Abstract
Immunoglobulin variable region exons are assembled from discontinuous variable (V), diversity (D), and joining (J) segments by the process of V(D)J recombination. V(D)J rearrangements of the immunoglobulin heavy chain (IgH) locus are tightly controlled in a tissue-specific, ordered and allele-specific manner by regulating accessibility of V, D, and J segments to the recombination activating gene proteins which are the specific components of the V(D)J recombinase. In this review we discuss recent advances and established models brought forward to explain the mechanisms underlying accessibility control of V(D)J recombination, including research on germline transcripts, spatial organization, and chromatin modifications of the immunoglobulin heavy chain (IgH) locus. Furthermore, we review the functions of well-described and potential new cis-regulatory elements with regard to processes such as V(D)J recombination, allelic exclusion, and IgH class switch recombination.
1. INTRODUCTION
An individual clone of mature B-cells expresses immunoglobulin (Ig) molecules as an antigen receptor. The typical sub-unit of an Ig molecule consists of two identical heavy chains (HC) and two identical light chains (LC). The N-terminal region of these chains contains the highly variable antigen binding site; whereas the C-terminal part is called constant region (C region). The C region of the IgH chain (CH) determines the effector functions of antibodies, which are the secreted form of Ig molecules.
Immunoglobulin (Ig) and T-cell receptor (TCR) variable region exons are assembled from large arrays of V (variable), D (diversity), and J (joining) gene segments during the development, respectively, of B and T lymphocytes. Once a functional immunoglobulin chain is expressed, allelic exclusion operates through a feedback mechanism to prevent further rearrangements of Ig heavy (IgH) and Ig light (IgL) chain genes. V(D)J recombination is mediated by a common recombinase complex that includes the recombination-activating gene products RAG1 and RAG2, which harbor endonuclease activity that introduces DNA double strand breaks (DSBs) at V, D, and J segments. The V(D)J reaction is completed by the ubiquitously expressed nonhomologous end-joining (NHEJ) factors that join the broken V, D, and J segments together. Still, Ig loci are only fully assembled in B lineage cells and TCR loci are only assembled in T lineage cells. Within a lineage, different loci are rearranged in a specific order. For example, IgH locus variable region exons are assembled before those of Ig light chains (IgL), and within the IgH locus D to JH recombination precedes VH to DJH recombination. Given such locus-specific regulation and a common V(D)J recombinase, accessibility of the different loci to the common V(D)J recombinase must underlie the cell-type and stage-dependent assembly of the different IgH and TCR gene families (Jung et al., 2006).
Activation of mature B-cells can alter their IgH loci through a separate form of genomic rearrangement which is termed IgH class switch recombination (CSR). CSR allows B-cells to express IgH chains with different constant regions which can change the effector functions of antibodies without altering variable region specificity. CSR is initiated by activation-induced cytosine deaminase (AID), the activity of which ultimately leads to DSBs in regions upstream of CH genes which are then joined by NHEJ or other end-joining pathways to complete the CSR reaction (Chaudhuri et al., 2007).
Ig and TCR loci contain a number of cis-regulatory elements which regulate V(D)J rearrangements, IgH CSR, and Ig gene expression at various levels. In this review, we will focus on the impact of cis-regulatory elements on genetic and epigenetic regulation of recombination events within the IgH locus.
2. THE IMMUNOGLOBULIN HEAVY CHAIN LOCUS
The murine IgH locus is a complex genomic region, spanning about 3 Mb close to the telomere of the long arm on chromosome 12. The IgH locus comprises arrays of V, D, and J segments upstream of several constant region exons (Fig. 1.1A). Different mouse strains carry varying numbers of VH and D elements. Some 150 VH segments are distributed over ~2.5 Mb in the 5′ part of the IgH locus and are classified in 16 VH gene families defined by sequence similarities (Johnston et al., 2006). These VH gene families are partially interspersed with one another but, depending on position, can be divided into proximal (3′ part of the VH cluster, close to IgH–D region, for example, VH7183), intermediate (e.g., VHS107), and distal (5′ part of the VH cluster, distant from IgH–D region, for example, VHJ558) families. 3′ of the VH elements, separated by ~90 kb, lie 10–15 D segments (Retter et al., 2007; Ye, 2004) followed by 4 JH elements. Because to the uniform transcriptional orientation of V, D, and J segments, V(D)J recombination events at the IgH locus result in deletion of the intervening sequence. The 3′ part of the IgH locus contains a series of sets of different constant (C) region exons Cμ, Cδ, Cγ3, Cγ1, Cγ2b, Cγ2a, Cε, Cα, which will be referred to as “CH genes” (Fig. 1.1B).
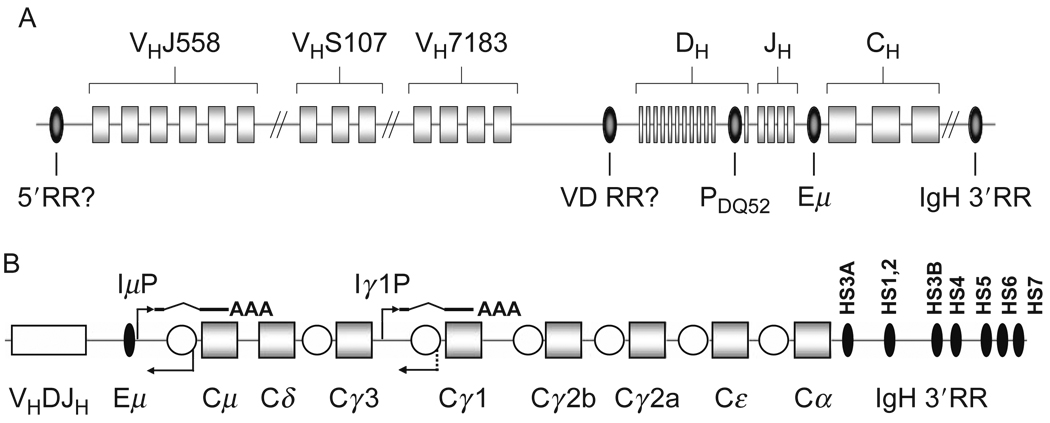
FIGURE 1.1.
Schematic depiction of the murine IgH locus. (A) VH, DH, JH gene segments and CH exons are shown as rectangles, known and potential regulatory elements as ovals. The VH families VHJ558, VHS107, and VH7183 are depicted as examples for distal, intermediate, and proximal VH families, respectively. The cis-regulatory elements PDQ52 (promoter of DQ52), Eμ (intronic enhancer), and IgH 3′RR (IgH 3′ regulatory region) are depicted. The potential regulatory elements 5′RR (5′ regulatory region) and VD RR (VH–DH intergenic regulatory region) are depicted with a question mark. Drawing not to scale. (B) The 3′ part of the IgH locus. An assembled VHDJH exon is shown as a white rectangle, CH genes as squares, Eμ and individual DNaseI hypersensitive sites within the IgH 3′RR are depicted as black ovals, switch regions as white circles. I promoters are located upstream of every switch region (Chaudhuri et al., 2007; Lennon and Perry, 1985; Lutzker and Alt, 1988), only μ and γ1 I promoters (IμP, Iγ1P) are depicted. Transcripts from I promoters get spliced and polyadenylated. Switch regions also get transcribed in the antisense orientation (Apel et al., 1992; Julius et al., 1988, Morrison et al., 1998; Perlot et al., 2008). Concomitant transcription from IμP and, for example, Iγ1P can target AID to μ and γ1 switch regions and thereby initiate CSR to Cγ1.
A large number of cis-regulatory elements were identified throughout the IgH locus. The intronic enhancer, Eμ, is located in the intron between JH4 and the CH exons (Fig. 1.1); the 3′ IgH regulatory region (IgH 3′RR) consists of several DNase hypersensitive sites and is located at the very 3′ end of the IgH locus (Fig. 1.1). Transcriptional promoters are present upstream of every VH segment (Fig. 1.2B), upstream of DH segments (Fig. 1.2A and C), and upstream of CH genes (Fig. 1.2A). In addition, antisense transcripts from less well-defined promoters were described in the VH, DH, JH regions, and upstream of CH genes. Section 7 of this chapter contains a detailed discussion of these IgH cis-regulatory elements.
FIGURE 1.2.
Transcripts within the IgH locus. VH, DH, JH gene segments and CH exons are shown as rectangles, enhancer and promoter elements as ovals. 12 bp and 23 bp RSSs are depicted as black and white triangles, respectively. Drawings not to scale. (A) The IgH locus in germline configuration is transcribed from the promoter of DQ52 (PDQ52) to produce the μ0 transcript (Alessandrini and Desiderio, 1991), and from within the Eμ enhancer to generate the Iμ transcript (Lennon and Perry, 1985; Su and Kadesch, 1990), both of which are getting spliced and polyadenylated (Kottmann et al., 1994, Su and Kadesch, 1990). DH and JH elements are transcribed in the antisense orientation (Bolland et al., 2007; Chakraborty et al., 2007), suggested start sites (dashed arrows) are located around PDQ52 (Chakraborty et al., 2007) and Eμ (Bolland et al., 2007). Sites of transcriptional termination of DH–JH antisense germline transcripts are unknown. (B) Unrearranged VH segments are transcribed in the sense orientation from the individual VH promoters (VHP) (Yancopoulos and Alt, 1985). The intron between the leader (L) and the VH exon (VH) is spliced out, and the VH sense germline transcript gets polyadenylated (Yancopoulos and Alt, 1985). The VH segments and VH intergenic regions can also get transcribed in the antisense orientation (Bolland et al., 2004). Start and termination sites of VH antisense germline transcripts are unknown. Therefore, individual antisense transcripts could comprise one VH segment and its adjacent regions or multiple VH segments including intergenic regions, shown as short and long solid arrows, respectively. (C) Upon D to JH recombination, the assembled DJH exon gets transcribed from the DH promoter (PDH) and spliced to the Cμ exons to generate the Dμ transcript (Alessandrini and Desiderio, 1991; Reth and Alt, 1984), which in one reading frame encodes for a short μHC molecule (Reth and Alt, 1984). DH antisense germline transcription is present throughout the remaining unrearranged DH segments (Chakraborty et al., 2007). Suggested origin of DH antisense germline transcripts is the region around the promoter of the recombined DH segment (depicted as PDH) (Chakraborty et al., 2007), transcriptional termination sites are unknown. (D) Upon VH to DJH recombination, the promoter of the rearranged VH segment (depicted as VHP) drives expression of mRNA encoding for the μHC. In addition to Iμ sense transcription, the Sμ switch region is also transcribed in the antisense orientation (Perlot et al., 2008) from promoters residing within Sμ (Apel et al., 1992; Morrison et al., 1998), the transcriptional termination site of the Sμ antisense transcript is unknown. The assembled VHDJH exon and the adjacent JH region are transcribed in the antisense orientation potentially from start sites within the JH cluster (dashed arrow) (Perlot et al., 2008), the transcriptional termination site of the VHDJH antisense transcript is unknown. Upstream unrearranged VH segments are transcriptionally silenced upon assembly of a functional VHDJH exon (Bolland et al., 2004; Yancopoulos and Alt, 1985).
3. V(D)J RECOMBINATION DURING B-CELL DEVELOPMENT
The IgH locus V(D)J exon is assembled at the pro-B-cell stage leading to the production of μ IgH heavy chains via splicing of the VHDJH exon onto the adjacent Cμ constant region exons. Functional μHC and surrogate Ig light chain proteins form a complex that is expressed on the surface of pre-B-cells and is known as the pre-B-cell receptor (pre-BCR) (Cobb et al., 2006). Signaling through the pre-BCR induces proliferation, signals cessation of further VH to DJH rearrangements at the IgH locus (i.e., allelic exclusion, see below), and promotes the onset of IgL variable region exon (VLJL) assembly. Thus, expression of the pre-BCR represents an important checkpoint at the pro- to pre-B-cell transition (Mårtensson et al., 2007). Subsequently, Igκ and Igλ LC variable regions are assembled during the pre-B-cell stage. Expression of a functional Igκ or Igλ LC along with μHC forms a complete Ig molecule which is expressed on the cell surface of the resulting immature B-cells (Gorman and Alt, 1998). Immature B-cells migrate to the periphery where mature naïve B-cells can be activated and undergo further modification of their IgH locus including IgH CSR and somatic hypermutation (SHM) (see below).
All V, D, and J segments are flanked by recombination signal sequences (RSSs) that consist of a conserved palindromic heptamer and a conserved AT-rich nonamer separated by a less conserved 12 bp or 23 bp spacer (Sakano et al., 1980). The RAG1/2 endonuclease recognizes and binds a pair of RSSs with different spacer lengths in the context of the 12/23 rule (Early et al., 1980; Sakano et al., 1980), which allows for efficient V(D)J recombination only between gene segments flanked by 12 bp and 23 bp RSSs (Fugmann et al., 2000). The 12/23 restriction provides some direction for which Ig gene segments can be assembled. For example, IgH D segments are flanked with 12 bp RSSs on both sides; whereas VH and JH segments are flanked with 23 bp RSSs, thus preventing direct VH–JH joining. In the TCRβ locus, however, direct Vβ to Jβ joints would be allowed according to the 12/23 rule but are denied by “beyond 12/23” restrictions (Bassing et al., 2000). Differential composition of RSSs implement “beyond 12/23” restriction at the nicking and pairing step of V(D)J recombination (Drejer-Teel et al., 2007; Jung et al., 2003).
RAG cutting precisely between RSSs and variable region gene segments results in formation of blunt RSS ends, and the formation of coding ends (CE) of the V, D, or J segments as closed hairpins. Coding joints (CJs) are formed through a joining reaction mediated by members of the NHEJ repair pathway. In this reaction, Ku proteins bind the free CEs and recruit DNA-dependent protein kinase catalytic subunit (DNA-PKcs), which activates the endonuclease activity of Artemis to open the hairpins. Subsequently, ends are joined by the XRCC4/DNA ligaseIV complex (Rooney et al., 2004). In contrast, the blunt SEs are precisely ligated to each other by NHEJ.
Tight regulation of V(D)J recombination is imperative to ensure proper lymphocyte development and genomic integrity. While V(D)J recombination is of enormous advantage in order to efficiently combat infections, erroneous V(D)J recombination can have adverse consequences including chromosomal translocations, which can contribute to neoplastic transformation and the development of leukemias and lymphomas.
4. CLASS SWITCH RECOMBINATION AND SOMATIC HYPERMUTATION
After a VHDJH variable exon is assembled upstream of the C region exons, a promoter 5′ of the rearranged VH segment drives expression of μ and δ HC molecules in mature B-cells. Upon antigen encounter and activation, a B-cell can switch to expression of downstream CH genes to generate antibodies with the same variable region specificity but a different CH effector function by CSR. Repetitive switch (S) regions are located upstream of every CH gene except Cδ. Introduction of DSBs in Sμ and a downstream switch region can result in joining of the two switch regions, deletion of the intervening sequence, and consequently expression of a downstream CH gene with the same variable region exon. AID is absolutely required for CSR. It appears to function by deaminating cytosines to uracils within the substrate S region DNA with the resulting mismatches somehow being processed into DSBs by cooption of normal repair pathways (Di Noia and Neuberger, 2007). As AID is a single-strand DNA-specific cytosine deaminase, its activity on duplex S region DNA is targeted by transcription (Chaudhuri et al., 2007). In this context, switch regions can be transcribed from an I (intervening) promoter located upstream of each S region, which allows for AID targeting to specific transcribed S regions (Fig. 1.2B). In addition to these sense germline transcripts, antisense transcripts were described in several S regions (Apel et al., 1992; Julius et al., 1988; Morrison et al., 1998; Perlot et al., 2008). AID initiated S region DSBs are joined by NHEJ or an alternative end-joining pathway to complete CSR.
AID is also required for SHM, a process during which the variable region exon gets mutated at a relatively high frequency in activated B-cells. SHM is initiated by transcription-dependent targeting of AID to assembled variable regions followed by error prone repair of the resulting mismatches (Di Noia and Neuberger, 2007). Through affinity maturation, B-cell clones producing higher affinity antibodies are selected and an efficient adaptive immune response is elicited.
5. IgH REARRANGEMENTS AND ALLELIC EXCLUSION
Expression of RAG1 and RAG2 is absolutely required for V(D)J recombination. In the haematopoietic lineage, RAG activity can first be demonstrated in common lymphoid progenitor (CLP) cells, which are precursor cells that can develop into B-cells, T-cells, natural killer (NK) cells, and dendritic cells (DC) (Borghesi et al., 2004). Together with the detection of D to JH rearrangements in non-B-cell lymphoid lineages (Borghesi et al., 2004; Born et al., 1988; Kurosawa et al., 1981), expression of RAG in CLPs suggests that the first IgH rearrangement step can occur, at least at low level, in CLPs. Thus, the IgH D to JH recombination step is not absolutely restricted to the B-lineage, in contrast to VH to DJH rearrangements which normally occur only in B-cells. Efficient D to JH rearrangement on both IgH alleles takes place after B lineage commitment in the pro-B-cell stage (Alt et al., 1984).
Once DJH segments are formed, one of the upstream VH elements can join to form a complete VHDJH exon. In the murine IgH locus, proximal VH segments are preferentially rearranged compared to distal VH elements throughout ontogeny (Malynn et al., 1990; Yancopoulos et al., 1984). However, peripheral B-cells do not show this preference as selection alters the B-cell repertoire (Yancopoulos et al., 1988). Both D to JH and VH to DJH recombination take place at the pro-B-cell stage, however, in an ordered manner such that D to JH rearrangement nearly always occurs before VH to DJH rearrangement (Alt et al., 1984). In this regard, VH to DJH recombination is the step that is regulated in the context of allelic exclusion to ensure expression of only one functional HC.
Successful VH to DJH recombination and expression of a μHC from one IgH allele, prevents a second DJH allele from undergoing VH to DJH rearrangement (Jung et al., 2006). Considering the junctional diversity generated during V(D)J recombination, only one out of three VHDJH exons will be in frame with the downstream Cμ exons; whereas two out of three will be out of frame and therefore unable to express a functional μHC (Mostoslavsky et al., 2004). The percentage of functional recombination events is further decreased by usage of VH pseudogenes containing stop codons, frameshifts, defective splice sites, or lacking an ATG translation start site, by stop codons in DH segments as well as through selection against certain reading frames of DJH joins (Gu et al., 1991). As a result, a substantial fraction of developing B-cells will not be able to generate a functional μHC from either IgH allele and will undergo apoptosis (Rajewsky, 1996). If a nonfunctional VH to DJH rearrangement occurs on the first allele, the second DJH allele can still undergo VH to DJH rearrangement (Alt et al., 1984).
Allelic exclusion of VH to DJH rearrangement is mediated by feedback regulation; a functional μHC together with surrogate light chains are assembled to a pre-BCR, which signals the cessation of further VH to DJH rearrangements (Alt et al., 1984; Jung et al., 2006). In this regard, endogenous IgH rearrangements are largely inhibited by the expression of a preassembled membrane-bound μHC transgene (Nussenzweig et al., 1988). Likewise, allelic exclusion was broken by targeted deletion of the μHC transmembrane exons (Kitamura and Rajewsky, 1992), by lack of a functional pre-BCR (Löffert et al., 1996), and by combined deletion of the downstream pre-BCR signaling molecules Syk and ZAP-70 (Schweighoffer et al., 2003). The combined data from these studies strongly support a feedback-mediated mechanism for allelic exclusion that is mediated by signaling through a functional μHC in the pre-BCR signaling complex.
The complete chain of events that leads to cessation of VH to DJH rearrangements and implementation of allelic exclusion is still elusive. However, it was shown that the onset of allelic exclusion after successful VH to DJH recombination is accompanied by the transient downregulation of RAG (Grawunder et al., 1995), decontraction of the IgH locus (Roldán et al., 2005), and loss of accessibility correlates such as VH germline transcripts and marks of active chromatin (see below). It has been estimated that only 1 in 10,000 wild-type B-lymphocytes actually escape allelic exclusion and express a functional μHC from both IgH alleles (Barreto and Cumano, 2000). Feedback regulation can explain cessation of VH to DJH rearrangement but would be ineffective if both IgH alleles would rearrange simultaneously. Therefore, it was suggested that the V(D)J recombination machinery targets one allele at a time (Alt et al., 1980). Supportive of this hypothesis was the observation that all Ig loci as well as the TCRβ locus undergo asynchronous replication (Mostoslavsky et al., 2001; Norio et al., 2005). At the allelically excluded Igκ locus it is thought that asynchronous replication facilitates allele specific chromatin changes (Mostoslavsky et al., 1998) that lead to the early replicating allele rearranging first (Mostoslavsky et al., 2001). A similar mechanism for VH to DJH recombination, the allelically excluded IgH rearrangement step, was speculated, but has not yet been demonstrated. Thus, asynchronous replication could conceivably play a role in the initiation phase of allelic exclusion. However, it does not provide an explanation for the maintenance of allelic exclusion during subsequent B-cell stages, which prevents further IgH rearrangements in the presence of RAG, which must be effected by feedback mechanisms that influence accessibility.
6. ACCESSIBILITY CONTROL
The accessibility hypothesis was proposed to explain how a single common V(D)J recombinase can target the different Ig and TCR loci in a lineage- and stage-specific manner (Yancopoulos and Alt, 1985). For example, Ig variable region exons are only fully assembled in B-cells while TCR variable region exons are only rearranged in T-cells. Similarly, IgH loci are rearranged during the pro-B-cell stage and not in pre-B-cells where IgL variable region assembly occurs. The accessibility hypothesis was first proposed based on the finding that germline VH gene segments are transcribed in pro-B-cells but not in subsequent B-cell stages, with germline VH transcription providing a potential correlate of accessibility (Yancopoulos and Alt, 1985). This hypothesis was proven by experiments that showed transfected TCR gene substrates could be rearranged by pro-B lines that do not rearrange endogenous TCR gene segments, first demonstrating a common V(D)J recombinase (Yancoupouls, 1986). This conclusion was confirmed and extended by other studies (Krangel, 2003; Stanhope-Baker et al., 1996). However, the precise mechanisms that mediate differential accessibility of Ig and TCR gene segments to V(D)J recombination are still not clear. Over the decades, several correlates of accessibility have been defined and a general picture is beginning to emerge as to how accessibility control might be regulated and implemented. Among the known correlates of accessibility are germline transcripts, chromatin modifications, DNase hypersensitivity, spatial organization, and positioning of Ig and TCR loci in the interphase nucleus.
6.1. Germline transcripts
Germline transcription is the production of transcripts from V, D, or J segments and adjacent regions before they undergo rearrangement (Fig. 1.2). Sense germline transcripts starting from promoters upstream of V, D, and J segments have been described in all Ig and TCR loci (Hesslein and Schatz, 2001), and their stage-specific expression patterns strongly correlate with accessibility of these transcribed elements (e.g., Yancopoulos and Alt, 1985). The precise role of sense germline transcripts is still not understood and has been debated (Krangel, 2003). Recent studies support the notion that active transcription mediates chromatin changes that render the transcribed regions accessible to the recombinase (Sen and Oltz, 2006). However, it has been debated whether germline transcripts are the cause or the effect of these chromatin changes, and neither possibility has been unequivocally proven or disproved. On one hand, the levels of germline transcripts exhibit a positive correlation with rearrangement efficiency (Sun and Storb, 2001), which could suggest that the process of transcription itself could promote RAG targeting. However, others have shown that the correlation between individual rearrangements and germline transcription is not absolute (Angelin-Duclos and Calame, 1998; Sikes et al., 2002).
The IgH locus in germline configuration is transcribed from the promoter of DQ52 (PDQ52), the 3′ most DH segment, towards Cμ, thereby producing the so-called μ0 transcript (Fig. 1.2A). After D to JH rearrangement, the recombined DJH element is transcribed (Fig. 1.2C) (Alessandrini and Desiderio, 1991; Reth and Alt, 1984); and at the same time, unrearranged VH segments are transcribed from their promoters (Fig. 1.2B). Germline VH transcription appears to be silenced upon a productive rearrangement (Corcoran, 2005; Yancopoulos et al., 1985).
More recently, antisense transcripts have been found to occur throughout the VH cluster (Fig. 1.2B) (Bolland et al., 2004), in the DH region (Fig. 1.2A and C) (Bolland et al., 2007; Chakraborty et al., 2007), and in the JH region (Fig. 1.2A) (Bolland et al., 2007; Perlot et al., 2008). VH antisense transcripts appear to be biallelic, and it has been argued that such transcripts are large and span several VH segments and the adjacent intergenic regions; but formal proof of their initiation sites is still lacking. VH antisense transcription was shown to initiate during D to JH recombination, and to be rapidly downregulated after VH to DJH recombination (Bolland et al., 2004). DH antisense transcripts were detected in RAG-deficient pro-B-cells as well as on the D–JH rearranged allele of B-cell lines with a functionally assembled IgH gene (Chakraborty et al., 2007). DH antisense transcripts have been suggested to originate from the 3′ most DH (Chakraborty et al., 2007) or from the JH region (Bolland et al., 2007).
The functional significance of antisense transcription in the context of V(D)J recombination has not been fully elucidated. It has been postulated that antisense transcription promotes an active chromatin state rendering the locus more accessible (Bolland et al., 2004), based on the observed correlation between antisense VH germline transcription and active VH to DJH recombination (Bolland et al., 2004). Similar conclusions were reached based on the observation of reduced antisense DH transcripts and reduced D to JH rearrangements in mice lacking the intronic enhancer, Eμ (Afshar et al., 2006; Bolland et al., 2007). Conversely, others have raised the possibility that antisense transcripts, at least in the DSP DH segments, could pair with low levels of postulated germline sense transcripts and elicit RNA interference-mediated transcriptional gene silencing (Chakraborty et al., 2007; Koralov et al., 2008). It should be noted that true germline sense transcripts have not been identified as yet in the germline DH segments, but their level may be as low as those originally identified in the S. pombe centromeric repeats and may only be revealed in an RNAi-deficient background (Volpe et al., 2002).
6.2. Spatial organization and nuclear positioning of the IgH locus
The spatial organization of the Ig and TCR loci was analyzed by three-dimensional fluorescence in situ hybridization (3D FISH), in which nuclear organization remains preserved. Several groups showed that before undergoing rearrangement, the IgH locus moves from its default position at the nuclear periphery to a more central compartment (Fuxa et al., 2004; Kosak et al., 2002), going along with the observation that the nuclear periphery has a repressive effect on transcription (Andrulis et al., 1998; Baxter et al., 2002; Reddy et al., 2008) and, therefore, might keep the IgH locus in an inaccessible state. These observations are consistent with the peripheral location of the IgH locus in thymocytes which have only low level of D to JH and no VH to DJH rearrangements (Fuxa et al., 2004; Kosak et al., 2002; Kurosawa et al., 1981). The centrally located IgH locus in pro-B-cells can undergo D to JH rearrangement; however, for rearrangements of the distant VH elements, long-range contraction and looping of the IgH locus (Jhunjhunwala et al., 2008) seems to be crucial as lack of IgH locus contraction in Pax5-deficient pro-B-cells does not allow for rearrangements of intermediate and distal VH families (Fuxa et al., 2004; Sayegh et al., 2005). After successful rearrangement, the IgH locus decontracts and, thereby, has been proposed to impede further VH to DJH rearrangements by increasing the distance between VH elements and the DJH region (Roldán et al., 2005). Therefore, it seems that one aspect of VH to DJH recombination accessibility might be influenced by spatial arrangement of the IgH locus within the nucleus.
In B lineage stages subsequent to the pro-B stage, one IgH allele is positioned in close proximity to centromeric heterochromatin (Roldán et al., 2005; Skok et al., 2001). This finding was interpreted as the monoallelic silencing of the nonproductive IgH allele, because transcriptionally silent genes have been shown to associate with centromeric heterochromatin (Brown et al., 1997). However, the VH cluster gets silenced on both alleles in the context of germline transcription, and considering the fact that rearrangements from both IgH alleles, productive and nonproductive in either DJH or VHDJH configuration, are expressed in all B-cell stages that were examined (Daly et al., 2007; Fukita et al., 1998; Ono and Nose, 2007), monoallelic silencing might be either a short-time transient phenomenon or recruitment to centromeric heterochromatin might have other implications in the process of allelic exclusion.
Studies of interactions between IgH and Igκ alleles demonstrated coordinated patterns of action of Ig loci during B-cell development. Interactions between IgH and Igκ, predominantly in pre-B-cells, were demonstrated to reposition the interacting IgH allele to centromeric heterochromatin and induce IgH locus decompaction (Hewitt et al., 2008). Therefore, this interchromosomal interaction could play a role in IgH allelic exclusion and the transition from accessible IgH alleles to accessible Igκ alleles.
6.3. Chromatin modifications
Eukaryotic DNA is packaged into nucleosomes in which genomic DNA is wrapped around histone octamers. The N-terminal ends of histones, called histone tails, can be marked by diverse modifications (e.g., acetylation, methylation, phosphorylation, ubiquitination, and others). The “histone code” (Jenuwein and Allis, 2001) translates patterns of histone modifications into repression or activation of chromatin. An extensive effort has been made to investigate the effects of histone modifications and also various other chromatin attributes such as DNA methylation, DNase sensitivity, and nucleosome remodeling on accessibility of Ig and TCR loci with hopes of shedding light on the epigenetic regulation of V(D)J recombination.
Posttranslational modifications of N-terminal histone tails can affect genome regulation in several ways. Histone modifications can directly affect chromatin structure, for example, through a change in charge. In this context, histone acetylation can loosen the association between DNA and the histone core or can alter higher order chromatin packaging. Alternatively chromatin modifications can disrupt or provide binding sites for chromatin remodeling complexes or other effector molecules. Prominent examples of such specialized binding domains are bromodomains specifically binding acetylated lysines, and chromodomains binding to dimethylated lysine 9 on histone 3 (Kouzarides and Berger, 2007).
Many studies showed that marks of active chromatin correlate with V(D)J rearrangements. For example, acetylated lysine 9 on histone 3 (H3K9ac), hyperacetylated histone 4, and dimethylated lysine 4 on histone 3 (H3K4me2) are active chromatin marks (Kouzarides and Berger, 2007). They are present in the D–JH region peaking around the 5′ most D segment, DFL16.1, and over the JH elements (Chakraborty et al., 2007; Morshead et al., 2003) in early pro-B-cells that are poised to undergo D to JH rearrangements. However, they are almost absent in thymocytes (Chakraborty et al., 2007). Following D to JH recombination, the proximal VH elements become hyperacetylated and, thereafter, in a manner that is dependent on IL-7R signaling and on its downstream effector STAT5 (signal transducer and activator of transcription 5), the distal VH segments become hyperacetylated (Bertolino et al., 2005; Chowdhury and Sen, 2001). Acetylation patterns seem to be narrowly confined to the VH segment, its promoter, and RSS (Johnson et al., 2003).
Histone hyperacetylation is lost after productive VH to DJH recombination, thereby contributing to rendering the VH cluster inaccessible in pre-B-cells (Chowdhury and Sen, 2003). Notably, an engineered locus that actively recruits an H3K9 methyltransferase shows downregulation of germline transcripts and impaired V(D)J recombination (Osipovich et al., 2004). H3K9me2 is absent in the D–JH region of pro-B-cells and present in thymocytes (Chakraborty et al., 2007), and removal of H3K9me2 from the VH region before VH to DJH recombination is dependent on Pax5 (Johnson et al., 2004), a transcription factor essential for B-cell commitment (Busslinger, 2004). In agreement with this data, loss of Pax5 leads to an inability to rearrange distal VH gene families (Hesslein et al., 2003; Nutt et al., 1997).
The antagonistic Polycomb (PcG) and Trithorax (trxG) groups of protein complexes establish and propagate a silenced or active chromatin state, respectively (Ringrose and Paro, 2004). Curiously, targeted deletion of the PcG protein Ezh2, an H3K27 methyltransferase, inhibits rearrangements of the distal VHJ558 family without affecting germline transcription (Su et al., 2003). H3K27 methylation was reported to be a mark of inactive chromatin (Kouzarides and Berger, 2007); therefore, it remains to be determined whether the results observed in the Ezh2 knock out are direct or indirect effects.
Recent studies reported that the PhD finger domain of RAG2 specifically binds to trimethylated H3K4 (Liu et al., 2007; Matthews et al., 2007), a histone modification associated with transcriptional start regions (Pokholok et al., 2005) also shown to be present in accessible IgH regions (Liu et al., 2007). Mutation of the conserved tryptophan residue W453 within the PhD finger domain of RAG2 abrogates RAG2 binding to H3K4me3 and impairs V(D)J recombination of chromosomal and extra-chromosomal substrates (Liu et al., 2007; Matthews et al., 2007). However, removal of the entire RAG2 noncore region, including the PhD domain, only leads to a partial impairment of V(D)J recombination (Liang et al., 2002). These seemingly contradicting data have been suggested to reflect the presence of an inhibitory function within the noncore region of RAG-2, which is relieved upon binding to H3K4me3, or can be circumvented by deleting the entire noncore region (Liu et al., 2007; Matthews et al., 2007). These recent studies provide the first direct link between epigenetic control of V(D)J rearrangement and RAG recombinase accessibility.
Chromatin remodeling complexes can change the composition, structure, or position of nucleosomes within chromatin. These changes are noncovalent and are dependent on ATP hydrolysis (Martens and Winston, 2003). The SWI/SNF chromatin remodeling complex contains a bromodomain that allows it to efficiently bind acetylated chromatin and mobilize nucleosomes or change nucleosome structure (Martens and Winston, 2003). In this regard, it was shown that unmodified or even hyperacetylated nucleosomes located directly on RSSs are inhibitory to RAG cleavage in vitro (Golding et al., 1999) and that addition of SWI/SNF improved substrate cleavage (Kwon et al., 2000). RSSs strongly attract nucleosomes and, thus, may implement some aspect of accessibility control (Baumann et al., 2003). Moreover, nucleosome positioning appears pivotal for V(D)J recombination in vivo (Cherry and Baltimore, 1999). Further supporting the importance of SWI/SNF complexes in V(D)J recombination, BRG1 (the ATPase subunit of SWI/SNF) was found to associate at Ig and TCR loci within hyperacetylated chromatin regions (Morshead et al., 2003). Functional targeting of BRG1 to a TCRβ minilocus lacking the essential Dβ promoter rescued V(D)J recombination of this substrate (Osipovich et al., 2007), substantiating the role of SWI/SNF complexes in V(D)J recombination and suggesting a role for transcriptional promoters in recruitment of chromatin remodeling complexes.
Another readout to assess chromatin structure is the DNase sensitivity assay. Less tightly packed chromatin- or nucleosome-free DNA is more sensitive to DNase or restriction enzyme digestion than heterochromatin regions. While cis-acting elements such as promoters and enhancers can be devoid of nucleosomes and, therefore, are DNase hypersensitive, accessible chromatin of Ig and TCR loci shows general DNase sensitivity (Yancopoulos et al., 1986). In this context, the region between DQ52 and Eμ is DNase sensitive before D to JH rearrangement and JH RSSs show enhanced sensitivity and seem to be nucleosome-free (Maës et al., 2006). The VH region becomes nuclease sensitive before VH to DJH rearrangement and reverts to a refractory state after successful VH to DJH recombination (Chowdhury and Sen, 2003).
Cytosines in mammalian DNA can be methylated in CpG dinucleotides. Generally, cytosine methylation corresponds to silenced genes (Stein et al., 1982; Vardimon et al., 1982) or silent regions throughout the genome; whereas promoter regions of expressed genes are found in an unmethylated state. Cytosine methylation can act by inhibiting regulatory proteins from binding to DNA (Watt and Molloy, 1988), or by recruiting methyl-CpG binding proteins which in turn can interact with HDACs to enforce a silent chromatin state (Jaenisch and Bird, 2003). In this regard, methylated V(D)J recombination substrates are refractory to active rearrangement (Cherry and Baltimore, 1999; Hsieh and Lieber, 1992); in particular, methylated RSSs can abolish RAG cleavage and V(D)J recombination (Whitehurst et al., 2000). Demethylation alone, however, is not sufficient to initiate V(D)J recombination (Cherry et al., 2000). The DH–JH cluster gets demethylated before the onset of D to JH recombination (Maës et al., 2001; Storb and Arp, 1983) characteristic of an accessible state. In this context, the JCκ region gets monoallelically demethylated and this demethylated allele undergoes rearrangement first (Mostoslavsky et al., 1998). The second allele stays in a repressive environment and somehow can get demethylated, if the rearrangement on the first Igκ allele is nonproductive (Goldmit and Bergman, 2004). More extensive studies on DNA methylation of the IgH locus could potentially help to elucidate aspects of accessibility control within this locus.
7. IgH LOCUS CONTROL THROUGH CIS-REGULATORY ELEMENTS
A formidable number of cis-regulatory elements have been identified throughout the IgH locus (Fig. 1.1). Enhancers are located in the JH–Cμ intronic region and at the very 3′ end of the locus. Promoters are found 5′ of VH and D segments as well as 5′ of most CH genes. Cis-elements in the IgH locus not only govern gene expression, but also play crucial roles in accessibility control in all its above-mentioned aspects and also control CSR. An extensive effort has been made to elucidate the many roles of these transcription elements. Ongoing research also aims at identifying missing regulatory elements and elucidating their role in IgH locus control.
7.1. Promoter of DQ52
DQ52 is the 3′ most D segment. Its promoter becomes active before D to JH rearrangement to generate the μ0 transcript (Fig. 1.2A) (Alessandrini and Desiderio, 1991; Kottmann et al., 1994; Schlissel et al., 1991a). This transcript runs all the way through the Cμ exons, which get spliced to the JH1 splice donor site (Schlissel et al., 1991b). The same promoter region also gives rise to a low-level antisense transcript (Chakraborty et al., 2007). It has been suggested that the repetitive nature of the DH region in combination with bidirectional transcription can elicit RNA interference-mediated transcriptional gene silencing that would lead to the observed inactive chromatin state of the DSP elements. However, as mentioned above, only antisense and no sense transcripts have been detected thus far in the germline DH region (Chakraborty et al., 2007).
Every DH element upstream of DQ52 has a bidirectional promoter which, upon D to JH rearrangement, potentially through approximation to the Eμ enhancer, gets activated to generate an antisense transcript and a sense transcript (Fig. 1.2C) (Alessandrini and Desiderio, 1991; Chakraborty et al., 2007). The sense transcript gets spliced in a way that the rearranged DJH segment is joined to the Cμ exons. In one reading frame this mRNA can encode for a shorter version of the μHC (Reth and Alt, 1984), which can inhibit subsequent VH to DJH rearrangements (Gu et al., 1991; Löffert et al., 1996; Malynn et al., 2002). Targeted deletion of the DQ52 promoter, which has both promoter and enhancer activity (Kottmann et al., 1994), in mice had no major impact on D to JH rearrangement, other than a slight shift in JH usage (Afshar et al., 2006; Nitschke et al., 2001). However, in these studies, μ0-like transcripts were still evident, suggesting that activity of the heterogeneous promoter of DQ52 was not entirely abrogated. In a different study, the intronic Eμ enhancer was replaced with a phosphoglycerate kinase promoter–neomycin resistance gene cassette (PGK-NeoR), which resulted in complete absence of μ0 transcripts and complete inhibition of D to JH rearrangement (Perlot et al., 2005). In this regard, targeted deletion of an analogous promoter element in the TCRβ locus, the promoter of Dβ1, led to diminished germline transcripts from this promoter and reduced Dβ1 rearrangements (Whitehurst et al., 1999), demonstrating an accessibility control function for this element in Dβ–Jβ recombination.
7.2. VH promoters
Every VH element has its own promoter that initiates VH germline transcripts before VH to DJH rearrangement (Fig. 1.2B), as well as transcripts of the assembled VHDJH exon after rearrangement (Fig. 1.2D). Most VH promoters can generate a germline transcript, in which a leader exon gets spliced to a VH exon (Fig. 1.2B). The transcript gets polyadenylated and contains an open reading frame (Yancopoulos and Alt, 1985); however, no VH protein or its function has thus far been demonstrated. The most conserved element across VH promoters is the octamer ATGCAAAT (Parslow et al., 1984). This sequence element has been shown to be necessary for VH transcription (Mason et al., 1985), and it binds the ubiquitously expressed Oct-1 and the B-cell-specific Oct-2, both POU family transcription factors. Most but not all VH promoters contain a TATA box, an initiator (Inr) element (Buchanan et al., 1997), a heptamer, and a pyrimidine stretch (Eaton and Calame, 1987). Additionally, binding sites for a number of mostly B-lineage-specific transcription factors and chromatin remodeling complexes have been identified in VH promoter regions (Johnston et al., 2006).
Germline transcripts from unrearranged VH promoters are generated upon D to JH rearrangement in pro-B-cells, and downregulated after completed VH to DJH recombination and assembly and expression of a functional VHDJH exon (Bolland et al., 2004; Hardy et al., 1991). The promoter of a recombined VH element stays active throughout B-cell development, and cell line experiments showed that the first upstream unrearranged VH segment can also be continuously expressed at reduced levels (Wang and Calame, 1985). Promoter activity of a functionally rearranged VH element was shown to be partially dependent on the 3′ regulatory region (Pinaud et al., 2001). Thus, VH promoters might fulfill a dual role: to help confer accessibility to germline VH segments and to drive expression of the assembled heavy chain gene.
7.3. Intronic enhancer
The IgH intronic enhancer (Eμ) was the first cellular (as opposed to viral) eukaryotic enhancer element described (Alt et al., 1982; Banerji et al., 1983; Gillies et al., 1983). Eμ comprises a 220 bp enhancer core (cEμ) and two flanking matrix attachment regions (MARs). Targeted deletion of both MARs shows that they are dispensable for efficient V(D)J recombination within the IgH locus (Sakai et al., 1999). Deletion of Eμ in B-cells (Chen et al., 1993; Sakai et al., 1999; Serwe and Sablitzky, 1993) and in the germline of mice (Afshar et al., 2006; Perlot et al., 2005) led to reduced D to JH rearrangement and severely impaired VH to DJH rearrangement. The residual V(D)J recombination activity in the IgH locus implies that activation of IgH rearrangements may also involve one or more additional enhancer type elements. One candidate for such a compensating element is the promoter/enhancer region PDQ52, which was speculated to promote D to JH recombination (Alessandrini and Desiderio, 1991). However, deletion of PDQ52 along with Eμ did not show increased impairment above that seen with deletion of Eμ alone (Afshar et al., 2006). However, since the deletion of PDQ52 appeared to be incomplete, this element can not yet be ruled out as having redundant functions with Eμ in conferring accessibility to the DH–JH region. Another candidate for cooperative function with Eμ is the 3′ IgH regulatory region, but the double knockout of Eμ and the IgH 3′RR has not been generated. By analogy, deletion of the intronic Igκ enhancer (iEκ) reduces Vk to Jκ rearrangements (Xu et al., 1996); whereas a double knockout of iEκ and the 3′Eκ enhancer completely blocks recombination of the Igκ locus (Inlay et al., 2002). The iEκ and 3′Eκ in the Igκ locus are the enhancer elements corresponding to the position of Eμ and IgH 3′RR in the IgH locus.
It has been puzzling why in Eμ knockout mice the VH to DJH step is more severely impaired than the D to JH step, even though the Eμ enhancer has no obvious effect on germline transcripts of intermediate and distal VH families (Perlot et al., 2005). One explanation could be significant underestimation of D to JH impairment in Eμ knockout mice. Initial very low levels of D to JH rearrangements could limit the crucial DJH substrates for subsequent VH to DJH rearrangements and, therefore, result in the observed strong reduction of VH to DJH recombination. After a productive rearrangement, feedback regulation inhibits further VH to DJH recombination, but does not block further D to JH rearrangements (Reth et al., 1987). Therefore, D to JH recombination might “catch up” over the course of B-cell development and mask a stronger impairment.
Notably, replacement of Eμ with a PGK-NeoR cassette (Chen et al., 1993; Perlot et al., 2005; Sakai et al., 1999) or introduction of PGK-NeoR cassette just 5′ of Eμ (Chen et al., 1993; Delpy et al., 2002) results in a much more severe impairment or a complete block of V(D)J recombination and concomitant complete loss of μ0 transcripts (Perlot et al., 2005). This phenomenon could be explained by a promoter competition/insulating mechanism. In such a scenario, the PGK-NeoR gene and its promoter might compete with PDQ52 for activity from a downstream cis-element such as the IgH 3′RR, which is known to act over long distances (Pinaud et al., 2001). Similar promoter competition for the IgH 3′RR has been observed between I promoters and the PGK-NeoR cassette introduced in the CH region (see below). Alternatively, the PGK-NeoR cassette could induce local chromatin changes that impede μ0 germline transcription and accessibility of D and JH segments.
Extensive studies revealed an array of binding sites for B-lineage-specific transcription factors and also for ubiquitously expressed proteins within the Eμ enhancer and the flanking MARs (Calame and Sen, 2004). The unique combination of these factors is likely to mediate the enhancer’s predominant activity in pro-B-cells (Inlay et al., 2006). In this context, replacement of iEκ with Eμ leads to premature Igκ rearrangement in pro-B-cells and absence of Igκ rearrangements in pre-B-cells, the stage when LC rearrangement normally takes place (Inlay et al., 2006), corroborating the pro-B-cell specificity of Eμ.
Eμ was suggested to play a role in regulating antisense transcripts through the JH and DH region (Afshar et al., 2006; Bolland et al., 2007), and additionally a promoter region within Eμ was identified that gives rise to the Iμ transcript (Lennon and Perry, 1985; Su and Kadesch, 1990). Starting at Eμ, this transcript extends through the μ switch region and Cμ. Transcription of switch regions was shown to be necessary for CSR, probably for targeting AID, and in this regard, deletion of Eμ leads to reduced Iμ transcript levels and reduced CSR (Bottaro et al., 1998; Perlot et al., 2005). Deletion of Eμ has no obvious effect on somatic hypermutation of VHDJH exons in mature B-cells (Perlot et al., 2005). An open question is how the activity of AID is specifically targeted to regions within Ig loci. It was speculated that cis-regulatory elements could determine this specificity, but neither cEμ nor the IgH 3′RR, alone (Morvan et al., 2003), seem to have a crucial role in targeting SHM to the IgH locus VHDJH segments. While an absolute requirement of cEμ or the IgH 3′RR for SHM can be excluded, there is a possibility that smaller defects of SHM in these mutants are masked by selection processes during affinity maturation. Also, a combined function of cEμ and the IgH 3′RR in promoting or targeting SHM is another possibility that needs to be tested.
7.4. 3′ IgH regulatory region and I promoters
I promoters are located upstream of all switch regions (Chaudhuri et al., 2007; Lennon and Perry, 1985; Lutzker and Alt, 1988). Transcripts initiating from I promoters are processed in such a way that an I (intervening) exon, located immediately downstream of the I promoter, is spliced to the associated CH exons. In this process, the intronic region including the S region is spliced out and the transcript gets polyadenylated. However, these transcripts appear “sterile,” as they do not contain an open reading frame and could not be shown to encode for a protein (Chaudhuri et al., 2007). Active transcription from I promoters is necessary for CSR as only transcribed S regions can become AID targets during CSR. In this context, deletion of I promoters abrogates efficient CSR to the associated CH genes; while replacement of I promoters with a constitutively active promoter directs CSR to the associated CH gene (Manis et al., 2002). Transcription from different I promoters prior to CSR can be induced upon stimulation with different activators or cytokines. Corresponding surface receptors for these molecules and their associated downstream signaling pathways effect different combinations of activating or repressive response elements within I promoter regions, which leads to CSR to different IgH isotypes under different stimulation conditions (Stavnezer, 2000). Most I promoters do not appear to act in isolation as efficient transcription from them also requires the IgH 3′RR (Pinaud et al., 2001) and physical interaction between the IgH 3′RR and specific I promoters has been implicated (Wuerffel et al., 2007).
The IgH 3′RR is located downstream of Cα at the very 3′ end of the IgH locus (Fig. 1.2B). This regulatory region consists of a number of DNaseI hypersensitive sites scattered over ~35 kb (Dariavach et al., 1991; Garrett et al., 2005; Lieberson et al., 1991; Matthias and Baltimore, 1993; Pettersson et al., 1990); up until now, none of them were shown to play a role in V(D)J recombination but more studies are needed (Khamlichi et al., 2000; Pinaud et al., 2001). The most striking function, control of IgH CSR, has been assigned to HS3b, HS4 within the IgH 3′RR. Targeted deletions in mice revealed severely reduced CSR to most IgH isotypes and reduced germline transcription from I promoters through the corresponding S regions (Pinaud et al., 2001), a process required for CSR (Jung et al., 1993; Zhang et al., 1993). Deletion of the more 5′ DNaseI hypersensitive sites within the IgH 3′RR HS3a and HS1,2 had no effect on CSR (Manis et al., 1998); however, replacement of HS3a or HS1,2 with a PGK-NeoR cassette resulted in a similar defect as in the HS3b, HS4 deletion (Cogné et al., 1994, Manis et al., 1998). The latter observations suggested a potential promoter competition/insulation between I promoters and the PGK-NeoR cassette for signals from within the IgH 3′RR. This hypothesis was strengthened by insertion of a PGK-NeoR cassette at the Iγ2b promoter or the Cε gene, respectively. In both cases, germline transcription and class switching to CH genes 3′ of the inserted PGK-NeoR cassette was unaffected; while germline transcription and class switching to CH genes 5′ of the inserted PGK-NeoR cassette was impaired (Seidl et al., 1999). These results suggest that the inserted PGK-NeoR cassette can interfere with the long-range control effect of IgH 3′RR on CSR in a position-dependent manner.
The IgH 3′RR is necessary for efficient expression of the rearranged HC from the promoter upstream of the assembled VHDJH exon (Pinaud et al., 2001), whereas the much more proximal Eμ enhancer is not required for HC expression (Perlot et al., 2005). Because it can influence expression of rearranged VHDJH segments, the IgH 3′ RR can function over a distance of at least 200 kb. Such long-range activity may be important for activating oncogenes translocated into the upstream portions of the CH locus in lymphomas. Not all of the seven described hypersensitivity sites in the spacious 3′ regulatory region have been knocked out yet, therefore, other potential functions still remain to be discovered. Apart from the above-mentioned effects on germline transcription, CSR, and IgH expression, it has been speculated that parts of the 3′ regulatory region might have a role in long-range chromatin organization. Finally, activity of the Iγ1 promoter does not appear to be dependent on the IgH 3′RR; suggesting that it carries sufficient regulatory elements itself or that there are other long range IgH locus elements that function in CSR to be defined.
7.5. Additional potential regulatory elements
Several laboratories suggested that the IgH locus can be associated with the nuclear periphery via its 5′ region (Kosak et al., 2002; Yang et al., 2005). The 5′ end of the IgH locus does not get deleted in the course of V(D)J recombination and as such is an attractive location for a missing regulatory element that controls processes such as accessibility control of the distal VH genes, positioning of the IgH locus, or feedback regulation. In fact, ~30 kb upstream of the most distal VH element an array of DNaseI hypersensitive sites has been identified (Pawlitzky et al., 2006). One of these sites, HS1, was reported to be pro-B-cell specific and potentially contain binding sites for the transcription factors PU.1, Pax5, and E2A. However, preliminary knockout experiments, in which HS1 was deleted, showed no effect on the IgH locus, as targeted alleles could still undergo efficient V(D)J recombination including all VH gene families. Furthermore, allelic exclusion was unaffected (Perlot, Pawlitzky, Brodeur, and Alt, unpublished data). Other potential functions of these sites, including acting as a boundary area as was suggested by DNA modifications confined to one side of 5′IgH hypersensitive sites (Reddy et al., 2008), are still being tested.
Another area that was speculated to harbor a regulatory element is the ~90 kb region between the VH and the DH clusters. This region could contain an element that ensures the ordered rearrangement of the D to JH and VH to DJH steps, such as a boundary element that influences activation of separate IgH locus domains. Moreover, the VH to DH intergenic region is deleted on a productively rearranged allele but remains in place on a DJH rearranged allele, suggesting an element might reside in this region that is responsible for shutting down the incompletely rearranged allele in the context of allelic exclusion. Potential support for such an element came from placement of a VH segment into the DH region, which resulted in breaking of lineage specificity, ordered rearrangement, and allelic exclusion of the introduced VH segment (Bates et al., 2007). Preliminary studies in which this intergenic region has been deleted have provided direct support for the notion that this region contains elements important for regulation of lineage specificity of VH to DJH rearrangement (Giallourakis, Franklin, and Alt, unpublished data).
7.6. Interplay between cis-regulatory elements
The transition from an inactive to an active chromatin state of the IgH locus is in part governed by Eμ. The intronic enhancer plays an important role in placing active chromatin marks throughout the DH–JH region (Chakraborty, Perlot, Subrahmanyam, Alt, and Sen, unpublished data). In addition, Eμ promotes transcripts from PDQ52 and supports formation of the DNaseI hypersensitive site at this promoter element (Perlot et al., 2005; Chakraborty, Perlot, Subrahmanyam, Alt, and Sen, unpublished data). These data argue in favor of a direct interaction between Eμ and PDQ52 reminiscent of a corresponding promoter/enhancer holocomplex described in the TCRβ locus (Oestreich et al., 2006). The fact that Eμ is not absolutely required for stimulation of PDQ52 suggests partial compensation by another cis-element. It was speculated that the IgH 3′RR can take over this role of interaction with PDQ52 and of activating the DH–JH region, because its ability to function over a long range to activate I region promoters and to influence expression of the promoter of a rearranged VHDJH segment (Pinaud et al., 2001).
No single cis-regulatory element has thus far been identified that is responsible for the bulk of VH germline transcription. It seems to be likely that VH promoters can be activated in trans by B-lineage and stage-specific factors. VH antisense transcripts might be a prerequisite of VH sense germline transcripts by initializing an active chromatin state and accessibility of the VH locus (Bolland et al., 2004). Start sites of these transcripts still remain elusive, which exacerbates the manipulation of such transcripts and a direct proof for this hypothesis.
After rearrangement of a complete VHDJH exon, the assembled VHDJH–Cμ gene is transcribed from the VH promoter 5′ of the rearranged VH. Transcription from the rearranged VH promoter is mainly supported by the IgH 3′RR (Pinaud et al., 2001) suggesting direct interaction of the two cis-elements. In this regard, the ability of the IgH 3′RR to form a direct complex with a region around Eμ and downstream I promoters was demonstrated during CSR (Wuerffel et al., 2007). However, cEμ does not appear to be involved in that interaction, since deletion of that element does not affect complex formation (Wuerffel et al., 2007). As mentioned above, the interaction of the IgH 3′RR with I region promoters likely underlies the cooperation of these two elements in I region transcription and regulation of IgH CSR.
8. CONCLUSIONS
Antigen receptor genes are assembled by the process of V(D)J recombination in a developmentally controlled manner. Differential accessibility at Ig and TCR loci is regulated at least in part by cooperative action of cis-regulatory elements. Tremendous progress has been made in identifying and elucidating the multiple layers of control of the IgH locus during V(D)J recombination; however, many important questions remain unanswered and new questions are emerging. Among these are processes involved in ordered IgH rearrangements, asynchronous VH to DJH rearrangements, enforcement of feedback regulation, the precise relevance and impact of chromosome positioning and movements, the role of antisense transcription throughout the IgH locus, and chromatin modifications. Likewise, a remarkable amount of progress has been made in elucidating the role of cis-acting elements in the regulation of IgH CSR, but again there are still many unanswered questions including precisely how these elements function to specifically target AID to S regions and the precise mechanisms by which the IgH 3′RR and I region promoters elements cooperate in response to external stimuli to specifically activate CSR to particular CH genes. To fully understand the genetic and epigenetic regulation of the IgH locus, all involved cis-regulatory elements and trans acting factors need to be identified and analyzed. More work also will need to be done to understand how these factors influence regulation at the level of chromatin structure and spatial organization. Understanding the mechanisms governing the IgH locus, a model system for gene expression and epigenetic regulation will also advance our understanding of various other unsolved biological problems.
ACKNOWLEDGMENTS
We thank Cosmas Giallourakis and John Manis for critically reviewing the manuscript and for discussions. T. P. received a Boehringer Ingelheim Fonds PhD scholarship. This work was supported by National Institutes of Health Grants PO1CA092625-05 and 2PO1AI031541-15 (to F.W.A.). F.W.A. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- Afshar R, Pierce S, Bolland DJ, Corcoran A, Oltz EM. Regulation of IgH gene assembly: Role of the intronic enhancer and 5′ Δθ52 region in targeting DHJH recombination. J. Immunol. 2006;176(4):2439–2447. doi: 10.4049/jimmunol.176.4.2439. [DOI] [PubMed] [Google Scholar]
- Alessandrini A, Desiderio SV. Coordination of immunoglobulin DJH transcription and D-to-JH rearrangement by promoter-enhancer approximation. Mol. Cell. Biol. 1991;11(4):2096–2107. doi: 10.1128/mcb.11.4.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt FW, Enea V, Bothwell AL, Baltimore D. Activity of multiple light chain genes in murine myeloma cells producing a single, functional light chain. Cell. 1980;21(1):1–12. doi: 10.1016/0092-8674(80)90109-9. [DOI] [PubMed] [Google Scholar]
- Alt FW, Rosenberg N, Casanova RJ, Thomas E, Baltimore D. Immunoglobulin heavy-chain expression and class switching in a murine leukaemia cell line. Nature. 1982;296(5855):325–331. doi: 10.1038/296325a0. [DOI] [PubMed] [Google Scholar]
- Alt FW, Yancopoulos GD, Blackwell TK, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984;3(6):1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394(6693):592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- Angelin-Duclos C, Calame K. Evidence that immunoglobulin VH-DJ recombination does not require germ line transcription of the recombining variable gene segment. Mol. Cell. Biol. 1998;18(11):6253–6264. doi: 10.1128/mcb.18.11.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel TW, Mautner J, Polack A, Bornkamm GW, Eick D. Two antisense promoters in the immunoglobulin mu-switch region drive expression of c-myc in the Burkitt’s lymphoma cell line BL67. Oncogene. 1992;7(7):1267–1271. [PubMed] [Google Scholar]
- Banerji J, Olson L, Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983;33(3):729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Barreto V, Cumano A. Frequency and characterization of phenotypic Ig heavy chain allelically included IgM-expressing B cells in mice. J. Immunol. 2000;164(2):893–899. doi: 10.4049/jimmunol.164.2.893. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Alt FW, Hughes MM, D’Auteuil M, Wehrly TD, Woodman BB, Gärtner F, White JM, Davidson L, Sleckman BP. Recombination signal sequences restrict chromosomal V(D)J recombination beyond the 12/23 rule. Nature. 2000;405(6786):583–586. doi: 10.1038/35014635. [DOI] [PubMed] [Google Scholar]
- Bates JG, Cado D, Nolla H, Schlissel MS. Chromosomal position of a VH gene segment determines its activation and inactivation as a substrate for V(D)J recombination. J. Exp. Med. 2007;204(13):3247–3256. doi: 10.1084/jem.20071787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann M, Mamais A, McBlane F, Xiao H, Boyes J. Regulation of V(D)J recombination by nucleosome positioning at recombination signal sequences. EMBO J. 2003;22(19):5197–5207. doi: 10.1093/emboj/cdg487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter J, Merkenschlager M, Fisher AG. Nuclear organisation and gene expression. Curr. Opin. Cell Biol. 2002;14(3):372–376. doi: 10.1016/s0955-0674(02)00339-3. [DOI] [PubMed] [Google Scholar]
- Bertolino E, Reddy K, Medina KL, Parganas E, Ihle J, Singh H. Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangements by transcription factor STAT5. Nat. Immunol. 2005;6(8):836–843. doi: 10.1038/ni1226. [DOI] [PubMed] [Google Scholar]
- Bolland DJ, Wood AL, Johnston CM, Bunting SF, Morgan G, Chakalova L, Fraser PJ, Corcoran AE. Antisense intergenic transcription in V(D)J recombination. Nat. Innunol. 2004;5(6):630–637. doi: 10.1038/ni1068. [DOI] [PubMed] [Google Scholar]
- Bolland DJ, Wood AL, Afshar R, Featherstone K, Oltz EM, Corcoran AE. Antisense intergenic transcription precedes Igh D-to-J recombination and is controlled by the intronic enhancer Emu. Mol. Cell. Biol. 2007;27(15):5523–5533. doi: 10.1128/MCB.02407-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghesi L, Hsu LY, Miller JP, Anderson M, Herzenberg L, Herzenberg L, Schlissel MS, Allman D, Gerstein RM. B lineage-specific regulation of V(D)J recombinase activity is established in common lymphoid progenitors. J. Exp. Med. 2004;199(4):491–502. doi: 10.1084/jem.20031800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Born W, White J, Kappler J, Marrack P. Rearrangement of IgH genes in normal thymocyte development. J. Immunol. 1988;140(9):3228–3232. [PubMed] [Google Scholar]
- Bottaro A, Young F, Chen J, Serwe M, Sablitzky F, Alt FW. Deletion of the IgH intronie enhancer and associated matrix-attachment regions decreases, but does not abolish, class switching at the mu locus. Int. Immunol. 1998;10(6):799–806. doi: 10.1093/intimm/10.6.799. [DOI] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91(6):845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Buchanan KL, Smith EA, Dou S, Corcoran LM, Webb CF. Family-specific differences in transcription efficiency of Ig heavy chain promoters. J. Immunol. 1997;159(3):1247–1254. [PubMed] [Google Scholar]
- Busslinger M. Transcriptional control of early B cell development. Annu. Rev. Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- Calame K, Sen R. Transcription of immunoglobulin genes. In: Honjo T, Alt FW, Neuberger MS, editors. Molecular Biology of B Cells. London: Elsevier Academic Press; 2004. pp. 83–100. [Google Scholar]
- Chakraborty T, Chowdhury D, Keyes A, Jani A, Subrahmanyam R, Ivanova I, Sen R. Repeat organization and epigenetic regulation of the DH-Cmu domain of the immunoglobulin heavy-chain gene locus. Mol. Cell. 2007;27(5):842–850. doi: 10.1016/j.molcel.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Basu U, Zarrin A, Yan C, Franco S, Perlot T, Vuong B, Wang J, Phan RT, Datta A, Manis J, Alt FW. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv. Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- Chen J, Young F, Bottaro A, Stewart V, Smith RK, Alt FW. Mutations of the intronic IgH enhancer and its flanking sequences differentially affect accessibility of the JH locus. EMBO J. 1993;12(12):4635–4645. doi: 10.1002/j.1460-2075.1993.tb06152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry SR, Baltimore D. Chromatin remodeling directly activates V(D)J recombination. Proc. Natl. Acad. Sci. USA. 1999;96(19):10788–10793. doi: 10.1073/pnas.96.19.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry SR, Beard C, Jaenisch R, Baltimore D. V(D)J recombination is not activated by demethylation of the kappa locus. Proc. Natl. Acad. Sci. USA. 2000;97(15):8467–8472. doi: 10.1073/pnas.150218497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D, Sen R. Stepwise activation of the immunoglobulin mu heavy chain gene locus. EMBO J. 2001;20(22):6394–6403. doi: 10.1093/emboj/20.22.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D, Sen R. Transient IL-7/IL-7R signaling provides a mechanism for feedback inhibition of immunoglobulin heavy chain gene rearrangements. Immunity. 2003;18(2):229–241. doi: 10.1016/s1074-7613(03)00030-x. [DOI] [PubMed] [Google Scholar]
- Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. Accessibility control of V(D)J recombination. Adv. Immunol. 2006;91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- Cogné M, Lansford R, Bottaro A, Zhang J, Gorman J, Young F, Cheng HL, Alt FW. A class switch control region at the 3′ end of the immunoglobulin heavy chain locus. Cell. 1994;77(5):737–747. doi: 10.1016/0092-8674(94)90057-4. [DOI] [PubMed] [Google Scholar]
- Corcoran AE. Immunoglobulin locus silencing and allelic exclusion. Semin. Immunol. 2005;17(2):141–154. doi: 10.1016/j.smim.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Daly J, Licence S, Nanou A, Morgan G, Mårtensson IL. Transcription of productive and nonproductive VDJ-recombined alleles after IgH allelic exclusion. EMBO J. 2007;26(19):4273–4282. doi: 10.1038/sj.emboj.7601846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dariavach P, Williams GT, Campbell K, Pettersson S, Neuberger MS. The mouse IgH 3′-enhancer. Eur. J. Immunol. 1991;21(6):1499–1504. doi: 10.1002/eji.1830210625. [DOI] [PubMed] [Google Scholar]
- Delpy L, Decourt C, Le Bert M, Cogné M. B cell development arrest upon insertion of a neo gene between JH and Emu: Promoter competition results in transcriptional silencing of germline JH and complete VDJ rearrangements. J. Immunol. 2002;169(12):6875–6882. doi: 10.4049/jimmunol.169.12.6875. [DOI] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Drejer-Teel AH, Fugmann SD, Schatz DG. The beyond 12/23 restriction is imposed at the nicking and pairing steps of DNA cleavage during V(D)J recombination. Mol. Cell. Biol. 2007;27(18):6288–6299. doi: 10.1128/MCB.00835-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early P, Huang H, Davis M, Calame K, Hood L. An immunoglobulin heavy chain variable region gene is generated from three segments of DNA: VH, D and JH. Cell. 1980;19(4):981–992. doi: 10.1016/0092-8674(80)90089-6. [DOI] [PubMed] [Google Scholar]
- Eaton S, Calame K. Multiple DNA sequence elements are necessary for the function of an immunoglobulin heavy chain promoter. Proc. Natl. Acad. Sci. USA. 1987;84(21):7634–7638. doi: 10.1073/pnas.84.21.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- Fukita Y, Jacobs H, Rajewsky K. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity. 1998;9(1):105–114. doi: 10.1016/s1074-7613(00)80592-0. [DOI] [PubMed] [Google Scholar]
- Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18(4):411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett FE, Emelyanov AV, Sepulveda MA, Flanagan P, Volpi S, Li F, Loukinov D, Eckhardt LA, Lobanenkov VV, Birshtein BK. Chromatin architecture near a potential 3′ end of the igh locus involves modular regulation of histone modifications during B-Cell development and in vivo occupancy at CTCF sites. Mol. Cell. Biol. 2005;25(4):1511–1525. doi: 10.1128/MCB.25.4.1511-1525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies SD, Morrison SL, Oi VT, Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Golding A, Chandler S, Ballestar E, Wolffe AP, Schlissel MS. Nucleosome structure completely inhibits in vitro cleavage by the V(D)J recombinase. EMBO J. 1999;18(13):3712–3723. doi: 10.1093/emboj/18.13.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmit M, Bergman Y. Monoallelic gene expression: A repertoire of recurrent themes. Immunol. Rev. 2004;200:197–214. doi: 10.1111/j.0105-2896.2004.00158.x. [DOI] [PubMed] [Google Scholar]
- Gorman JR, Alt FW. Regulation of immunoglobulin light chain isotype expression. Adv. Immunol. 1998;69:113–181. doi: 10.1016/s0065-2776(08)60607-0. [DOI] [PubMed] [Google Scholar]
- Grawunder U, Leu TM, Schatz DG, Werner A, Rolink AG, Melchers F, Winkler TH. Down-regulation of RAG1 and RAG2 gene expression in preB cells after functional immunoglobulin heavy chain rearrangement. Immunity. 1995;3(5):601–608. doi: 10.1016/1074-7613(95)90131-0. [DOI] [PubMed] [Google Scholar]
- Gu H, Kitamura D, Rajewsky K. B cell development regulated by gene rearrangement: Arrest of maturation by membrane-bound D mu protein and selection of DH element reading frames. Cell. 1991;65(1):47–54. doi: 10.1016/0092-8674(91)90406-o. [DOI] [PubMed] [Google Scholar]
- Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 1991;173(5):1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesslein DG, Schatz DG. Factors and forces controlling V(D)J recombination. Adv. Immunol. 2001;78:169–232. doi: 10.1016/s0065-2776(01)78004-2. [DOI] [PubMed] [Google Scholar]
- Hesslein DG, Pflugh DL, Chowdhury D, Bothwell AL, Sen R, Schatz DG. Pax5 is required for recombination of transcribed, acetylated, 5′ IgH V gene segments. Genes Dev. 2003;17(1):37–42. doi: 10.1101/gad.1031403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt SL, Farmer D, Marszalek K, Cadera E, Liang HE, Xu Y, Schlissel MS, Skok JA. Association between the Igk and Igh immunoglobulin loci mediated by the 3′ Igk enhancer induces ‘decontraction’ of the Igh locus in pre-B cells. Nat. Immunol. 2008;9(4):396–404. doi: 10.1038/ni1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CL, Lieber MR. CpG methylated minichromosomes become inaccessible for V(D)J recombination after undergoing replication. EMBO J. 1992;11(1):315–325. doi: 10.1002/j.1460-2075.1992.tb05054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inlay M, Alt FW, Baltimore D, Xu Y. Essential roles of the kappa light chain intronic enhancer and 3′ enhancer in kappa rearrangement and demethylation. Nat. Immunol. 2002;3(5):463–468. doi: 10.1038/ni790. [DOI] [PubMed] [Google Scholar]
- Inlay MA, Lin T, Gao HH, Xu Y. Critical roles of the immunoglobulin intronic enhancers in maintaining the sequential rearrangement of IgH and Igk loci. J. Exp. Med. 2006;203(7):1721–1732. doi: 10.1084/jem.20052310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33 Suppl:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA, Murre C. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133(2):265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Angelin-Duclos C, Park S, Calame KL. Changes in histone acetylation are associated with differences in accessibility of V(H) gene segments to V-DJ recombination during B-cell ontogeny and development. Mol. Cell. Biol. 2003;23(7):2438–2450. doi: 10.1128/MCB.23.7.2438-2450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Pflugh DL, Yu D, Hesslein DG, Lin KI, Bothwell AL, Thomas-Tikhonenko A, Schatz DG, Calame K. B cell-specific loss of histone 3 lysine 9 methylation in the V(H) locus depends on Pax5. Nat. Immunol. 2004;5(8):853–861. doi: 10.1038/ni1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CM, Wood AL, Bolland DJ, Corcoran AE. Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J. Immunol. 2006;176(7):4221–4234. doi: 10.4049/jimmunol.176.7.4221. [DOI] [PubMed] [Google Scholar]
- Julius MA, Street AJ, Fahrlander PD, Yang JQ, Eisenman RN, Marcu KB. Translocated c-myc genes produce chimeric transcripts containing antisense sequences of the immunoglobulin heavy chain locus in mouse plasmacytomas. Oncogene. 1988;2(5):469–476. [PubMed] [Google Scholar]
- Jung S, Rajewsky K, Radbruch A. Shutdown of class switch recombination by deletion of a switch region control element. Science. 1993;259(5097):984–987. doi: 10.1126/science.8438159. [DOI] [PubMed] [Google Scholar]
- Jung D, Bassing CH, Fugmann SD, Cheng HL, Schatz DG, Alt FW. Extrachromosomal recombination substrates recapitulate beyond 12/23 restricted VDJ recombination in nonlymphoid cells. Immunity. 2003;18(1):65–74. doi: 10.1016/s1074-7613(02)00507-1. [DOI] [PubMed] [Google Scholar]
- Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- Khamlichi AA, Pinaud E, Decourt C, Chauveau C, Cogné M. The 3′ IgH regulatory region: A complex structure in a search for a function. Adv. Immunol. 2000;75:317–345. doi: 10.1016/s0065-2776(00)75008-5. [DOI] [PubMed] [Google Scholar]
- Kitamura D, Rajewsky K. Targeted disruption of mu chain membrane exon causes loss of heavy-chain allelic exclusion. Nature. 1992;356(6365):154–156. doi: 10.1038/356154a0. [DOI] [PubMed] [Google Scholar]
- Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132(5):860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296(5565):158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Kottmann AH, Zevnik B, Welte M, Nielsen PJ, Köhler G. A second promoter and enhancer element within the immunoglobulin heavy chain locus. Eur. J. Immunol. 1994;24(4):817–821. doi: 10.1002/eji.1830240407. [DOI] [PubMed] [Google Scholar]
- Kouzarides T, Berger SL. Chromatin modifications and their mechanism of action. In: Allis CD, Jenuwein T, Reinberg D, Caparros ML, editors. Epigenetics. Cold Spring Harbor, NY: Cold Spring Harbor Academic Press; 2007. pp. 191–209. [Google Scholar]
- Krangel MS. Gene segment selection in V(D)J recombination: Accessibility and beyond. Nat. Immunol. 2003;4(7):624–630. doi: 10.1038/ni0703-624. [DOI] [PubMed] [Google Scholar]
- Kurosawa Y, von Boehmer H, Haas W, Sakano H, Trauneker A, Tonegawa S. Identification of D segments of immunoglobulin heavy-chain genes and their rearrangement in T lymphocytes. Nature. 1981;290(5807):565–570. doi: 10.1038/290565a0. [DOI] [PubMed] [Google Scholar]
- Kwon J, Morshead KB, Guyon JR, Kingston RE, Oettinger MA. Histone acetylation and hSWI/SNF remodeling act in concert to stimulate V(D)J cleavage of nucleosomal DNA. Mol. Cell. 2000;6(5):1037–1048. doi: 10.1016/s1097-2765(00)00102-7. [DOI] [PubMed] [Google Scholar]
- Lennon GG, Perry RP. C mu-containing transcripts initiate heterogeneously within the IgH enhancer region and contain a novel 5′-nontranslatable exon. Nature. 1985;318(6045):475–478. doi: 10.1038/318475a0. [DOI] [PubMed] [Google Scholar]
- Liang HE, Hsu LY, Cado D, Cowell LG, Kelsoe G, Schlissel MS. The “dispensable” portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity. 2002;17(5):639–651. doi: 10.1016/s1074-7613(02)00448-x. [DOI] [PubMed] [Google Scholar]
- Lieberson R, Giannini SL, Birshtein BK, Eckhardt LA. An enhancer at the 3′ end of the mouse immunoglobulin heavy chain locus. Nucleic Acids Res. 1991;19(4):933–937. doi: 10.1093/nar/19.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27(4):561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffert D, Ehlich A, Müller W, Rajewsky K. Surrogate light chain expression is required to establish immunoglobulin heavy chain allelic exclusion during early B cell development. Immunity. 1996;4(2):133–144. doi: 10.1016/s1074-7613(00)80678-0. [DOI] [PubMed] [Google Scholar]
- Lutzker S, Alt FW. Structure and expression of germ line immunoglobulin gamma 2b transcripts. Mol. Cell. Biol. 1988;8(4):1849–1852. doi: 10.1128/mcb.8.4.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maës J, O’Neill LP, Cavelier P, Turner BM, Rougeon F, Goodhardt M. Chromatin remodeling at the Ig loci prior to V(D)J recombination. J. Immunol. 2001;167(2):866–874. doi: 10.4049/jimmunol.167.2.866. [DOI] [PubMed] [Google Scholar]
- Maës J, Chappaz S, Cavelier P, O’Neill L, Turner B, Rougeon F, Goodhardt M. Activation of V(D)J recombination at the IgH chain JH locus occurs within a 6-kilobase chromatin domain and is associated with nucleosomal remodeling. J. Immunol. 2006;176(9):5409–5417. doi: 10.4049/jimmunol.176.9.5409. [DOI] [PubMed] [Google Scholar]
- Malynn BA, Yancopoulos GD, Barth JE, Bona CA, Alt FW. Biased expression of JH-proximal VH genes occurs in the newly generated repertoire of neonatal and adult mice. J. Exp. Med. 1990;171(3):843–859. doi: 10.1084/jem.171.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malynn BA, Shaw AC, Young F, Stewart V, Alt FW. Truncated immunoglobulin Dmu causes incomplete developmental progression of RAG-deficient pro-B cells. Mol. Immunol. 2002;38(7):547–556. doi: 10.1016/s0161-5890(01)00085-2. [DOI] [PubMed] [Google Scholar]
- Manis JP, van der Stoep N, Tian M, Ferrini R, Davidson L, Bottaro A, Alt FW. Class switching in B cells lacking 3′ immunoglobulin heavy chain enhancers. J. Exp. Med. 1998;188(8):1421–1431. doi: 10.1084/jem.188.8.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis JP, Tian M, Alt FW. Mechanism and control of class-switch recombination. Trends Immunol. 2002;23(1):31–39. doi: 10.1016/s1471-4906(01)02111-1. [DOI] [PubMed] [Google Scholar]
- Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 2003;13(2):136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- Mårtensson IL, Keenan RA, Licence S. The pre-B-cell receptor. Curr. Opin. Immunol. 2007;19(2):137–142. doi: 10.1016/j.coi.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Mason JO, Williams GT, Neuberger MS. Transcription cell type specificity is conferred by an immunoglobulin VH gene promoter that includes a functional consensus sequence. Cell. 1985;41(2):479–487. doi: 10.1016/s0092-8674(85)80021-0. [DOI] [PubMed] [Google Scholar]
- Matthews AG, Kuo AJ, Ramón-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450(7172):1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthias P, Baltimore D. The immunoglobulin heavy chain locus contains another B-cell-specific 3′ enhancer close to the alpha constant region. Mol. Cell. Biol. 1993;13(3):1547–1553. doi: 10.1128/mcb.13.3.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison AM, Jager U, Chott A, Schebesta M, Haas OA, Busslinger M. Deregulated PAX-5 transcription from a translocated IgH promoter in marginal zone lymphoma. Blood. 1998;92(10):3865–3878. [PubMed] [Google Scholar]
- Morshead KB, Ciccone DN, Taverna SD, Allis CD, Oettinger MA. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc. Natl. Acad. Sci. USA. 2003;100(20):11577–11582. doi: 10.1073/pnas.1932643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morvan CL, Pinaud E, Decourt C, Cuvillier A, Cogné M. The immunoglobulin heavy-chain locus hs3b and hs4 3′ enhancers are dispensable for VDJ assembly and somatic hypermutation. Blood. 2003;102(4):1421–1427. doi: 10.1182/blood-2002-12-3827. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Singh N, Kirillov A, Pelanda R, Cedar H, Chess A, Bergman Y. Kappa chain monoallelic demethylation and the establishment of allelic exclusion. Genes Dev. 1998;12(12):1801–1811. doi: 10.1101/gad.12.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Singh N, Tenzen T, Goldmit M, Gabay C, Elizur S, Qi P, Reubinoff BE, Chess A, Cedar H, Bergman Y. Asynchronous replication and allelic exclusion in the immune system. Nature. 2001;414(6860):221–225. doi: 10.1038/35102606. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Alt FW, Rajewsky K. The lingering enigma of the allelic exclusion mechanism. Cell. 2004;118(5):539–544. doi: 10.1016/j.cell.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Nitschke L, Kestler J, Tallone T, Pelkonen S, Pelkonen J. Deletion of the DQ52 element within the Ig heavy chain locus leads to a selective reduction in VDJ recombination and altered D gene usage. J. Immunol. 2001;166(4):2540–2552. doi: 10.4049/jimmunol.166.4.2540. [DOI] [PubMed] [Google Scholar]
- Norio P, Kosiyatrakul S, Yang Q, Guan Z, Brown NM, Thomas S, Riblet R, Schildkraut CL. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol. Cell. 2005;20(4):575–587. doi: 10.1016/j.molcel.2005.10.029. [DOI] [PubMed] [Google Scholar]
- Nussenzweig MC, Shaw AC, Sinn E, Campos-Torres J, Leder P. Allelic exclusion in transgenic mice carrying mutant human IgM genes. J. Exp. Med. 1988;167(6):1969–1974. doi: 10.1084/jem.167.6.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Urbánek P, Rolink A, Busslinger M. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 1997;11(4):476–491. doi: 10.1101/gad.11.4.476. [DOI] [PubMed] [Google Scholar]
- Oestreich KJ, Cobb RM, Pierce S, Chen J, Ferrier P, Oltz EM. Regulation of TCRbeta gene assembly by a promoter/enhancer holocomplex. Immunity. 2006;24(4):381–391. doi: 10.1016/j.immuni.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Ono M, Nose M. Persistent expression of an unproductive immunoglobulin heavy chain allele with DH-JH-gamma configuration in peripheral tissues. APMIS. 2007;115(12):1350–1356. doi: 10.1111/j.1600-0463.2007.apm_870.xml.x. [DOI] [PubMed] [Google Scholar]
- Osipovich O, Milley R, Meade A, Tachibana M, Shinkai Y, Krangel MS, Oltz EM. Targeted inhibition of V(D)J recombination by a histone methyltransferase. Nat. Immunol. 2004;5(3):309–316. doi: 10.1038/ni1042. [DOI] [PubMed] [Google Scholar]
- Osipovich O, Cobb RM, Oestreich KJ, Pierce S, Ferrier P, Oltz EM. Essential function for SWI-SNF chromatin-remodeling complexes in the promoter-directed assembly of Terb genes. Nat. Immunol. 2007;8(8):809–816. doi: 10.1038/ni1481. [DOI] [PubMed] [Google Scholar]
- Parslow TG, Blair DL, Murphy WJ, Granner DK. Structure of the 5 ends of immunoglobulin genes: A novel conserved sequence. Proc. Natl. Acad. Sci. USA. 1984;81(9):2650–2654. doi: 10.1073/pnas.81.9.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlitzky I, Angeles CV, Siegel AM, Stanton ML, Riblet R, Brodeur PH. Identification of a candidate regulatory element within the 5 flanking region of the mouse Igh locus defined by pro-B cell-specific hypersensitivity associated with binding of PU.1, Pax5, and E2A. J. Immunol. 2006;176(11):6839–6851. doi: 10.4049/jimmunol.176.11.6839. [DOI] [PubMed] [Google Scholar]
- Perlot T, Alt FW, Bassing CH, Suh H, Pinaud E. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc. Natl. Acad. Sci. USA. 2005;102(40):14362–14367. doi: 10.1073/pnas.0507090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlot T, Li G, Alt FW. Antisense transcripts from immunoglobulin heavy-chain locus V(D)J and switch regions. Proc. Natl. Acad. Sci. USA. 2008;105(10):3843–3848. doi: 10.1073/pnas.0712291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson S, Cook GP, Brüggemann M, Williams GT, Neuberger MS. A second B cell-specific enhancer 3′ of the immunoglobulin heavy-chain locus. Nature. 1990;344(6262):165–168. doi: 10.1038/344165a0. [DOI] [PubMed] [Google Scholar]
- Pinaud E, Khamlichi AA, Le Morvan C, Drouet M, Nalesso V, Le Bert M, Cogné M. Localization of the 3′ IgH locus elements that effect long-distance regulation of class switch recombination. Immunity. 2001;15(2):187–199. doi: 10.1016/s1074-7613(01)00181-9. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122(4):517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381(6585):751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008 doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Reth MG, Alt FW. Novel immunoglobulin heavy chains are produced from DJH gene segment rearrangements in lymphoid cells. Nature. 1984;312(5993):418–423. doi: 10.1038/312418a0. [DOI] [PubMed] [Google Scholar]
- Reth M, Petrac E, Wiese P, Lobel L, Alt FW. Activation of V kappa gene rearrangement in pre-B cells follows the expression of membrane-bound immunoglobulin heavy chains. EMBO J. 1987;6(11):3299–3305. doi: 10.1002/j.1460-2075.1987.tb02649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retter I, Chevillard C, Scharfe M, Conrad A, Hafner M, Im TH, Ludewig M, Nordsiek G, Severitt S, Thies S, Mauhar A, Blöcker H, et al. Sequence and characterization of the Ig heavy chain constant and partial variable region of the mouse strain 129S1. J. Immunol. 2007;179(4):2419–2427. doi: 10.4049/jimmunol.179.4.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Roldán E, Fuxa M, Chong W, Martinez D, Novatchkova M, Busslinger M, Skok JA. Locus ‘decontraction’ and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat. Immunol. 2005;6(1):31–41. doi: 10.1038/ni1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney S, Chaudhuri J, Alt FW. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol. Rev. 2004;200:115–131. doi: 10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- Sakai E, Bottaro A, Davidson L, Sleckman BP, Alt FW. Recombination and transcription of the endogenous Ig heavy chain locus is effected by the Ig heavy chain intronic enhancer core region in the absence of the matrix attachment regions. Proc. Natl. Acad. Sci. USA. 1999;96(4):1526–1531. doi: 10.1073/pnas.96.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H, Maki R, Kurosawa Y, Roeder W, Tonegawa S. Two types of somatic recombination are necessary for the generation of complete immunoglobulin heavy-chain genes. Nature. 1980;286(5774):676–683. doi: 10.1038/286676a0. [DOI] [PubMed] [Google Scholar]
- Sayegh C, Jhunjhunwala S, Riblet R, Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 2005;19(3):322–327. doi: 10.1101/gad.1254305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel MS, Corcoran LM, Baltimore D. Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. J. Exp. Med. 1991a;173(3):711–720. doi: 10.1084/jem.173.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel M, Voronova A, Baltimore D. Helix-loop-helix transcription factor E47 activates germ-line immunoglobulin heavy-chain gene transcription and rearrangement in a pre-T-cell line. Genes Dev. 1991b;5(8):1367–1376. doi: 10.1101/gad.5.8.1367. [DOI] [PubMed] [Google Scholar]
- Schweighoffer E, Vanes L, Mathiot A, Nakamura T, Tybulewicz VL. Unexpected requirement for ZAP-70 in pre-B cell development and allelic exclusion. Immunity. 2003;18(4):523–533. doi: 10.1016/s1074-7613(03)00082-7. [DOI] [PubMed] [Google Scholar]
- Seidl KJ, Manis JP, Bottaro A, Zhang J, Davidson L, Kisselgof A, Oettgen H, Alt FW. Position-dependent inhibition of class-switch recombination by PGK-neor cassettes inserted into the immunoglobulin heavy chain constant region locus. Proc. Natl. Acad. Sci. USA. 1999;96(6):3000–3005. doi: 10.1073/pnas.96.6.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R, Oltz E. Genetic and epigenetic regulation of IgH gene assembly. Curr. Opin. Immunol. 2006;18(3):237–242. doi: 10.1016/j.coi.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Serwe M, Sablitzky F. V(D)J recombination in B cells is impaired but not blocked by targeted deletion of the immunoglobulin heavy chain intron enhancer. EMBO J. 1993;12(6):2321–2327. doi: 10.1002/j.1460-2075.1993.tb05886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikes ML, Meade A, Tripathi R, Krangel MS, Oltz EM. Regulation of V(D)J recombination: A dominant role for promoter positioning in gene segment accessibility. Proc. Natl. Acad. Sci. USA. 2002;99(19):12309–12314. doi: 10.1073/pnas.182166699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skok JA, Brown KE, Azuara V, Caparros ML, Baxter J, Takacs K, Dillon N, Gray D, Perry RP, Merkenschlager M, Fisher AG. Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat. Immunol. 2001;2(9):848–854. doi: 10.1038/ni0901-848. [DOI] [PubMed] [Google Scholar]
- Stanhope-Baker P, Hudson KM, Shaffer AL, Constantinescu A, Schlissel MS. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 1996;85(6):887–897. doi: 10.1016/s0092-8674(00)81272-6. [DOI] [PubMed] [Google Scholar]
- Stavnezer J. Molecular processes that regulate class switching. Curr. Top. Microbiol. Immunol. 2000;245(2):127–168. doi: 10.1007/978-3-642-59641-4_6. [DOI] [PubMed] [Google Scholar]
- Stein R, Razin A, Cedar H. In vitro methylation of the hamster adenine phosphoribosyltransferase gene inhibits its expression in mouse L cells. Proc. Natl. Acad. Sci. USA. 1982;79(11):3418–3422. doi: 10.1073/pnas.79.11.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storb U, Arp B. Methylation patterns of immunoglobulin genes in lymphoid cells: correlation of expression and differentiation with undermethylation. Proc. Natl. Acad. Sci. USA. 1983;80(21):6642–6646. doi: 10.1073/pnas.80.21.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su LK, Kadesch T. The immunoglobulin heavy-chain enhancer functions as the promoter for I mu sterile transcription. Mol. Cell. Biol. 1990;10(6):2619–2624. doi: 10.1128/mcb.10.6.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 2003;4(2):124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- Sun T, Storb U. Insertion of phosphoglycerine kinase (PGK)-neo 5′ of Jlambdal dramatically enhances VJlambdal rearrangement. J. Exp. Med. 2001;193(6):699–712. doi: 10.1084/jem.193.6.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardimon L, Kressmann A, Cedar H, Maechler M, Doerfler W. Expression of a cloned adenovirus gene is inhibited by in vitro methylation. Proc. Natl. Acad. Sci. USA. 1982;79(4):1073–1077. doi: 10.1073/pnas.79.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297(5588):1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wang XF, Calame K. The endogenous immunoglobulin heavy chain enhancer can activate tandem VH promoters separated by a large distance. Cell. 1985;43(3 Pt 2):659–665. doi: 10.1016/0092-8674(85)90238-7. [DOI] [PubMed] [Google Scholar]
- Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2(9):1136–1143. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- Whitehurst CE, Chattopadhyay S, Chen J. Control of V(D)J recombinational accessibility of the D beta 1 gene segment at the TCR beta locus by a germline promoter. Immunity. 1999;10(3):313–322. doi: 10.1016/s1074-7613(00)80031-x. [DOI] [PubMed] [Google Scholar]
- Whitehurst CE, Schlissel MS, Chen J. Deletion of germline promoter PD beta 1 from the TCR beta locus causes hypermethylation that impairs D beta 1 recombination by multiple mechanisms. Immunity. 2000;13(5):703–714. doi: 10.1016/s1074-7613(00)00069-8. [DOI] [PubMed] [Google Scholar]
- Wuerffel R, Wang L, Grigera F, Manis J, Seising E, Perlot T, Alt FW, Cogne M, Pinaud E, Kenter AL. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;27(5):711–722. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Davidson L, Alt FW, Baltimore D. Deletion of the Ig kappa light chain intronic enhancer/matrix attachment region impairs but does not abolish V kappa J kappa rearrangement. Immunity. 1996;4(4):377–385. doi: 10.1016/s1074-7613(00)80251-4. [DOI] [PubMed] [Google Scholar]
- Yancopoulos GD, Alt FW. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40(2):271–281. doi: 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]
- Yancopoulos GD, Desiderio SV, Paskind M, Kearney JF, Baltimore D, Alt FW. Preferential utilization of the most JH-proximal VH gene segments in pre-B-cell lines. Nature. 1984;311(5988):727–733. doi: 10.1038/311727a0. [DOI] [PubMed] [Google Scholar]
- Yancopoulos GD, Blackwell TK, Suh H, Hood L, Alt FW. Introduced T cell receptor variable region gene segments recombine in pre-B cells: evidence that B and T cells use a common recombinase. Cell. 1986;44(2):251–259. doi: 10.1016/0092-8674(86)90759-2. [DOI] [PubMed] [Google Scholar]
- Yancopoulos GD, Malynn BA, Alt FW. Developmentally regulated and strain-specific expression of murine VH gene families. J. Exp. Med. 1988;168(1):417–435. doi: 10.1084/jem.168.1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Riblet R, Schildkraut CL. Sites that direct nuclear compartmentalization are near the 5 end of the mouse immunoglobulin heavy-chain locus. Mol. Cell. Biol. 2005;25(14):6021–6030. doi: 10.1128/MCB.25.14.6021-6030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J. The immunoglobulin IGHD gene locus in C57BL/6 mice. Immunogenetics. 2004;56(6):399–404. doi: 10.1007/s00251-004-0712-z. [DOI] [PubMed] [Google Scholar]
- Zhang J, Bottaro A, Li S, Stewart V, Alt FW. A selective defect in IgG2b switching as a result of targeted mutation of the I gamma 2b promoter and exon. EMBO J. 1993;12(9):3529–3537. doi: 10.1002/j.1460-2075.1993.tb06027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]