed to identify IN cellular cofactors and their roles in viral replication will provide a wealth of possible protein sites for the design of allosteric inhibitors effective at disrupting critical protein-protein interactions; similar to the successful design of the 2-(quinolin-3-yl) acetic acids for the IN-LEDGF/p75 interaction.
Figure 9.
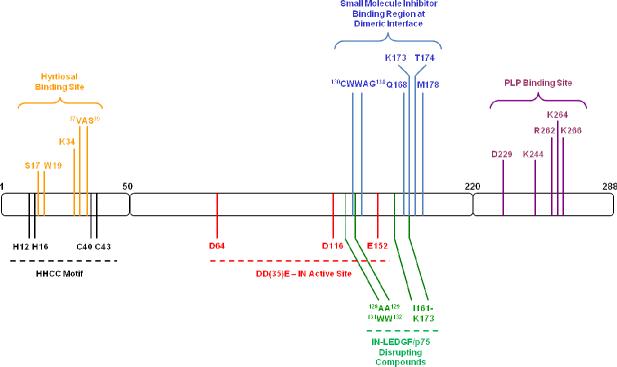
IN allosteric binding sites for inhibitor development map to regions removed from the enzyme active site. Domain organization of IN with the HHCC motif (black) and DD(35)E active site (red) residues highlighted. IN residues involved in binding compounds of each allosteric inhibitory approach are color coded: N-terminal hyrtiosal (yellow), oligomerization disruption (blue), IN-LEDGF/p75 disruption (green), and C-terminal PLP (purple).
Acknowledgements
This study was supported through funds provided the National Institutes of Allergy and Infectious Diseases, National Institutes of Health (R21AI081610 grant).
Abbreviations
- (IN)
Integrase
- (IBD)
integrase binding domain
- (LEDGF)
lens epithelium-derived growth factor
- (LTR)
long terminal repeat
- (MS)
mass spectrometric
- (NNRTI)
non-nucleoside reverse transcriptase inhibitor
- (NRTI)
nucleoside/nucleotide reverse transcriptase inhibitor
- (PLP)
pyridoxal 5'-phosphate
- (RT)
reverse transcriptase
- (SPR)
surface plasmon resonance
- (SC)
synaptic complex
- (TIBO)
tetrahydroimidazo-[4,5,1-jk] [1,4]-benzodiazepin-2(1H)-one and –thione
- (WT)
wild type
References
- [1].Asante-Appiah E, Skalka AM. Adv Virus Res. 1999;52:351–369. doi: 10.1016/s0065-3527(08)60306-1. [DOI] [PubMed] [Google Scholar]
- [2].Bushman FD, Craigie R. Proc Natl Acad Sci U S A. 1991;88(4):1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Al-Mawsawi LQ, Al-Safi RI, Neamati N. Expert Opin Emerg Drugs. 2008;13(2):213–225. doi: 10.1517/14728214.13.2.213. [DOI] [PubMed] [Google Scholar]
- [4].Dayam R, Al-Mawsawi LQ, Neamati N. Drugs R D. 2007;8(3):155–168. doi: 10.2165/00126839-200708030-00003. [DOI] [PubMed] [Google Scholar]
- [5].Croxtall JD, Scott LJ. Drugs. 2010;70(5):631–642. doi: 10.2165/11204590-000000000-00000. [DOI] [PubMed] [Google Scholar]
- [6].Markowitz M, Nguyen BY, Gotuzzo E, Mendo F, Ratanasuwan W, Kovacs C, Prada G, Morales-Ramirez JO, Crumpacker CS, Isaacs RD, Campbell H, Strohmaier KM, Wan H, Danovich RM, Teppler H. J Acquir Immune Defic Syndr. 2009;52(3):350–356. doi: 10.1097/QAI.0b013e3181b064b0. [DOI] [PubMed] [Google Scholar]
- [7].Lennox JL, DeJesus E, Lazzarin A, Pollard RB, Madruga JV, Berger DS, Zhao J, Xu X, Williams-Diaz A, Rodgers AJ, Barnard RJ, Miller MD, DiNubile MJ, Nguyen BY, Leavitt R, Sklar P. Lancet. 2009;374(9692):796–806. doi: 10.1016/S0140-6736(09)60918-1. [DOI] [PubMed] [Google Scholar]
- [8].Markowitz M, Nguyen BY, Gotuzzo E, Mendo F, Ratanasuwan W, Kovacs C, Prada G, Morales-Ramirez JO, Crumpacker CS, Isaacs RD, Gilde LR, Wan H, Miller MD, Wenning LA, Teppler H. J Acquir Immune Defic Syndr. 2007;46(2):125–133. doi: 10.1097/QAI.0b013e318157131c. [DOI] [PubMed] [Google Scholar]
- [9].Shimura K, Kodama EN. Antivir Chem Chemother. 2009;20(2):79–85. doi: 10.3851/IMP1397. [DOI] [PubMed] [Google Scholar]
- [10].Zolopa AR, Berger DS, Lampiris H, Zhong L, Chuck SL, Enejosa JV, Kearney BP, Cheng AK. J Infect Dis. 2010;201(6):814–822. doi: 10.1086/650698. [DOI] [PubMed] [Google Scholar]
- [11].Mascolinli M, Kort R. J Int AIDS Soc. 2010;13(Suppl 1):S3. doi: 10.1186/1758-2652-13-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Metifiot M, Maddali K, Naumova A, Zhang X, Marchand C, Pommier Y. Biochemistry. 2010;49(17):3715–3722. doi: 10.1021/bi100130f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Serrao E, Odde S, Ramkumar K, Neamati N. Retrovirology. 2009;6:25. doi: 10.1186/1742-4690-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Goethals O, Clayton R, Van Ginderen M, Vereycken I, Wagemans E, Geluykens P, Dockx K, Strijbos R, Smits V, Vos A, Meersseman G, Jochmans D, Vermeire K, Schols D, Hallenberger S, Hertogs K. J Virol. 2008;82(21):10366–10374. doi: 10.1128/JVI.00470-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Marinello J, Marchand C, Mott BT, Bain A, Thomas CJ, Pommier Y. Biochemistry. 2008;47(36):9345–9354. doi: 10.1021/bi800791q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Canducci F, Barda B, Ceresola E, Spagnuolo V, Sampaolo M, Boeri E, Nozza S, Cossarin F, Galli A, Gianotti N, Castagna A, Lazzarin A, Clementi M. Clin Microbiol Infect. 2010 doi: 10.1111/j.1469-0691.2010.03375.x. [DOI] [PubMed] [Google Scholar]
- [17].Van Wesenbeeck L, Rondelez E, Feyaerts M, Verheyen A, Van der Borght K, Smits V, Cleyberg C, De Wolf H, Van Baelen K, Stuyver LJ. Antimicrob Agents Chemother. 2010 doi: 10.1128/AAC.01733-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Debyser Z, Pauwels R, Andries K, Desmyter J, Kukla M, Janssen PA, De Clercq E. Proc Natl Acad Sci U S A. 1991;88(4):1451–1455. doi: 10.1073/pnas.88.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Esnouf R, Ren J, Ross C, Jones Y, Stammers D, Stuart D. Nat Struct Biol. 1995;2(4):303–308. doi: 10.1038/nsb0495-303. [DOI] [PubMed] [Google Scholar]
- [20].Hopkins AL, Ren J, Esnouf RM, Willcox BE, Jones EY, Ross C, Miyasaka T, Walker RT, Tanaka H, Stammers DK, Stuart DI. J Med Chem. 1996;39(8):1589–1600. doi: 10.1021/jm960056x. [DOI] [PubMed] [Google Scholar]
- [21].Ren J, Esnouf R, Garman E, Somers D, Ross C, Kirby I, Keeling J, Darby G, Jones Y, Stuart D, et al. Nat Struct Biol. 1995;2(4):293–302. doi: 10.1038/nsb0495-293. [DOI] [PubMed] [Google Scholar]
- [22].Spence RA, Kati WM, Anderson KS, Johnson KA. Science. 1995;267(5200):988–993. doi: 10.1126/science.7532321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Basavapathruni A, Bailey CM, Anderson KS. J Biol Chem. 2004;279(8):6221–6224. doi: 10.1074/jbc.C300523200. [DOI] [PubMed] [Google Scholar]
- [24].Cruchaga C, Odriozola L, Andreola M, Tarrago-Litvak L, Martinez-Irujo JJ. Biochemistry. 2005;44(9):3535–3546. doi: 10.1021/bi048129z. [DOI] [PubMed] [Google Scholar]
- [25].Odriozola L, Cruchaga C, Andreola M, Dolle V, Nguyen CH, Tarrago-Litvak L, Perez-Mediavilla A, Martinez-Irujo JJ. J Biol Chem. 2003;278(43):42710–42716. doi: 10.1074/jbc.M212673200. [DOI] [PubMed] [Google Scholar]
- [26].Grobler JA, Stillmock K, Hu B, Witmer M, Felock P, Espeseth AS, Wolfe A, Egbertson M, Bourgeois M, Melamed J, Wai JS, Young S, Vacca J, Hazuda DJ. Proc Natl Acad Sci U S A. 2002;99(10):6661–6666. doi: 10.1073/pnas.092056199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Nature. 2010;464(7286):232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cooper DA, Steigbigel RT, Gatell JM, Rockstroh JK, Katlama C, Yeni P, Lazzarin A, Clotet B, Kumar PN, Eron JE, Schechter M, Markowitz M, Loutfy MR, Lennox JL, Zhao J, Chen J, Ryan DM, Rhodes RR, Killar JA, Gilde LR, Strohmaier KM, Meibohm AR, Miller MD, Hazuda DJ, Nessly ML, DiNubile MJ, Isaacs RD, Teppler H, Nguyen BY. N Engl J Med. 2008;359(4):355–365. doi: 10.1056/NEJMoa0708978. [DOI] [PubMed] [Google Scholar]
- [29].Kobayashi M, Nakahara K, Seki T, Miki S, Kawauchi S, Suyama A, Wakasa-Morimoto C, Kodama M, Endoh T, Oosugi E, Matsushita Y, Murai H, Fujishita T, Yoshinaga T, Garvey E, Foster S, Underwood M, Johns B, Sato A, Fujiwara T. Antiviral Res. 2008;80(2):213–222. doi: 10.1016/j.antiviral.2008.06.012. [DOI] [PubMed] [Google Scholar]
- [30].Shimura K, Kodama E, Sakagami Y, Matsuzaki Y, Watanabe W, Yamataka K, Watanabe Y, Ohata Y, Doi S, Sato M, Kano M, Ikeda S, Matsuoka M. J Virol. 2008;82(2):764–774. doi: 10.1128/JVI.01534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Maignan S, Guilloteau JP, Zhou-Liu Q, Clement-Mella C, Mikol V. J Mol Biol. 1998;282(2):359–368. doi: 10.1006/jmbi.1998.2002. [DOI] [PubMed] [Google Scholar]
- [32].Mazumder A, Engelman A, Craigie R, Fesen M, Pommier Y. Nucleic Acids Res. 1994;22(6):1037–1043. doi: 10.1093/nar/22.6.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Delelis O, Parissi V, Leh H, Mbemba G, Petit C, Sonigo P, Deprez E, Mouscadet JF. PLoS One. 2007;2(7):e608. doi: 10.1371/journal.pone.0000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. Nucleic Acids Res. 2005;33(3):977–986. doi: 10.1093/nar/gki241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Guiot E, Carayon K, Delelis O, Simon F, Tauc P, Zubin E, Gottikh M, Mouscadet JF, Brochon JC, Deprez E. J Biol Chem. 2006;281(32):22707–22719. doi: 10.1074/jbc.M602198200. [DOI] [PubMed] [Google Scholar]
- [36].Li M, Mizuuchi M, Burke TR, Jr., Craigie R. EMBO J. 2006;25(6):1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ren G, Gao K, Bushman FD, Yeager M. J Mol Biol. 2007;366(1):286–294. doi: 10.1016/j.jmb.2006.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bera S, Pandey KK, Vora AC, Grandgenett DP. J Mol Biol. 2009;389(1):183–198. doi: 10.1016/j.jmb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Van Maele B, Busschots K, Vandekerckhove L, Christ F, Debyser Z. Trends Biochem Sci. 2006;31(2):98–105. doi: 10.1016/j.tibs.2005.12.002. [DOI] [PubMed] [Google Scholar]
- [40].Al-Mawsawi LQ, Neamati N. Trends Pharmacol Sci. 2007;28(10):526–535. doi: 10.1016/j.tips.2007.09.005. [DOI] [PubMed] [Google Scholar]
- [41].Johnson MS, McClure MA, Feng DF, Gray J, Doolittle RF. Proc Natl Acad Sci U S A. 1986;83(20):7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Burke CJ, Sanyal G, Bruner MW, Ryan JA, LaFemina RL, Robbins HL, Zeft AS, Middaugh CR, Cordingley MG. J Biol Chem. 1992;267(14):9639–9644. [PubMed] [Google Scholar]
- [43].Zheng R, Jenkins TM, Craigie R. Proc Natl Acad Sci U S A. 1996;93(24):13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lee SP, Xiao J, Knutson JR, Lewis MS, Han MK. Biochemistry. 1997;36(1):173–180. doi: 10.1021/bi961849o. [DOI] [PubMed] [Google Scholar]
- [45].Cai M, Huang Y, Caffrey M, Zheng R, Craigie R, Clore GM, Gronenborn AM. Protein Sci. 1998;7(12):2669–2674. doi: 10.1002/pro.5560071221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cai M, Zheng R, Caffrey M, Craigie R, Clore GM, Gronenborn AM. Nat Struct Biol. 1997;4(7):567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- [47].Carayon K, Leh H, Henry E, Simon F, Mouscadet JF, Deprez E. Nucleic Acids Res. 2010;38(11):3692–3708. doi: 10.1093/nar/gkq087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cannon PM, Wilson W, Byles E, Kingsman SM, Kingsman AJ. J Virol. 1994;68(8):4768–4775. doi: 10.1128/jvi.68.8.4768-4775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Engelman A, Craigie R. J Virol. 1992;66(11):6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Engelman A, Englund G, Orenstein JM, Martin MA, Craigie R. J Virol. 1995;69(5):2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].LaFemina RL, Schneider CL, Robbins HL, Callahan PL, LeGrow K, Roth E, Schleif WA, Emini EA. J Virol. 1992;66(12):7414–7419. doi: 10.1128/jvi.66.12.7414-7419.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Masuda T, Planelles V, Krogstad P, Chen IS. J Virol. 1995;69(11):6687–6696. doi: 10.1128/jvi.69.11.6687-6696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wiskerchen M, Muesing MA. J Virol. 1995;69(1):376–386. doi: 10.1128/jvi.69.1.376-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wu X, Liu H, Xiao H, Conway JA, Hehl E, Kalpana GV, Prasad V, Kappes JC. J Virol. 1999;73(3):2126–2135. doi: 10.1128/jvi.73.3.2126-2135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yung E, Sorin M, Pal A, Craig E, Morozov A, Delattre O, Kappes J, Ott D, Kalpana GV. Nat Med. 2001;7(8):920–926. doi: 10.1038/90959. [DOI] [PubMed] [Google Scholar]
- [56].Wang JY, Ling H, Yang W, Craigie R. EMBO J. 2001;20(24):7333–7343. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Du L, Shen L, Yu Z, Chen J, Guo Y, Tang Y, Shen X, Jiang H. ChemMedChem. 2008;3(1):173–180. doi: 10.1002/cmdc.200700223. [DOI] [PubMed] [Google Scholar]
- [58].Khan E, Mack JP, Katz RA, Kulkosky J, Skalka AM. Nucleic Acids Res. 1991;19(4):851–860. doi: 10.1093/nar/19.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lodi PJ, Ernst JA, Kuszewski J, Hickman AB, Engelman A, Craigie R, Clore GM, Gronenborn AM. Biochemistry. 1995;34(31):9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- [60].Lutzke RA, Vink C, Plasterk RH. Nucleic Acids Res. 1994;22(20):4125–4131. doi: 10.1093/nar/22.20.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Vink C, Oude Groeneger AM, Plasterk RH. Nucleic Acids Res. 1993;21(6):1419–1425. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Woerner AM, Marcus-Sekura CJ. Nucleic Acids Res. 1993;21(15):3507–3511. doi: 10.1093/nar/21.15.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Williams KL, Zhang Y, Shkriabai N, Karki RG, Nicklaus MC, Kotrikadze N, Hess S, Le Grice SF, Craigie R, Pathak VK, Kvaratskhelia M. J Biol Chem. 2005;280(9):7949–7955. doi: 10.1074/jbc.M413579200. [DOI] [PubMed] [Google Scholar]
- [64].Chen JC, Krucinski J, Miercke LJ, Finer-Moore JS, Tang AH, Leavitt AD, Stroud RM. Proc Natl Acad Sci U S A. 2000;97(15):8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Li HY, Zawahir Z, Song LD, Long YQ, Neamati N. J Med Chem. 2006;49(15):4477–4486. doi: 10.1021/jm060307u. [DOI] [PubMed] [Google Scholar]
- [66].Maroun RG, Gayet S, Benleulmi MS, Porumb H, Zargarian L, Merad H, Leh H, Mouscadet JF, Troalen F, Fermandjian S. Biochemistry. 2001;40(46):13840–13848. doi: 10.1021/bi011328n. [DOI] [PubMed] [Google Scholar]
- [67].Zhao L, O'Reilly MK, Shultz MD, Chmielewski J. Bioorg Med Chem Lett. 2003;13(6):1175–1177. doi: 10.1016/s0960-894x(03)00040-4. [DOI] [PubMed] [Google Scholar]
- [68].Molteni V, Greenwald J, Rhodes D, Hwang Y, Kwiatkowski W, Bushman FD, Siegel JS, Choe S. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 4):536–544. doi: 10.1107/s0907444901001652. [DOI] [PubMed] [Google Scholar]
- [69].Shkriabai N, Patil SS, Hess S, Budihas SR, Craigie R, Burke TR, Jr., Le Grice SF, Kvaratskhelia M. Proc Natl Acad Sci U S A. 2004;101(18):6894–6899. doi: 10.1073/pnas.0400873101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kessl JJ, Eidahl JO, Shkriabai N, Zhao Z, McKee CJ, Hess S, Burke TR, Jr., Kvaratskhelia M. Mol Pharmacol. 2009;76(4):824–832. doi: 10.1124/mol.109.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Du L, Zhao YX, Yang LM, Zheng YT, Tang Y, Shen X, Jiang HL. Acta Pharmacol Sin. 2008;29(10):1261–1267. doi: 10.1111/j.1745-7254.2008.00863.x. [DOI] [PubMed] [Google Scholar]
- [72].Al-Mawsawi LQ, Fikkert V, Dayam R, Witvrouw M, Burke TR, Jr., Borchers CH, Neamati N. Proc Natl Acad Sci U S A. 2006;103(26):10080–10085. doi: 10.1073/pnas.0511254103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Ellison V, Brown PO. Proc Natl Acad Sci U S A. 1994;91(15):7316–7320. doi: 10.1073/pnas.91.15.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Hazuda DJ, Felock PJ, Hastings JC, Pramanik B, Wolfe AL. J Virol. 1997;71(9):7005–7011. doi: 10.1128/jvi.71.9.7005-7011.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Al-Mawsawi LQ, Hombrouck A, Dayam R, Debyser Z, Neamati N. Virology. 2008;377(2):355–363. doi: 10.1016/j.virol.2008.04.030. [DOI] [PubMed] [Google Scholar]
- [76].Wielens J, Headey SJ, Jeevarajah D, Rhodes DI, Deadman J, Chalmers DK, Scanlon MJ, Parker MW. FEBS Lett. 2010;584(8):1455–1462. doi: 10.1016/j.febslet.2010.03.016. [DOI] [PubMed] [Google Scholar]
- [77].Busschots K, De Rijck J, Christ F, Debyser Z. Mol Biosyst. 2009;5(1):21–31. doi: 10.1039/b810306b. [DOI] [PubMed] [Google Scholar]
- [78].Shinohara T, Singh DP, Fatma N. Prog Retin Eye Res. 2002;21(3):341–358. doi: 10.1016/s1350-9462(02)00007-1. [DOI] [PubMed] [Google Scholar]
- [79].Hombrouck A, De Rijck J, Hendrix J, Vandekerckhove L, Voet A, De Maeyer M, Witvrouw M, Engelborghs Y, Christ F, Gijsbers R, Debyser Z. PLoS Pathog. 2007;3(3):e47. doi: 10.1371/journal.ppat.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. Science. 2006;314(5798):461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- [81].Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M, Poeschla EM. J Virol. 2004;78(17):9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. J Biol Chem. 2003;278(35):33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- [83].Busschots K, Voet A, De Maeyer M, Rain JC, Emiliani S, Benarous R, Desender L, Debyser Z, Christ F. J Mol Biol. 2007;365(5):1480–1492. doi: 10.1016/j.jmb.2006.10.094. [DOI] [PubMed] [Google Scholar]
- [84].Cherepanov P, Sun ZY, Rahman S, Maertens G, Wagner G, Engelman A. Nat Struct Mol Biol. 2005;12(6):526–532. doi: 10.1038/nsmb937. [DOI] [PubMed] [Google Scholar]
- [85].Rahman S, Lu R, Vandegraaff N, Cherepanov P, Engelman A. Virology. 2007;357(1):79–90. doi: 10.1016/j.virol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- [86].Ciuffi A, Bushman FD. Trends Genet. 2006;22(7):388–395. doi: 10.1016/j.tig.2006.05.006. [DOI] [PubMed] [Google Scholar]
- [87].Engelman A, Cherepanov P. PLoS Pathog. 2008;4(3):e1000046. doi: 10.1371/journal.ppat.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Meehan AM, Poeschla EM. Biochim Biophys Acta. 2010;1799(3–4):182–191. doi: 10.1016/j.bbagrm.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Poeschla EM. Cell Mol Life Sci. 2008;65(9):1403–1424. doi: 10.1007/s00018-008-7540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Du L, Zhao Y, Chen J, Yang L, Zheng Y, Tang Y, Shen X, Jiang H. Biochem Biophys Res Commun. 2008;375(1):139–144. doi: 10.1016/j.bbrc.2008.07.139. [DOI] [PubMed] [Google Scholar]
- [91].Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. Proc Natl Acad Sci U S A. 2005;102(48):17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].De Luca L, Barreca ML, Ferro S, Christ F, Iraci N, Gitto R, Monforte AM, Debyser Z, Chimirri A. ChemMedChem. 2009;4(8):1311–1316. doi: 10.1002/cmdc.200900070. [DOI] [PubMed] [Google Scholar]
- [93].Al-Mawsawi LQ, Christ F, Dayam R, Debyser Z, Neamati N. FEBS Lett. 2008;582(10):1425–1430. doi: 10.1016/j.febslet.2008.02.076. [DOI] [PubMed] [Google Scholar]
- [94].Christ F, Voet A, Marchand A, Nicolet S, Desimmie BA, Marchand D, Bardiot D, Van der Veken NJ, Van Remoortel B, Strelkov SV, De Maeyer M, Chaltin P, Debyser Z. Nat Chem Biol. 2010;6(6):442–448. doi: 10.1038/nchembio.370. [DOI] [PubMed] [Google Scholar]
- [95].Tsantrizos YS, Boes M, Brochu C, Fenwick C, Malenfant E, Mason S, Pesant M. Boehringer Ingelheim Pharma. 2007 PCT/CA2007/000845. [Google Scholar]