SUMMARY
Kinases execute pivotal cellular functions and are therefore widely investigated as potential targets in anticancer treatment. Here we analyze the kinase gene expression profiles of various tumor types and reveal the wee1 kinase to be overexpressed in glioblastomas. We demonstrate that WEE1 is a major regulator of the G2 checkpoint in glioblastoma cells. Inhibition of WEE1 by siRNA or small molecular compound in cells exposed to DNA damaging agents results in abrogation of the G2 arrest, premature termination of DNA repair, and cell death. Importantly, we show that the small-molecule inhibitor of WEE1 sensitizes glioblastoma to ionizing radiation in vivo. Our results suggest that inhibition of WEE1 kinase holds potential as a therapeutic approach in treatment of glioblastoma.
INTRODUCTION
Glioblastoma (GBM) is one of the most aggressive human cancers and the most common primary brain tumor. The median survival of GBM patients is <15 months because this tumor is inherently resistant to conventional therapy (Stupp et al., 2009). Although the conventional treatment with surgery, irradiation (IR), and temozolomide (TMZ) postpones tumor progression and extends patients survival to some extent, these tumors universally recur and unrelentingly result in patient death. Despite recent advances in understanding the underlying molecular mechanisms, there has been little improvement in clinical outcome (Wen and Kesari, 2008).
Manipulation of oncogenic kinase activity has become a therapeutic concept in human cancer because kinases regulate crucial cellular functions such as proliferation, apoptosis, cell metabolism, migration, DNA damage repair, and responses to the microenvironment (Manning et al., 2002). Several human cancers are considered to be kinase-driven (Weinstein, 2002; Krause and Van Etten, 2005), and inhibitors of several cancer-driving kinases are under evaluation as potential therapeutic agents (Cohen, 2002; Vieth et al., 2005). Most of these inhibitors aim at stagnation of tumor growth by interrupting the replicative cycle of cancer cells. Examples include inhibitors of epidermal growth factor receptor (EGFR), polo-like kinase 1 (PLK1), v-akt murine thymoma viral oncogene homologue (AKT), mitogen-activated protein kinase (MAPK), protein kinase C (PKC), vascular endothelial growth factor receptor (VEGFR), and platelet-derived growth factor receptor (PDGFR) (reviewed by Zhang et al., 2009). These therapeutic kinase targets are mainly deregulated by either mutation, protein fusion or gene overexpression (Krause and Van Etten, 2005). Thus far, results from clinical trials testing the efficacy of kinase inhibitors in patients with GBM have been disappointing (Omuro et al., 2007; De Witt Hamer, 2010).
In this study we focused on the kinase gene expression profile of GBM using publicly available gene expression data sets to identify additional putative therapeutic targets.
Significance.
The resistance of glioblastoma cells to irradiation and chemotherapy is partly due to their proficient ability to repair treatment-induced DNA damage during the G2 cell-cycle arrest. Many kinase inhibitors intend to prolong cell-cycle arrest to halt cancer cell division. Here, we inhibit the WEE1 kinase, a gatekeeper of the DNA damage-induced G2 arrest, “pushing” glioblastoma cells through the G2 arrest phase and thereby inducing mitotic catastrophe and cell death. This strategy results in extensive cytotoxicity in vitro and, more importantly, eradication of irradiated brain tumors in mice, without showing adverse side effects. Therefore, manipulation of WEE1 activity may prove therapeutically attractive as a sensitizing approach for glioblastoma treatment.
RESULTS
In Silico Analysis of Microarray Data Identifies Cancer-Specific Kinase Expression Profiles
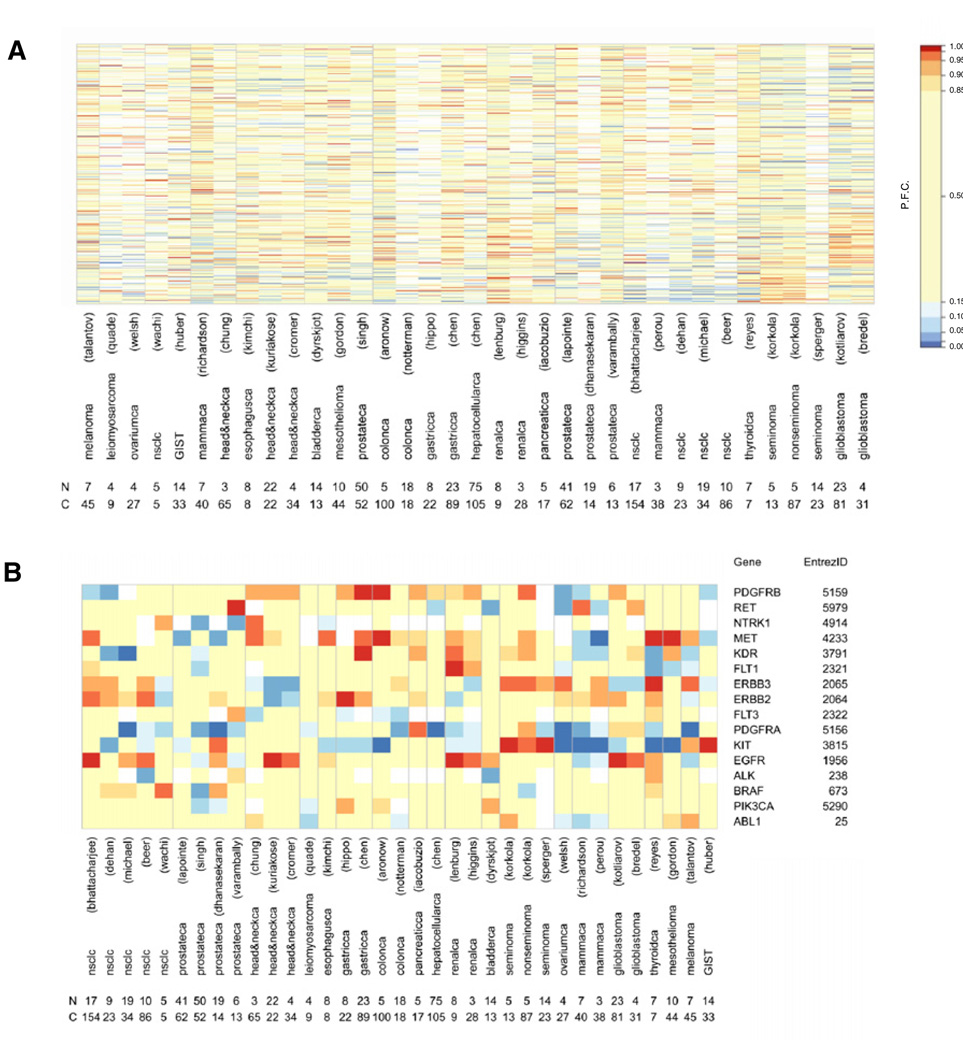
To identify kinase targets, we used a cancer-wide approach and selected 34 cancer-versus-normal data sets (Table S1 available online), including two GBM data sets obtained by different microarray platforms in distinct laboratories (Bredel et al., 2005; Kotliarov et al., 2006). Two parameters of gene expression were determined, details of which are described in Supplemental Experimental Procedures. First, to compare the differential kinase gene expression of cancer-versus-normal samples among data sets, the fold change was determined for all genes within a data set. Then, the percentile of fold change was determined for each kinase within a data set to relate its expression value to all genes in that data set. This allowed comparison of expression levels of each kinase between data sets. Second, to compare the frequency at which overexpression occurs in cancer samples between data sets, the frequency of overexpression was determined for each kinase within each cancer-versus-normal data set. To recover the human kinome from the genes present in the data sets, the Entrez GeneIDs of the human protein kinase family (518 kinases) and the human lipid kinase family (33 kinases) were retrieved (Figures 1A, Figure S1, and Table S2) (Manning et al., 2002; Peri et al., 2003). The vast majority of kinase genes were represented on the microarray platforms (Table S1 and Table S2). We found distinct kinase expression profiles in various cancer types (Figure 1A). Thus, potential therapeutic targets overexpressed in a specific type of cancer may not necessarily be valid in other cancer types.
Figure 1. Kinase Expression Profiles of Cancer Data Sets.
(A) Heatmap of percentile fold change of gene expression of kinases (rows) in cancer data sets (columns). Number of normal tissue samples (N) and cancer samples (C) per data set are shown. (B) Selected list of kinases expressed in various types of cancer. Number of normal tissue samples (N) and cancer samples (C) per data set are shown. Percentile fold change values as in color legend, missing values in white. P.F.C., percentile fold change.
See also Figure S1, Table S1, and Table S2.
To validate our approach, we selected kinases previously demonstrated to be overexpressed in specific cancers and determined their expression across our kinase profiles (Figure 1B). Our analysis revealed v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (kit) to be selectively overexpressed in seminomas and gastrointestinal stromal tumors, which show clinical response to anti-KIT treatment (Pedersini et al., 2007; Demetri et al., 2002). Moreover, egfr overexpression was confirmed in head and neck cancers, non-small cell lung cancers, and renal cell cancers, for which anti-EGFR treatment can prolong overall survival (Lynch et al., 2004; Agulnik et al., 2007; Ravaud et al., 2008).
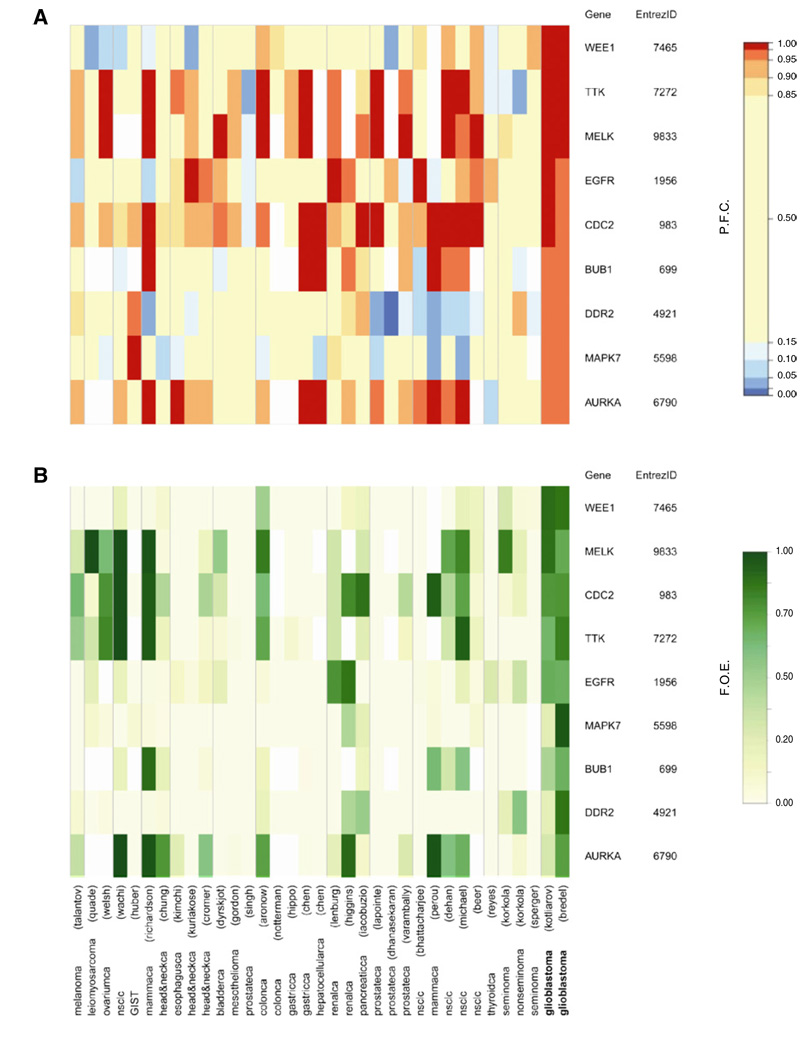
Although we retrieved the kinase expression profiles of several tumor types, our prime focus was to identify potential therapeutic targets in glioblastoma. We considered overexpression at a level surpassing 95% of all genes in a data set as substantial. Additionally, we considered overexpression in >20% of the patient population as frequent. A listing of nine kinases was obtained that were both substantially (Figure 2A) and frequently (Figure 2B) overexpressed in the two available GBM data sets. In previous investigations four of these nine selected kinases, aurora kinase A (aurka) (Klein et al., 2004), egfr (Nicholas et al., 2006), maternal embryonic leucine zipper kinase (melk) (Nakano et al., 2008), and cyclin-dependent kinase 1 (cdc2) (Hodgson et al., 2009), were described as relevantly overexpressed in human GBM samples, emphasizing the biological plausibility of our results. The other five kinases were not previously associated with human GBM. pdgfr, kinase insert domain receptor (kdr), fms-related tyrosine kinase 4 (flt4), transforming growth factor, beta receptor 1 (tgfbr1), and transforming growth factor, beta receptor 2 (tgfbr2) had been described to be overexpressed in GBM (Rich and Bigner, 2004) but they were not included in our list. Despite marked overexpression in the two GBM data sets (Table S3 and Table S4), these kinases did not meet our selection criteria. Given the noise levels in microarray data, we preferred high specificity over sensitivity.
Figure 2. Glioblastoma-Specific Kinases.
Heatmaps of percentile fold change gene expression (A) and frequency of overexpression (B) of nine glioblastoma-specific kinases (rows) in data sets (columns). F.O.E., frequency of overexpression; P.F.C., percentile fold change. See also Table S3 and Table S4.
wee1 mRNA and Protein Are Overexpressed in GBM and Its Expression Level Correlates with Survival
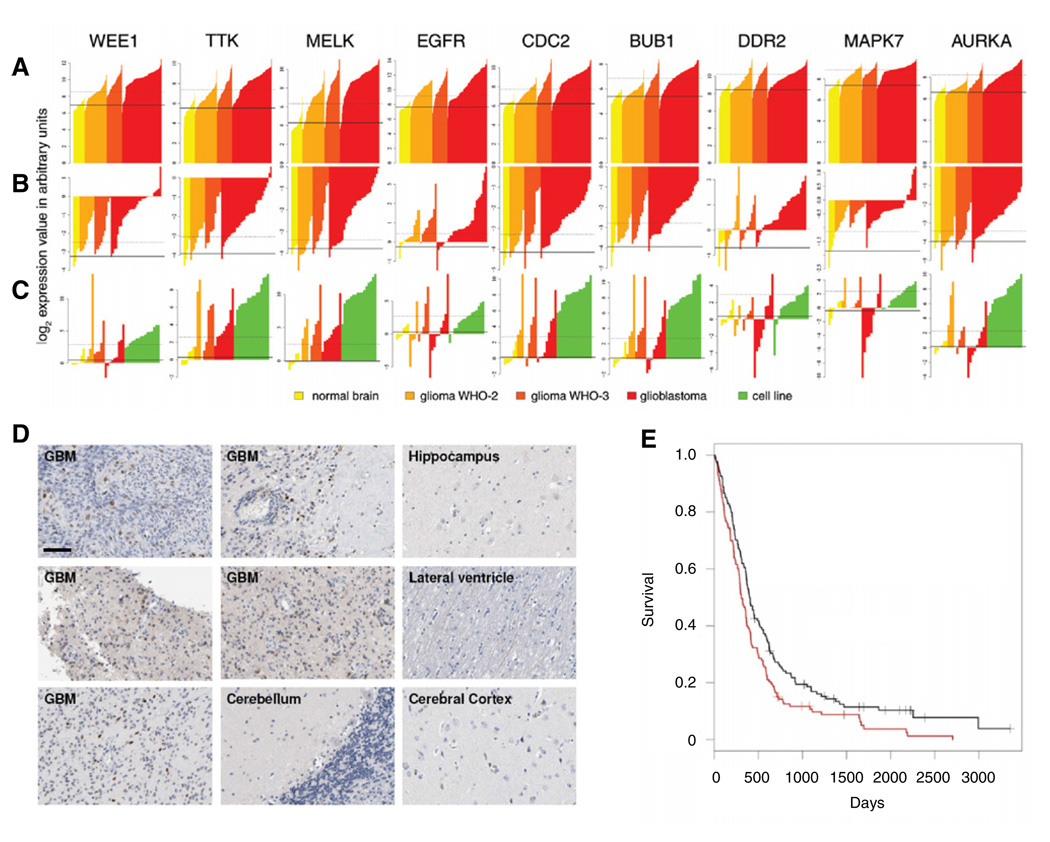
The individual data for the nine overexpressed kinases retrieved from the GBM studies is presented in Figures 3A and 3B and Figure S2, according to glioma grade and histological subtype. To validate the results obtained in silico, we determined mRNA expression of these kinases in GBM cell lines and primary samples of GBM and normal brain using quantitative reverse transcription polymerase chain reaction (RT-PCR) (Figure 3C, Table S5, and Table S6).
Figure 3. Gene Expression Levels Per Sample in Two GBM Array Data Sets and Quantitative PCR Data for the Nine Glioblastoma-Specific Kinases.
(A–C) Bar charts of expression intensities in arbitrary units for the nine glioblastoma-specific kinases in samples from two GBM data sets, (A) Kotliarov et al. (2006),
(B) Bredel et al. (2005), and (C) confirming quantitative RT-PCR data in human samples and GBM cell lines (see Table S5 for details and ordering of samples). Each bar represents one sample. Color coding is according to grade. The solid line represents the mean of expression intensities for the normal tissue samples; the dotted line represents the threshold for overexpression (see Supplemental Experimental Procedures).
(D) Immunohistochemical analysis of WEE1 in GBMs and non-neoplastic brain tissues. Scale bar represents 200 µm.
(E) Kaplan-Meier survival curves of GBM patients divided on basis of wee1 expression, red indicates high and black indicates low wee1 expression.
See also Figure S2 and Tables S5–S7.
Because WEE1 kinase plays an important role in controlling the cell-cycle progression (Russell and Nurse, 1987) and because it topped the ranking of overexpressed kinases (Figure 2, Figure 3, and Figure S2), we further studied its role in GBM. We first determined the levels of WEE1 protein expression in normal brain and GBM tissue sections using immunohistochemistry. Consistent with the gene expression data, GBM cell nuclei contained high levels of WEE1 protein, as compared to non-neoplastic brain regions (Figure 3D).
We then investigated the correlation between wee1 mRNA level and patient survival using published data from 267 GBM patients (Lee et al., 2008). Expression of wee1 was classified as high or low, based on whether the values were above or below the median wee1 expression levels, respectively. This showed high tumor wee1 expression to be significantly correlated with worse patient survival (log rank p < 0.0059), patients with high wee1-expressing GBM had a median survival time of 308 days and a 2-year survival rate of 14% versus a median survival time of 402 days and a 2-year survival rate of 26% for patients with low wee1-expressing GBM (Figure 3E). wee1 expression level was significantly associated with patient survival after correction for confounding by age, gender, tumor recurrence, and prior treatment modalities, in a multivariate Cox proportional-hazards regression model (hazard ratio: 0.69, 95% confidence interval: 0.53–0.89, p = 0.0047 and Table S7).
Inhibition of WEE1 Abrogates the G2 Checkpoint and Sensitizes GBM Cells to TMZ and IR In Vitro
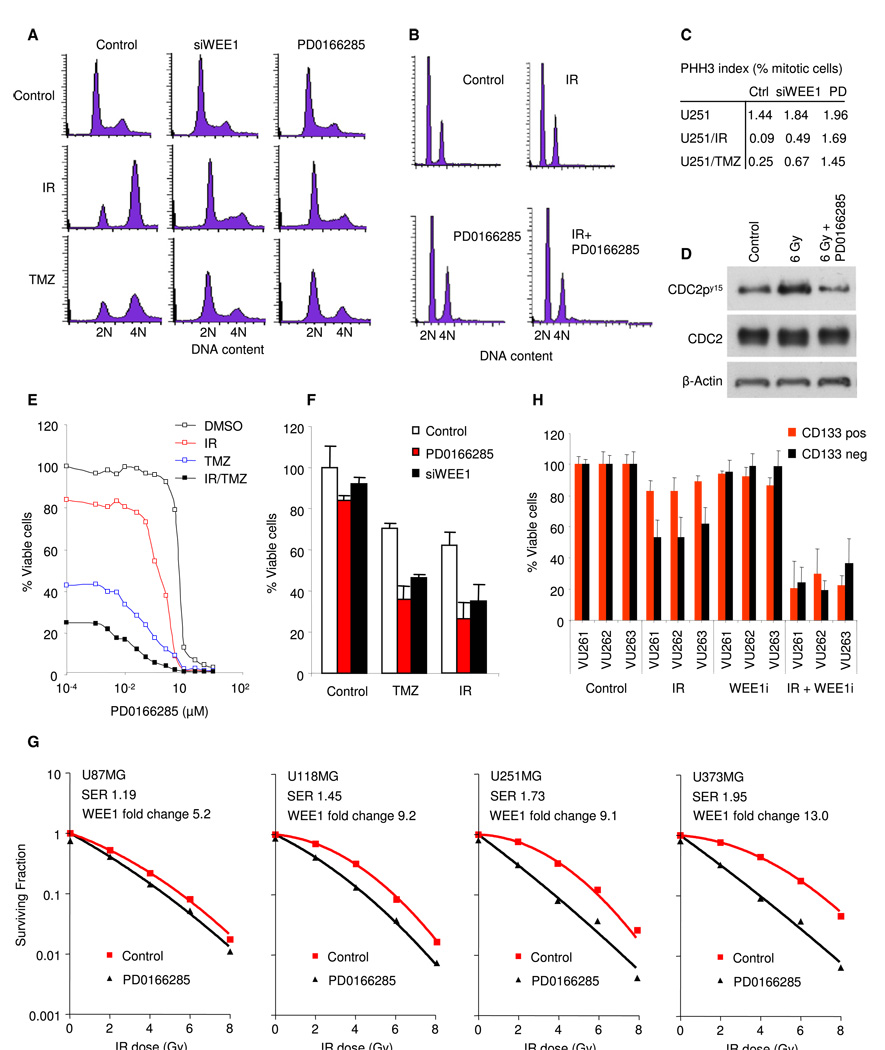
After DNA damage induction, GBM cells mainly arrest in the G2 phase, due to an impaired G1 checkpoint (Hirose et al., 2001). Indeed, U251MG GBM cells arrested in the G2 phase after IR and TMZ treatment (Figures 4A and 4C). We then focused on the role of WEE1 in the G2 checkpoint activation in response to DNA damage. To specifically inhibit WEE1 function, we transfected GBM cells with siRNA directed against the wee1 mRNA. As determined by quantitative RT-PCR, the wee1 siRNA efficiently knocked down wee1 mRNA levels, as compared to nontargeting control siRNA (Figure S3A). After 48 hr the transfected cells were either exposed to 6 Gray (Gy) of IR or treated with 100 µMTMZ and 16 hr later cell-cycle distribution was monitored by fluorescence activated cell sorting. DNA content was monitored by propidium iodide staining and phospho-histone-H3 staining was used as an indicator of mitotic entry. Knock down of wee1 resulted in significant abrogation of the IR- and TMZ-induced G2 arrest and decrease in the amount of mitotic cells (Figures 4A and 4C and Figures S3B and S3C). Similar abrogation of the DNA damage-induced arrest in U251MG cells was achieved by using the WEE1 inhibitor PD0166285 (Wang et al., 2001) (Figures 4A and4C, Figure S3D, and Figure S4E). Comparative cell-cycle results were obtained using additional established GBM cell lines (U118MG, U87MG, U373MG) and primary GBM cells (VU147, VU148, E98) (Figures S3D and S3E). These results indicate that WEE1 is a major determinant of the DNA damage-inducedG2 arrest in GBM cells. Importantly, no adverse effects of WEE1 inhibition on cell cycle were observed in primary human fibroblasts and astrocytes, showing no loss of G1 arrest after exposure to IR or TMZ in the presence or absence of WEE1 inhibitor or siRNA against wee1 (Figure 4B and Figures S3D and S3E). Furthermore, no effects of PD0166285 on fibroblasts and astrocytes were observed by WST-1 cell viability measurements and microscopical monitoring of cellular morphology (data not shown). Inhibition of WEE1 also reduces phosphorylation of its downstream substrate CDC2 (Figure 4D and Figure S3F). This suggests that the inhibitory CDC2 phosphorylation is reduced by inhibition of WEE1 and thereby the CDC2-mediated G2 arrest is abolished in GBM cells.
Figure 4. Inhibition of WEE1 Abrogates the G2 Checkpoint and Enhances Chemo- and Radiation-Sensitivity In Vitro.
(A) Cell-cycle analysis of U251MG GBM cells at 16 hr after treatment with 6 Gy of IR, 100 µM of TMZ, and 1 µM of PD0166285.
(B) Cell-cycle analysis of human primary fibroblasts at 16 hr after treatment.
(C) Quantitation of PHH3 expression in U251MG cells treated with WEE1 siRNA, WEE1 inhibitor, or nontreated.
(D) Western blot analysis of CDC2, phosphorylated CDC2, and β-actin in U251MG GBM cells at 16 hr after treatment.
(E) Representative cell counts of U251MG GBM cells at 4 days after treatment.
(F) WST-1 viability analysis of U251MG cells at 4 days after treatment.
(G) Colony formation of U87MG, U251MG, U118MG, and U373MG, at 7–14 days after treatment.
(H) WST-1 viability analysis of CD133 sorted primary GBM cells at 4 days after treatment. Shown are averages of experiments carried out in triplicate, error bars indicate standard deviation. ***p < 0.001, t test.
See also Figure S3.
We then investigated the effects of WEE1 inhibition on the viability of cells treated with IR, TMZ, or both. wee1 silencing by siRNA or WEE1 inhibition by PD0166285 had a significant sensitizing effect in combination with 6 Gy of IR, 100 µM of TMZ, or both (Figures 4E and 4F and Figure S3G). At concentrations up to 1 µm, PD0166285 had no effects on the viability of GBM cells in the absence of IR and TMZ (Figure 4E). In addition, radiosensitizing effects of PD0166285 in GBM cells could be measured by colony formation assay (Figure 4G), which correlated with wee1 gene expression levels in the GBM cells. U373MG cells with a fold change log2 wee1 expression (FC) compared to six non-neoplastic brain samples of 13.0 (see Figure 3C) showed the largest sensitization, with a sensitizing enhancement ratio (SER) of 1.95, U251MG cells (FC: 9.1, SER: 1.73) and U118MG cells (FC: 9.2, SER: 1.45) showed intermediate sensitization, whereas U87MG cells (FC 5.2 that is within the upper threshold for normal expression of FC 6.4 and SER of 1.19) showed the least sensitization (Figure 3C and Figure 4G). In contrast, no correlation was observed between p53 mutation status and WEE1 inhibitor sensitivity in the GBM cells analyzed here. However, U87 cells, which express wee1 mRNA levels within the normal range (Figure 3C and Figure 4G) and express the wild-type p53 (Figure S3H), showed the least PD0166285-mediated radiosensitization (Figure 4G). There was no significant effect of PD0166285 on colony formation in non-irradiated GBM cells.
A subpopulation of GBM cells, the so-called cancer stem-like cells, has been described to be particularly resistant to chemoand radiotherapy (Bao et al., 2006; Liu et al., 2006). In silico analysis using the ONCOMINE database (Rhodes et al., 2004) showed that wee1 was overexpressed to an even higher extent in a subpopulation of GBM cells cultured in neural basal medium (Lee et al., 2006) (Figure S3I). GBM neurospheres formed by such cells expressed CD133, Nestin, and SOX2 (Figure S3J), putative GBM stem-like cell markers (Lee et al., 2006). Next, the radiosensitizing effect of WEE1 inhibitor PD0166285 on primary GBM neurospheres was determined. GBM neurospheres failed to respond to exposure to IR alone, confirming their radio-resistance (Bao et al., 2006). Importantly, dramatic cell death was measured when primary GBM neurospheres were exposed to IR in the presence of PD0166285. Although GBM stem-like cell markers are still subject of continuous debate (Chen et al., 2010), we isolated primary CD133 positive and negative GBM cells with high wee1 expression and intermediate wee1 expression, respectively, as determined by qRT-PCR (Figure S3I). Subsequently we treated the sorted cells with 0.5 µM of PD0166285 and 6 Gy of IR. After 4 days the radiation-resistant CD133 positive GBM cells were efficiently killed as determined by WST-1 cell viability assay (Figure 4H). These results suggest that radiation resistance of GBM stem-like cell populations can be overcome by WEE1 inhibition in vitro.
Inhibition of WEE1 in Irradiated GBM Cells Leads to Mitotic Catastrophe Due to Residual DNA Damage
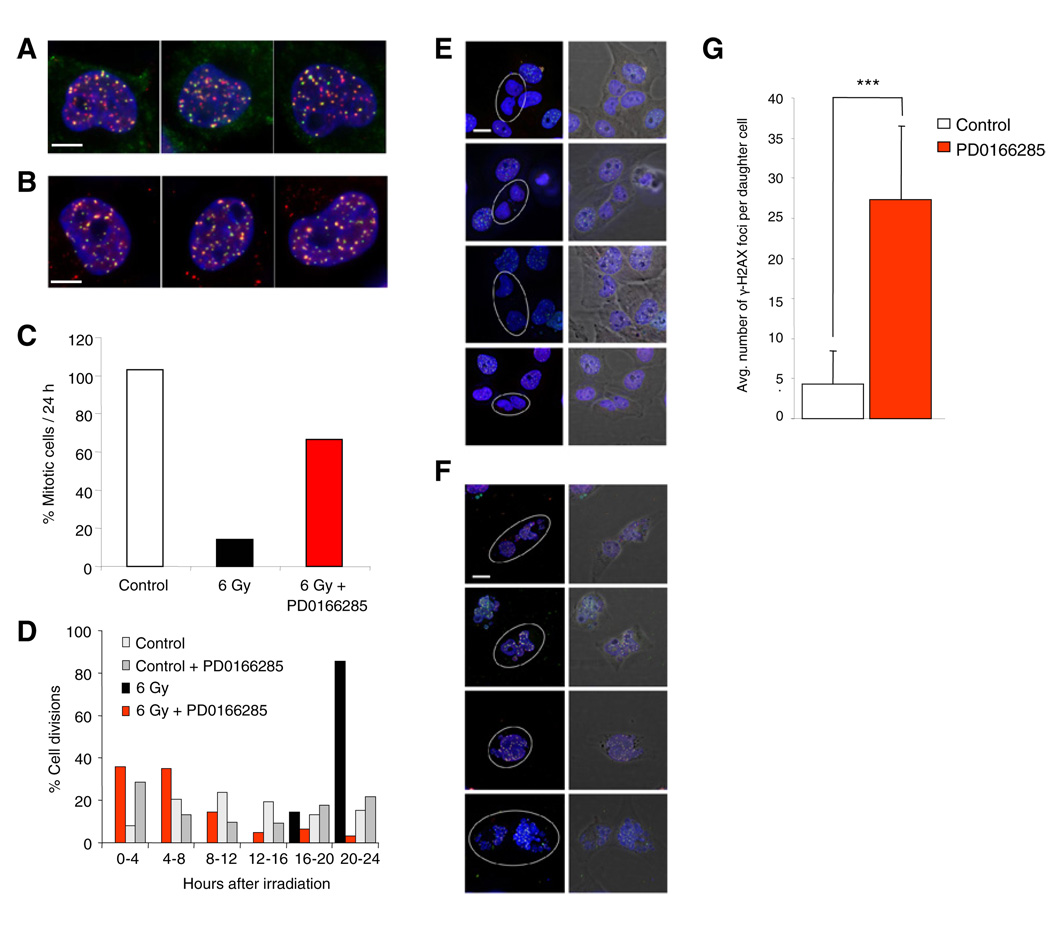
After induction of DNA damage, WEE1 activates and sustains the G2 cell-cycle arrest until DNA damage is sufficiently repaired (Rowley et al., 1992). Therefore, inhibition of WEE1 should abrogate G2 arrest activation and result in increased amounts of unrepaired DNA damage in cells prematurely entering mitosis. To determine the effect of WEE1 inhibition on the rejoining of double-strand breaks (DSBs) and on DSB-induced cell-cycle arrest, we exposed GBM cells to IR in the presence or absence of the WEE1 inhibitor. IR resulted in induction of DSBs as evidenced by formation of ionizing radiation-induced foci (IRIF) by a DSB marker γ-H2AX (Rogakou et al., 1999), and by repair factors ATMp1981, MDC1, and MRE11 (Goldberg et al., 2003; Lavin, 2007) (Figures 5A and 5B). Phase-contrast time-lapse imaging of the U251MG cells at 10-min intervals showed that IR induced a considerable delay in the onset of mitosis (Figures 5C and 5D). In contrast, cells irradiated in the presence of PD0166285 failed to delay their mitotic entry (Figures 5C and 5D), in agreement with the role of WEE1 in activating and maintaining the G2 arrest.
Figure 5. Inhibition of WEE1 in Irradiated GBM Cells Leads to Mitotic Catastrophe.
(A and B) Immunofluorescence visualization of IRIF in irradiated U251MG cells in triplicate. Cells were exposed to 2 Gy, fixed 30 min later and stained for (A) total DNA (blue), MDC1 (red) and ATMp1981 (green) and (B) total DNA (blue), MRE11 (red) γ-H2AX (green). Scale bar represents 5 µm.
(C) Quantitation of the percentage of cells that entered mitosis during the first 24 hr of imaging after sham-irradiation (Control) or exposure to 6 Gy in the presence or absence of 500 nM of WEE1 inhibitor. At least 300 cells were scored per data point.
(D) Distribution of the percentage of cells entering mitosis from (C) in the different periods after indicated treatment.
(E and F) Immunofluorescence analysis of IRIF in postmitotic cells. Cells were exposed to 6 Gy, imaged for 16 hr in the absence (E) or presence (F) of 500 nM WEE1 inhibitor, fixed and stained for total DNA (blue), MDC1 (red), and ATMp1981 (green). Postmitotic cells are indicated by circles. Scale bar represents 10 µm.
(G) Quantitation of the number of IRIF in postmitotic cells treated as in (E) and (F). IRIF in at least 50 cells were scored per data point. Error bars indicate standard deviation. ***p < 0.001, t test.
See also Movie S1.
In the absence of a functional G2 arrest, irradiated GBM cells may not be capable of repairing the DNA damage before entering mitosis, leading to mitotic catastrophe. We irradiated GBM cells and then time-lapse imaged the cells in the presence or absence of WEE1 inhibitor (Movie S1). Cells were fixed and stained for γ-H2AX and MDC1 and analyzed for completion of mitosis by fluorescence microscopy. Exposure to IR alone resulted in only a few residual DSBs in the daughter cell nuclei, indicating near-complete DSB repair before the cells entered mitosis (Figures 5E and 5G). In contrast, cells irradiated in the presence of WEE1 inhibitor presented their daughter cells with considerable amount of unrepaired DSBs (Figures 5F and 5G). Additionally, the presence of PD0166285 resulted in largely fragmented nuclei of the daughter cells (Figure 5F and Movie S1), indicative of mitotic catastrophe due to DNA fragments devoid of centromeres, which can not be properly segregated during mitosis (Loffler et al., 2006). Together, these results support the concept that G2 abrogation by WEE1 inhibition results in mitotic catastrophe due to unrepaired DSBs that explains the radiosensitizing properties of the WEE1 inhibitor.
WEE1 Inhibition Enhances Radiosensitivity in Orthotopic GBM Models
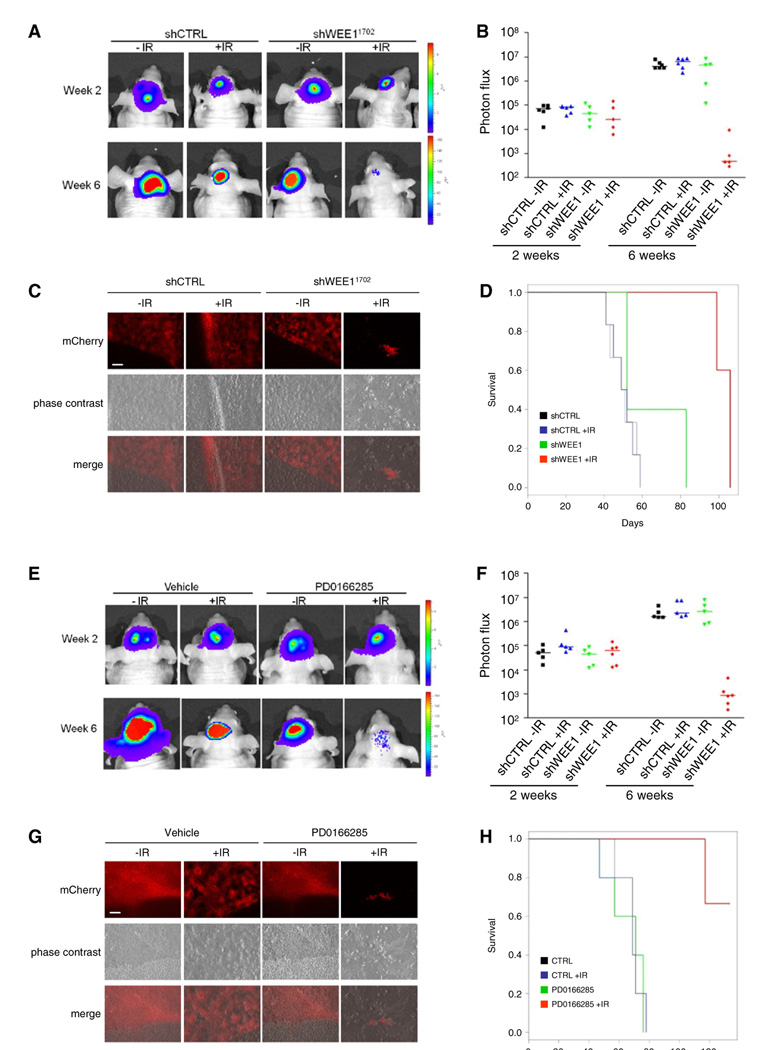
To compare the antitumor efficacy of short hairpin-mediated wee1 knock down alone or in combination with IR in vivo, we first used a bioluminescent orthotopic model of human U251-FlucmCherry cells (U251-FM) (Candolfi et al., 2007). U251-FM cells were transduced with lentivectors encoding a control shRNA or a shRNA directed against wee1 (shWEE11702). First, the efficiency of wee1 knock down and reduction of cell viability in response to wee1 knock down after IR-mediated DNA damage was determined in these cells (Figure S4A). Second, control or shWEE1 transduced U251-FM cells were implanted in the brain of nude mice. The growth of tumors was monitored weekly by bioluminescence imaging. Two weeks after intracranial injection of the transduced cells, 50% of the mice in each group were exposed to a single dose of 6 Gy. In mice injected with U251-FM cells expressing shControl strong tumor progression was observed in both irradiated and non-irradiated groups at 6 weeks after injection of the cells (Figures 6A and 6B). Similarly, the nonirradiated U251-FM-shWEE1 cells showed strong increase in bioluminescence signal after 6 weeks, whereas mice injected with U251-FM-shWEE1 cells showed significant tumor regression after IR at 6 weeks after injection (Figures 6A and 6B). Tumor burden was markedly reduced in irradiated mice carrying shWEE1 transduced cells (Figure 6C). Survival analysis showed a significant (p = 0.001) advantage for combining irradiation with wee1 knock down (Figure 6D). These results indicate that wee1 knockdown sensitizes U251-FM GBMs to IR in vivo.
Figure 6. In Vivo Analysis of WEE1 Inhibition in an Orthotopic U251-FM Mouse Model.
(A) U251-FM cells stably expressing shWEE1 or shControl were injected intracranially. The mice were divided in two groups of which one was irradiated with 6 Gy. Representative bioluminescence imaging results are shown 2 and 6 weeks after cancer cell injections.
(B) Relative photon flux at 2 and 6 weeks after cancer cell injections.
(C) Representative mCherry images of the brain tumors after treatment. Size bar represents 200 µm.
(D) wee1 silencing enhances radiosensitivity in vivo, as depicted by survival curves (n = 5/group).
(E) U251-FM cells were injected intracranially, and mice were irradiated with 6 Gy, and injected intraperitoneally with 500 µl of 20 µM of PD0166285 or PBS control for 5 consecutive days. Representative bioluminescence imaging results are shown 2 and 6 weeks after cancer cell injections.
(F) Relative photon flux 2 and 6 weeks after cancer cell injections.
(G) Representative mCherry images of the brain tumors after treatment. Size bar represents 200 µm.
(H) PD0166285 enhances radiosensitivity in vivo, depicted is the survival of the mice (n = 5/group).
See also Figure S4.
To determine the pharmacological characteristics and anti-tumor efficacy of PD0166285 on GBM outcome in vivo, we first implanted GBM cells subcutaneously (SC) into nude mice and determine the PD0166285 dosage sufficient to inhibit WEE1 activity in vivo. Mice were injected with various doses of PD0166285 (0, 20, 100, 200, or 400 µM in 100 µl) 20 days after tumor cells implantation. No adverse side effects were observed at the concentrations used. Tumors were removed 24 hr later and analyzed for inhibition of WEE1-mediated CDC2 phosphorylation by western blotting (Figure S4B). A single injection of 20 µM of the WEE1 inhibitor considerably reduced the CDC2 phosphorylation and was used in further experiments. Next, U251-FM human GBM cells were injected intracranially into nude mice. Mice with established GBMs were treated daily with PD0166285 or phosphate buffer solution (PBS) control via an intraperitoneal (IP) injection starting at day 14 after injection of the GBM cells. At day 15, the mice were sham irradiated or exposed to a single dose of 6 Gy. The results showed strong tumor progression in both irradiated and non-irradiated mock treated mice at 6 weeks after injection of the cells (Figures 6E and 6F). Similarly, the non-irradiated PD0166285 treated mice showed strong increase in tumor signal after 6 weeks. In contrary, irradiated mice treated with PD0166285 showed significant tumor regression 6 weeks after tumor injection (Figures 6E and 6F). Additionally, tumor burden was markedly reduced in this animal group (Figure 6G). Survival analysis showed a significant (p = 0.001) advantage for combining irradiation with PD0166285 (Figure 6H). These results indicate that pharmacological targeting of WEE1 sensitizes U251-FM GBM tumors to IR in vivo.
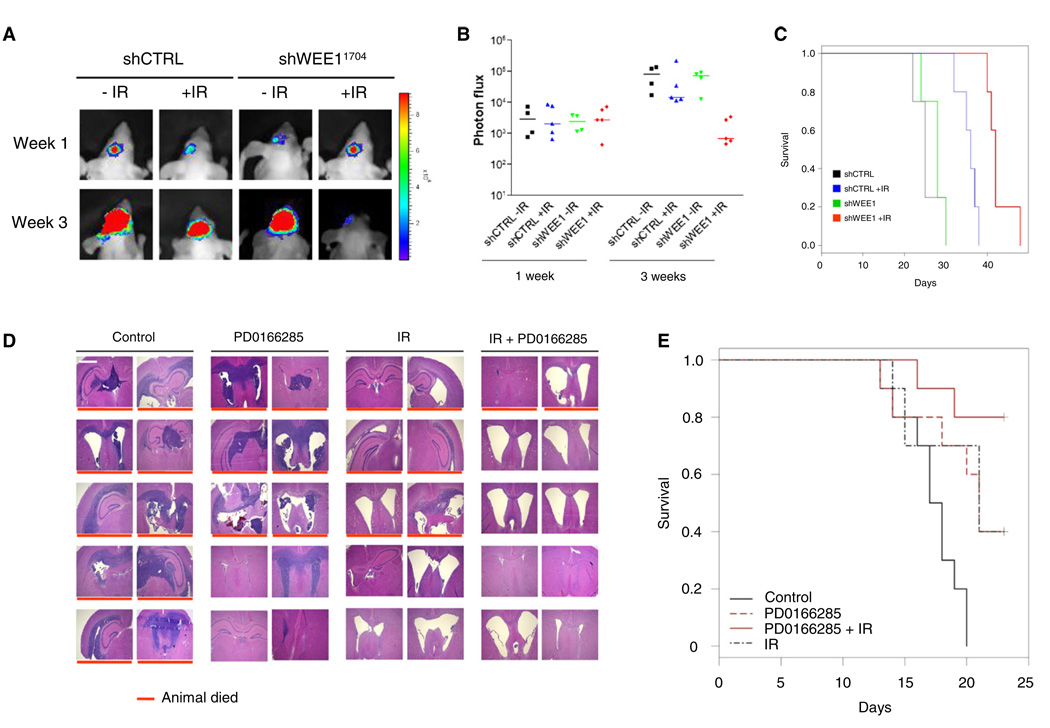
To confirm these results in an independent orthotopic GBM model we used the E98 mouse model (Claes et al., 2008) because it displays a highly invasive phenotype characteristic for human GBM. We verified that WEE1 protein was overexpressed in this model by immunohistochemistry on tumor sections (Figure S5A). To improve tumor detection, the E98 cells were transduced to stably express Fluc and mCherry (E98-FM). To determine whether wee1 knock down by shRNAs caused radiosensitization in vivo, E98-FM cells were transduced with lentivectors encoding shControl, or shWEE11704 directed against wee1. The efficiency of wee1 knock down and reduction of cell viability in response to WEE1 knock down combined with IR was verified in these cells (Figure S5B). We then stereotactically injected E98-FM-shControl or E98-FM-shWEE1 cells into nude mice. After 10 days the mice were divided into two treatment groups, with and without IR. In line with the U251-FM model, irradiated mice carrying shWEE1 transduced E98-FM tumors showed markedly reduced tumor burden (Figures 7A and 7B), and their survival was significantly prolonged as compared to other treatment groups (Figure 7C).
Figure 7. In Vivo Analysis of WEE1 Inhibition in an Invasive E98 GBM Mouse Model.
(A) E98-FM cells stably expression shWEE1 or shControl were grown in the brains of nude mice. The mice were divided in two arms of which one was irradiated with 3.5 Gy. Representative bioluminescence imaging results are shown 7 days and 3 weeks after onset of tumor growth.
(B) Relative photon flux at 7 days and 3 weeks after onset of tumor growth. (n = 4/group for control and n = 5 for IR treated).
(C) Survival curves of the mice in (B).
(D) E98 GBMs were grown in the brain of nude mice, irradiated with 8 Gy, and injected intraperitoneally with 500 µl of 20 µM of PD0166285 or PBS control for 5 consecutive days. Depicted are hematoxylin and eosin staining of irradiated E98 GBM tumors treated with PD0166285 or PBS control. Red lines indicate nonsurvivors. Size bar represents 1000 µm.
(E) PD0166285 enhances radiosensitivity in vivo. Depicted is survival (n = 10/group), log rank p = 0.001.
See also Figure S5.
Next we determined the pharmacological effects of PD0166285 in the E98 GBM model. In line with the U251-FM model, mice treated with PD0166285 and irradiation showed considerably reduced tumor burden (Figure 7D) and their survival was significantly prolonged as compared to other treatment groups (Figure 7E). Both groups of mice subjected to IR or PD0166285 monotherapy showed intermediate results. On day 23, all remaining mice were sacrificed and histological analysis of the brains was performed. Importantly, E98 GBM tumors were undetectable in any mice that were alive after the IR/PD0166285 combination treatment. This could not be explained by lack of tumor growth before the treatment, given the effects in mice without treatment and clear changes of brain structure indicating that tumor growth had in fact occurred (Figure 7C). The sham-treated mice and the mice treated with IR or WEE1 inhibitor alone, showed typical infiltrative GBM growth, although a significant beneficial effect of irradiation was observed (Figure 7D). This in vivo therapeutic response to PD0166285 in combination with irradiation and the lack of adverse side effects underline the potential of WEE1 kinase inhibition in GBM treatment.
Analysis of “Push” and “Pull” Kinase Inhibitors on DNA Damage Repair in GBM Cells
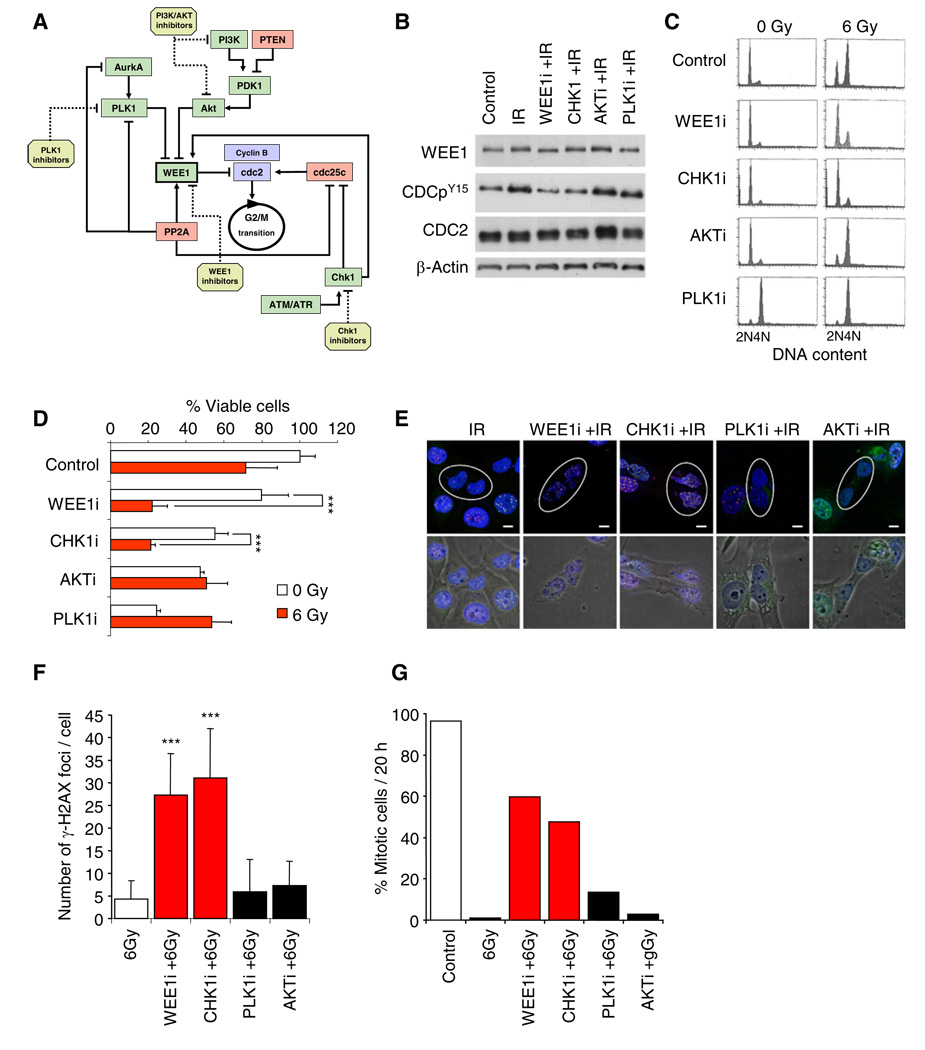
WEE1 is part of an intricate network of kinases and phosphatases regulating the G2 checkpoint (Figure 8A) (Elder et al., 2001; Katayama et al., 2005; Kim and Ferrell, 2007; O’Connell et al., 1997; van Vugt et al., 2004; Watanabe et al., 2004). Several kinase inhibitors were developed to “pull back” the cancer cells during cell-cycle arrest, aiming at stagnation of tumor growth, examples include inhibitors targeting PLK1 and AKT. In contrast, other kinase inhibitors, such as those targeting CHK1 and WEE1, can abrogate the G2 arrest and push cancer cells into premature mitosis.
Figure 8. Analysis of the Effects of G2 Checkpoint Prolongation and Abrogation on DNA Repair and on DNA Damage-Induced Mitotic Catastrophe in GBM Cells.
(A) Schematic overview of kinases, phosphatases, and inhibitors involved in the G2 cell-cycle arrest. Kinases (green) and phosphatases (red) and their interactions are indicated.
(B) Western blot analysis of CDC2, CDC2py15, WEE1, and β-actin, in control or irradiated U251MG cells treated with the indicated inhibitors for 6 hr.
(C) Cell-cycle analysis of irradiated or non-irradiated U251MG cells treated with different inhibitors for 6 hr. At 16 hr after IR the cells were analyzed by fluorescence-activated cell sorting.
(D) WST-1 viability analysis of irradiated or non-irradiated U251MG cells treated with different inhibitors at 4 days after treatment.
(E) Immunofluorescence analysis of IRIF in postmitotic cells. Cells were exposed to 6 Gy, imaged for 16 hr in standard medium or in medium containing one of the following kinase inhibitors, CHK1 (200 nM), WEE1 (500 nM), PLK1 (100 nM), AKT (50 µM), fixed and stained for total DNA (blue), MDC1 (red), and ATMp1981 (green). Inhibitors of AKT and PLK1 were removed from the medium at 6 hr after irradiation because continuous exposure completely abrogated cell division. Postmitotic cells are indicated by ellipsoids. Scale bar represents 5 µm.
(F) Quantitation of the number of IRIF in postmitotic cells treated as in (A). IRIF in at least 50 cells were scored per data point.
(G) Quantitation of the percentage of cells treated as in (A) that entered mitosis during the first 20 hr of imaging. At least 300 cells were scored per data point.
Experiments were carried out in triplicate. Error bars represent standard deviation. ***p < 0.001, t test.
See also Movie S2.
We first determined the effect of the selected inhibitors on CDC2 phosphorylation by western blotting (Figure 8B). Irradiation of GBM cells resulted in increased phosphorylation of CDC2, which was not affected by inhibitors of PLK1 or AKT, but significantly reduced by inhibitors of CHK1 and WEE1 (Figure 8B). We then analyzed the effects of these inhibitors on the cell cycle and radiosensitization of GBM cells (Figures 8C and 8D). The inhibition of CHK1 and WEE1 kinases resulted in abrogation of IR-induced G2 arrest (Figure 8C). In contrast, the inhibitors of PLK1 and AKT increased the fraction of cells in the G2 phase as compared to cells treated with IR alone (Figure 8C). We then determined the effects of the different kinase inhibitors on radiosensitivity of GBM cells (Figure 8D). A significant reduction in viability was observed when cells were irradiated in the presence of CHK1 and WEE1 inhibitors. In marked contrast, inhibition of AKT failed to increase sensitivity of GBM cells to IR and inhibition of PLK1 exhibited radioprotective properties.
G2 checkpoint abrogation by WEE1 inhibition resulted in mitotic catastrophe due to unrepaired DSBs (Figure 5 and Movie S1). Similar results were obtained when GBM cells were irradiated in the presence of CHK1 inhibitor (Movie S2). In contrast, incubation of cells in the presence of inhibitors of PLK1 and AKT for 6 hr after irradiation failed to affect the mitotic entry, as compared to cells exposed to IR alone (Movie S2).
We then assessed the amount of residual DNA damage in cells that underwent mitosis after exposure to IR and the different inhibitors (Figures 8E and 8F). Daughter cells originating from cells irradiated in the presence of CHK1 and WEE1 inhibitors contained a large number of unrepaired DSBs indicating incomplete DNA repair (Figures 8E and 8F). In contrast, the combination of IR with PLK1 and AKT inhibitors did not result in an increase of unrepaired DSBs in the daughter cells, suggesting that DNA repair was near-complete before the cells entered mitosis (Figures 8E and 8F). In agreement with the effects of inhibitors of PLK1, AKT, CHK1, and WEE1 on CDC2 phosphorylation (Figure 8B), a considerable delay in the onset of mitosis was measured in the GBM cells treated with inhibitors of PLK1 and AKT—the “cell-cycle pullers” (Figure 8G and Movie S2). In contrast, cells irradiated in the presence of inhibitors of CHK1 and WEE1—the cell-cycle pushers—failed to delay the mitotic entry (Figure 8G and Movie S2). Thus, the strategy assuming abrogation rather than prolongation of the G2 checkpoint may be more effective in increasing sensitivity of GBM cells to DNA damage in vitro.
DISCUSSION
Three main findings are put forward by the present study. First, we have identified a GBM-specific kinase expression profile, providing a list of substantially and frequently overexpressed kinases involved in GBM signaling pathways. In addition, analysis of other cancer kinase expression profiles verified previously known overexpressed kinases and provided a platform for retrieval of kinase treatment targets. Implications consist of a further advancement of our understanding of oncogenic kinase signaling and a possibility for rational planning of future kinase inhibitory strategies, although, clearly, kinase overexpression is not the only mode of deregulation. Indeed WEE1 activity has been described to be regulated by protein ubiquitination, proteolytic degradation, and phosphorylation (Watanabe et al., 2004; Rothblum-Oviatt et al., 2001; Kim and Ferrell, 2007; van Vugt et al., 2004; O’Connell et al., 1997; Katayama et al., 2005; McGowan and Russell, 1993). Second, the list of differentially expressed kinases in GBM was spearheaded by wee1 that we now present as a potential treatment target. Third, we postulate that the profound overexpression of wee1 in treatment-resistant GBM contributes to efficient DNA repair at the G2 checkpoint. Moreover, the subpopulation of so-called GBM stem-like cells, that are more resistant to irradiation and chemotherapeutics (Bao et al., 2006; Liu et al., 2006) and were described to reside in a low proliferative state in vascular niches (Calabrese et al., 2007), were sensitized by WEE1 inhibition, indicating a major role for WEE1 in conferring radio-resistance of these cells. Our findings support the concept that cancer cells with sub-lethal DNA damage induced by conventional radio- and chemotherapy can be catastrophically pushed into premature mitosis by inhibition of WEE1 activity.
The kinases identified as most frequently overexpressed are often involved in cell-cycle regulation. DNA damaging agents, such as those used during standard GBM treatment, induce arrest in G1 or G2 phases (Iliakis et al., 2003; Hirose et al., 2001). These arrests are brought about by checkpoint mechanisms that provide time for repair of sub-lethal DNA damage before resuming the cell cycle (Branzei and Foiani, 2008). Due to defective TP53 signaling, many cancer cells lack the functional G1 arrest and depend to a greater extent on the G2 checkpoint in response to DNA damage (Kawabe, 2004). Therefore, abrogation of the G2 checkpoint, inducing premature mitotic entry and subsequent cell death, has emerged as a potential therapeutic strategy (Kawabe, 2004; Vogelstein et al., 2000). Inhibitors of the G2 checkpoint activation such as caffeine, pentoxifylline and the CHK1 kinase inhibitor UCN-01 have been shown to push cells containing residual unrepaired DNA damage into premature mitosis, resulting in sensitization to DNA-damaging agents (Reinhardt et al., 2007; Russell et al., 1996; Sarkaria et al., 1999).
WEE1 kinase is involved in the G2 checkpoint control by acting directly on CDC2, the driving force for G2 progression (McGowan and Russell, 1993; O’Connell et al., 1997; Parker and Piwnica-Worms, 1992; Rowley et al., 1992; Russell and Nurse, 1987). WEE1 was identified through genetic studies of cell size control and cell-cycle progression in Schizosaccharomyces pombe (Nurse, 1975). Subsequent work established WEE1 as a tyrosine kinase belonging to the Ser/Thr family of protein kinases (Russell and Nurse, 1987; Featherstone and Russell, 1991). WEE1 catalyzes the inhibitory tyrosine (Y15) phosphorylation of CDC2/cyclin B kinase, so that the G2/M transition is prevented (Russell and Nurse, 1987; Parker and Piwnica-Worms, 1992; McGowan and Russell, 1993).
From several arguments it seems reasonable that inhibition of WEE1 can be a safe therapeutic strategy. First, many cancer cells, and GBM cells in particular, harbor p53 mutations, which render these cells more dependent on the G2 checkpoint (Kawabe, 2004; Wang et al., 2001; Li et al., 2002). Second, inhibition of WEE1 affected neither normal human fibroblasts nor astrocytes in terms of DNA cytometry and cellular morphology. Third, no acute adverse effects were detected in mice after high dose injection of WEE1 inhibitor in our dose-effect experiment.
Deregulation of the TP53 pathway occurs in 87% of GBMs by various mechanisms, including cdkn2a deletion or mutation, mdm2 and mdm4 amplification and P53 deletion or mutation (The Cancer Genome Atlas Research Network, 2008). Although IR sensitization mediated by WEE1 inhibition has been described to be dependent on TP53 activity (Wang et al., 2001; Li et al., 2002), we did not see such a correlation. However, a functional G1 arrest with concurrent wild-type TP53 expression in fibroblasts and astrocytes is likely to explain the absence of cell death after induction of DNA damage by IR and WEE1 inhibition in these non-neoplastic cells, whereas the level of WEE1 inhibitor sensitivity in G2-dependent GBM cells depends on the level of wee1 expression.
Overexpression of wee1 was described previously for several types of cancer. The expression of wee1 was found to be upregulated in poorly differentiated and more advanced hepatocellular carcinoma (Masaki et al., 2003). Further, WEE1 inhibition has been suggested as anticancer therapy based on findings in cell lines from melanoma, cervical carcinoma, colon carcinoma, and ovarian carcinoma (Hashimoto et al., 2006; Wang et al., 2001; Wang et al., 2004). Of note, our stringent in silico analysis on data of primary normal versus cancer tissues indicates overexpression of wee1 in most cancer types (for 27 of 35 data sets; Figure 2A). However, the top-ranking wee1 expression levels in GBM were not reached by any of the other cancer types analyzed. Nonsmall cell lung carcinoma, (non-)seminoma, and colon carcinoma also showed high wee1 mRNA expression levels, whereas the other cancer types mostly showed moderate overexpression as compared to the relevant non-neoplastic control tissues.
Intriguingly, monotherapeutic inhibition of WEE1 also resulted in a beneficial response of GBMs in mice. An explanation for this effect can be that DNA damage in GBM cells is not only induced by cancer therapy but that the DNA integrity of these cells is continuously threatened by inherent genomic instability (The Cancer Genome Atlas Research Network, 2008).
Like the PLK1 and AKT inhibitors, many of the small molecule inhibitors or antibodies have been developed to restrain cancer growth by halting cell replication. Efficacy of this strategy has been demonstrated (Agulnik et al., 2007; Lynch et al., 2004; Ravaud et al., 2008; Vieth et al., 2005; Zhang et al., 2009). However, results of the present study stress the importance of proper treatment timing when combining such agents with DNA damage-induction. Inhibition of CHK1 and WEE1 has clear radiosensitizing effects, suggesting that promoting, rather than halting the cell-cycle progression should be considered as radio-sensitizing strategy in glioblastoma. Effects of CHK1 inhibitors were analyzed in various tumor models (Bao et al., 2006; Hirose et al., 2001; Reinhardt et al., 2007; Sidi et al., 2008), revealing significant toxicity to non-neoplastic cells, which might be explained by involvement of CHK1 in activation of both the G2 and the G1 cell-cycle checkpoints (Kortmansky et al., 2005; Shieh et al., 2000). Here we show that inhibition of WEE1 by PD0166285 does not result in measurable toxic side effects at therapeutically relevant doses in mice, which may be explained by its G2-specific cell-cycle interference.
In summary, we revealed a dominant role of the WEE1 kinase in controlling the G2 checkpoint in GBM and designated this protein as a possible target in treatment-resistant glioblastoma. We showed that GBM cells can be pushed into mitotic catastrophe by WEE1 inhibition after DNA damaging radio- or chemotherapy. Our results suggest that inhibition of WEE1 activity may prove effective in enhancing GBM treatment.
EXPERIMENTAL PROCEDURES
Compounds
The WEE1 inhibitor PD0166285 (Pfizer, Ann Arbor, MI) was diluted in PBS. Inhibitors of PLK1 (BI-2536) and PI3K/AKT (LY-294002) (Axon Medchem, Groningen, the Netherlands) were dissolved in dimethyl sulfoxide (DMSO) (Sigma-Aldrich, St. Louis, MO). Inhibitors (Calbiochem, La Jolla, CA) were diluted in DMSO. Temozolomide (Schering Plough, Kenilworth, NJ) was diluted in DMSO, prepared freshly each time, and used at a final concentration of 100 µM.
Tissue Samples and Cells
Surgical samples were obtained from four glioma patients with World Health Organization (WHO) grade 2, five patients with WHO grade 3, and 14 patients with glioblastoma multiforme (GBM) after informed consent and approval by the Hospital Medical Ethical Committee (Table S5). Furthermore, 16 GBM cell lines were included (Table S6). Cancer stem-like cell cultures were derived from patients with GBM (VUmc, the Netherlands). The stem-like cells were isolated and cultured as described elsewhere (Lee et al., 2006).
RNA Isolation and Interference
Total RNA was isolated using the RNAeasy kit (QIAGEN, Valencia, CA), according the manufacturer’s protocol. The quality and quantity of the RNA was analyzed by photospectrometry. RNA interference was carried out by transfecting siRNAs (50 nM, QIAGEN). Lentivectors encoding validated wee1 shRNAs (Sigma-Aldrich; WEE1 Reference Sequence NM_003390; shRNA reference numbers TRCN0000001702 and TRCN0000001704, referred to as shWEE11702 and shWEE11704, respectively), were produced and used to transduce U251-FM and E98-FM cells. The SHC002 scrambled shRNA construct (Sigma-Aldrich) was used as a negative control.
Immunofluorescence Staining, Immunohistochemistry, and Western Blotting
Cell cultures were fixed using 2% paraformaldehyde in PBS for 15 min at room temperature (RT), permeabilized using 1% Triton X-100 in PBS for 30 min at RT. The following antibodies were used: rabbit anti-MDC1 (A300-051A, Bethyl Laboratories), mouse anti-γ-H2AX (05-636, Millipore), rabbit anti-MRE11 (de Jager et al., 2001), mouse anti-ATMpthr1981 (4526S, Cell Signaling Technology) goat anti-mouse-Cy3 (115-165-166), and goat anti-rabbit-FITC (111-095-144) (Jackson Immunoresearch). Immunohistochemical staining was carried out as described previously (Yoshida et al., 2004). Western blotting was carried out with the following primary antibodies: mouse anti-WEE1 (1:1,000, Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti-CDC2 (1:1000, Abcam), rabbit anti-CDC2pY15 (1:2000, Abcam), mouse anti-β-actin (1:5000, Santa Cruz), and mouse anti-p53 (1:1000, NeoMarkers).
Irradiation, Live-Cell Imaging, and Fluorescence Imaging
Approximately 1.5 × 105 cells were plated in 25 cm2 culture flasks and incubated for 24 hr in standard medium. Next, required inhibitors were added for 30 min and cells were exposed to gamma-irradiation. Next, cells were mounted under Leica IRBE inverted fluorescence microscope (Leica Microsystems) in an incubator with adjustable CO2 concentration, at 37°C. Positions of imaged cells were saved using custom-written software. Phase-contrast, time-lapse movies were acquired at 10-min intervals for required period of time. If required, after imaging, cells were fixed, stained for indicated proteins, and imaged using fluorescence microscopy.
In Vivo Analysis Using the U251 and E98 Orthotopic GBM Mouse Models
All experiments on mice were approved by the Subcommittee on Research Animal Care at Radboud University (Nijmegen, the Netherlands), VUmc (Amsterdam, the Netherlands), or Massachusetts General Hospital (Boston, MA), and were carried out in accordance to their guidelines and regulations. E98 tumor fragments (8 mm3) were grafted subcutaneously in the flank of Balb/C nude mice (n = 10). For the experiments with the orthotopic tumors, U251MG and E98 cells were transduced to express Fluc and mCherry, to generate U251-FM and E98-FM cells. One million cells were injected intracranially. Starting at 12 days after tumor inoculation, mice received the WEE1 inhibitor PD0166285 via intraperitoneal injections (500 µl of a 20 µM solution diluted in NaCl) or vehicle for 5 consecutive days. On days 10–15, mice were irradiated with a single dose of irradiation.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from the Accelerate Brain Cancer Cure and the Dutch Cancer Society. We thank Z. Kwidama, L. Wedekind, M. Ubels, and W. Tigchelaar for technical assistance, S. Heukelom and P. Koken from the Department of Radiotherapy, VUmc, for use of the irradiation source, and J. Skog for CD133 analysis.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures, seven tables, Supplemental Experimental Procedures, and two movies and can be found with this article online at doi:10.1016/j.ccr.2010.08.011.
REFERENCES
- Agulnik M, da Cunha Santos G, Hedley D, Nicklee T, Dos Reis PP, Ho J, Pond GR, Chen H, Chen S, Shyr Y, et al. Predictive and pharmacodynamic biomarker studies in tumor and skin tissue samples of patients with recurrent or metastatic squamous cell carcinoma of the head and neck treated with erlotinib. J. Clin. Oncol. 2007;25:2184–2190. doi: 10.1200/JCO.2006.07.6554. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- Bredel M, Bredel C, Juric D, Harsh GR, Vogel H, Recht LD, Sikic BI. Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res. 2005;65:8679–8689. doi: 10.1158/0008-5472.CAN-05-1204. [DOI] [PubMed] [Google Scholar]
- Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Candolfi M, Curtin JF, Nichols WS, Muhammad AG, King GD, Pluhar GE, McNiel EA, Ohlfest JR, Freese AB, Moore PF, et al. Intracranial glioblastoma models in preclinical neuro-oncology: neuropathological characterization and tumor progression. J. Neurooncol. 2007;85:133–148. doi: 10.1007/s11060-007-9400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Nishimura MC, Bumbaca SM, Kharbanda S, Forrest WF, Kasman IM, Greve JM, Soriano RH, Gilmour LL, Rivers CS, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17:362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- Claes A, Schuuring J, Boots-Sprenger S, Hendriks-Cornelissen S, Dekkers M, van der Kogel AJ, Leenders WP, Wesseling P, Jeuken JW. Phenotypic and genotypic characterization of orthotopic human glioma models and its relevance for the study of anti-glioma therapy. Brain Pathol. 2008;18:423–433. doi: 10.1111/j.1750-3639.2008.00141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. Protein kinases—the major drug targets of the twenty-first century? Nat. Rev. Drug Discov. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- de Jager M, Dronkert ML, Modesti M, Beerens CE, Kanaar R, van Gent DC. DNA-binding and strand-annealing activities of human Mre11: implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Res. 2001;29:1317–1325. doi: 10.1093/nar/29.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Witt Hamer PC. Small molecule kinase inhibitors in glioblastoma: a systematic review of clinical studies. Neuro-oncol. 2010;12:304–316. doi: 10.1093/neuonc/nop068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetri GD, von Mehren M, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, Heinrich MC, Tuveson DA, Singer S, Janicek M, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N. Engl. J. Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- Elder RT, Yu M, Chen M, Zhu X, Yanagida M, Zhao Y. HIV-1 Vpr induces cell cycle G2 arrest in fission yeast (Schizosaccharomyces pombe) through a pathway involving regulatory and catalytic subunits of PP2A and acting on both Wee1 and Cdc25. Virology. 2001;287:359–370. doi: 10.1006/viro.2001.1007. [DOI] [PubMed] [Google Scholar]
- Featherstone C, Russell P. Fission yeast p107wee1 mitotic inhibitor is a tyrosine/serine kinase. Nature. 1991;349:808–811. doi: 10.1038/349808a0. [DOI] [PubMed] [Google Scholar]
- Goldberg M, Stucki M, Falck J, D’Amours D, Rahman D, Pappin D, Bartek J, Jackson SP. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- Hashimoto O, Shinkawa M, Torimura T, Nakamura T, Selvendiran K, Sakamoto M, Koga H, Ueno T, Sata M. Cell cycle regulation by the Wee1 inhibitor PD0166285, pyrido [2,3-d] pyrimidine, in the B16 mouse melanoma cell line. BMC Cancer. 2006;6:292. doi: 10.1186/1471-2407-6-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Berger MS, Pieper RO. p53 effects both the duration of G2/M arrest and the fate of temozolomide-treated human glioblastoma cells. Cancer Res. 2001;61:1957–1963. [PubMed] [Google Scholar]
- Hodgson JG, Yeh RF, Ray A, Wang NJ, Smirnov I, Yu M, Hariono S, Silber J, Feiler HS, Gray JW, et al. Comparative analyses of gene copy number and mRNA expression in GBM tumors and GBM xenografts. Neuro. Oncol. 2009;11:477–487. doi: 10.1215/15228517-2008-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliakis G, Wang Y, Guan J, Wang H. DNA damage checkpoint control in cells exposed to ionizing radiation. Oncogene. 2003;22:5834–5847. doi: 10.1038/sj.onc.1206682. [DOI] [PubMed] [Google Scholar]
- Katayama K, Fujita N, Tsuruo T. Akt/protein kinase B-dependent phosphorylation and inactivation of WEE1Hu promote cell cycle progression at G2/M transition. Mol. Cell. Biol. 2005;25:5725–5737. doi: 10.1128/MCB.25.13.5725-5737.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe T. G2 checkpoint abrogators as anticancer drugs. Mol. Cancer Ther. 2004;3:513–519. [PubMed] [Google Scholar]
- Kim SY, Ferrell JEJ. Substrate competition as a source of ultrasensitivity in the inactivation of Wee1. Cell. 2007;128:1133–1145. doi: 10.1016/j.cell.2007.01.039. [DOI] [PubMed] [Google Scholar]
- Klein A, Reichardt W, Jung V, Zang KD, Meese E, Urbschat S. Overexpression and amplification of STK15 in human gliomas. Int. J. Oncol. 2004;25:1789–1794. [PubMed] [Google Scholar]
- Kortmansky J, Shah MA, Kaubisch A, Weyerbacher A, Yi S, Tong W, Sowers R, Gonen M, O’Reilly E, Kemeny N, et al. Phase I trial of the cyclin-dependent kinase inhibitor and protein kinase C inhibitor 7-hydroxystaurosporine in combination with Fluorouracil in patients with advanced solid tumors. J. Clin. Oncol. 2005;23:1875–1884. doi: 10.1200/JCO.2005.03.116. [DOI] [PubMed] [Google Scholar]
- Kotliarov Y, Steed ME, Christopher N, Walling J, Su Q, Center A, Heiss J, Rosenblum M, Mikkelsen T, Zenklusen JC, et al. High-resolution global genomic survey of 178 gliomas reveals novel regions of copy number alteration and allelic imbalances. Cancer Res. 2006;66:9428–9436. doi: 10.1158/0008-5472.CAN-06-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy. N. Engl. J. Med. 2005;353:172–187. doi: 10.1056/NEJMra044389. [DOI] [PubMed] [Google Scholar]
- Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–7758. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Lee Y, Scheck AC, Cloughesy TF, Lai A, Dong J, Farooqi HK, Liau LM, Horvath S, Mischel PS, Nelson SF. Gene expression analysis of glioblastomas identifies the major molecular basis for the prognostic benefit of younger age. BMC Med. Genomics. 2008;1:52. doi: 10.1186/1755-8794-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang Y, Sun Y, Lawrence TS. Wild-type TP53 inhibits G(2)-phase checkpoint abrogation and radiosensitization induced by PD0166285, a WEE1 kinase inhibitor. Radiat. Res. 2002;157:322–330. doi: 10.1667/0033-7587(2002)157[0322:wttigp]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, Lu L, Irvin D, Black KL, Yu JS. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol. Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffler H, Lukas J, Bartek J, Kramer A. Structure meets function-centrosomes, genome maintenance and the DNA damage response. Exp. Cell Res. 2006;312:2633–2640. doi: 10.1016/j.yexcr.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Masaki T, Shiratori Y, Rengifo W, Igarashi K, Yamagata M, Kurokohchi K, Uchida N, Miyauchi Y, Yoshiji H, Watanabe S, et al. Cyclins and cyclin-dependent kinases: comparative study of hepatocellular carcinoma versus cirrhosis. Hepatology. 2003;37:534–543. doi: 10.1053/jhep.2003.50112. [DOI] [PubMed] [Google Scholar]
- McGowan CH, Russell P. Human Wee1 kinase inhibits cell division by phosphorylating p34cdc2 exclusively on Tyr15. EMBO J. 1993;12:75–85. doi: 10.1002/j.1460-2075.1993.tb05633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano I, Masterman-Smith M, Saigusa K, Paucar AA, Horvath S, Shoemaker L, Watanabe M, Negro A, Bajpai R, Howes A, et al. Maternal embryonic leucine zipper kinase is a key regulator of the proliferation of malignant brain tumors, including brain tumor stem cells. J. Neurosci. Res. 2008;86:48–60. doi: 10.1002/jnr.21471. [DOI] [PubMed] [Google Scholar]
- Nicholas MK, Lukas RV, Jafri NF, Faoro L, Salgia R. Epidermal growth factor receptor - mediated signal transduction in the development and therapy of gliomas. Clin. Cancer Res. 2006;12:7261–7270. doi: 10.1158/1078-0432.CCR-06-0874. [DOI] [PubMed] [Google Scholar]
- Nurse P. Genetic control of cell size at cell division in yeast. Nature. 1975;256:547–551. doi: 10.1038/256547a0. [DOI] [PubMed] [Google Scholar]
- O’Connell MJ, Raleigh JM, Verkade HM, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omuro AMP, Faivre S, Raymond E. Lessons learned in the development of targeted therapy for malignant gliomas. Mol. Cancer Ther. 2007;6:1909–1919. doi: 10.1158/1535-7163.MCT-07-0047. [DOI] [PubMed] [Google Scholar]
- Parker LL, Piwnica-Worms H. Inactivation of the p34cdc2-cyclin B complex by the human WEE1 tyrosine kinase. Science. 1992;257:1955–1957. doi: 10.1126/science.1384126. [DOI] [PubMed] [Google Scholar]
- Pedersini R, Vattemi E, Mazzoleni G, Graiff C. Complete response after treatment with imatinib in pretreated disseminated testicular seminoma with overexpression of c-KIT. Lancet Oncol. 2007;8:1039–1040. doi: 10.1016/S1470-2045(07)70344-3. [DOI] [PubMed] [Google Scholar]
- Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TKB, Gronborg M, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravaud A, Hawkins R, Gardner JP, von der Maase H, Zantl N, Harper P, Rolland F, Audhuy B, Machiels JP, Petavy F, et al. Lapatinib versus hormone therapy in patients with advanced renal cell carcinoma: a randomized phase III clinical trial. J. Clin. Oncol. 2008;26:2285–2291. doi: 10.1200/JCO.2007.14.5029. [DOI] [PubMed] [Google Scholar]
- Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell. 2007;11:175–189. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich JN, Bigner DD. Development of novel targeted therapies in the treatment of malignant glioma. Nat. Rev. Drug Discov. 2004;3:430–446. doi: 10.1038/nrd1380. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothblum-Oviatt CJ, Ryan CE, Piwnica-Worms H. 14-3-3 binding regulates catalytic activity of human Wee1 kinase. Cell Growth Differ. 2001;12:581–589. [PubMed] [Google Scholar]
- Rowley R, Hudson J, Young PG. The wee1 protein kinase is required for radiation-induced mitotic delay. Nature. 1992;356:353–355. doi: 10.1038/356353a0. [DOI] [PubMed] [Google Scholar]
- Russell KJ, Wiens LW, Demers GW, Galloway DA, Le T, Rice GC, Bianco JA, Singer JW, Groudine M. Preferential radiosensitization of G1 checkpoint-deficient cells by methylxanthines. Int. J. Radiat. Oncol. Biol. Phys. 1996;36:1099–1106. doi: 10.1016/s0360-3016(96)00432-4. [DOI] [PubMed] [Google Scholar]
- Russell P, Nurse P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 1987;49:559–567. doi: 10.1016/0092-8674(87)90458-2. [DOI] [PubMed] [Google Scholar]
- Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM, Abraham RT. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- Sidi S, Sanda T, Kennedy RD, Hagen AT, Jette CA, Hoffmans R, Pascual J, Imamura S, Kishi S, Amatruda JF, et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell. 2008;133:864–877. doi: 10.1016/j.cell.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vugt MATM, Bras A, Medema RH. Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol. Cell. 2004;15:799–811. doi: 10.1016/j.molcel.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Vieth M, Sutherland JJ, Robertson DH, Campbell RM. Kinomics: characterizing the therapeutically validated kinase space. Drug Discov. Today. 2005;10:839–846. doi: 10.1016/S1359-6446(05)03477-X. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Wang Y, Decker SJ, Sebolt-Leopold J. Knockdown of Chk1, Wee1 and Myt1 by RNA interference abrogates G2 checkpoint and induces apoptosis. Cancer Biol. Ther. 2004;3:305–313. doi: 10.4161/cbt.3.3.697. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li J, Booher RN, Kraker A, Lawrence T, Leopold WR, Sun Y. Radiosensitization of p53 mutant cells by PD0166285, a novel G(2) checkpoint abrogator. Cancer Res. 2001;61:8211–8217. [PubMed] [Google Scholar]
- Watanabe N, Arai H, Nishihara Y, Taniguchi M, Watanabe N, Hunter T, Osada H. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc. Natl. Acad. Sci. USA. 2004;101:4419–4424. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB. Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- Wen PY, Kesari S. Malignant gliomas in adults. N. Engl. J. Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Tanaka S, Mogi A, Shitara Y, Kuwano H. The clinical significance of Cyclin B1 and Wee1 expression in non-small-cell lung cancer. Ann. Oncol. 2004;15:252–256. doi: 10.1093/annonc/mdh073. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.