Abstract
Purpose
To identify mutations in 6 genes of 9 Chinese families with congenital cataract and microcornea.
Methods
Nine unrelated families with congenital cataract and microcornea were collected. Cycle sequencing was used to detect variants in the coding and adjacent regions of the crystallin alpha A (CRYAA), crystallin beta B1 (CRYBB1), crystallin beta A4 (CRYBA4), crystallin gamma C (CRYGC), crystallin gamma D (CRYGD), and gap junction protein alpha 8 (GJA8) genes.
Results
Upon complete analysis of the 6 genes, three mutations in 2 genes were detected in 3 families, respectively. These mutations were not present in 96 normal controls. Of the three mutations, two novel heterozygous mutations in GJA8, c.136G>A (p.Gly46Arg) and c.116C>G (p.Thr39Arg), were found in two families with congenital cataract and microcornea. The rest one, a heterozygous c.34C>T (p.Arg12Cys) mutation in CRYAA, was identified in three patients from a family with nuclear cataract, microcornea with axial elongation. No mutation in the 6 genes was detected in the remaining 6 families.
Conclusions
Mutations in GJA8 and CRYAA were identified in three families with cataract and microcornea. Elongation of axial length accompanied with myopia was a novel phenotype in the family with the c.34C>T mutation in CRYAA. Our results expand the spectrum of GJA8 mutations as well as their associated phenotypes.
Introduction
Congenital cataract is a leading cause of childhood blindness accounting for about 10%~38% of blindness in children [1], with a prevalence around 0.006%~0.06% in live births [2,3]. It may occur alone or associated with other ocular or systemic abnormalities. Microcornea, one of the most frequent abnormalities associated with congenital cataract, results from the secondary damage of the lens maldevelopment or from mutations in some growth or transcription factors [4]. To date, around 200 genes and loci have been associated with cataracts [4,5]. Of these genes, mutations in at least 9 genes were reported to be responsible for congenital cataract associated with microcornea, including genes encoding crystallins (crystallin alpha-A [CRYAA], OMIM 123580; crystallin beta-A4 [CRYBA4], OMIM 123631; crystallin beta-B1 [CRYBB1], OMIM 600929; crystallin beta-B2 [CRYBB2], OMIM 123620; crystallin gamma-C [CRYGC], OMIM 123680; and crystallin gamma-D [CRYGD], OMIM 123690) [6-14], gap junction protein alpha 8 (GJA8, OMIM 600897) [6,15], v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog (MAF, OMIM 177075) [16,17], and solute carrier family 16 member 12 (SLC16A12, OMIM 611910) [18]. Analyses of individual gene in patients with cataract and microcornea have been frequently reported [8-16,18-21] but comprehensive analysis of all these genes in the same set of families is rare [6].
In this study, we performed mutational screening of 6 genes (CRYAA, CRYBB1, CRYBA4, CRYGC, CRYGD, and GJA8) in 9 Chinese families with congenital cataract and microcornea. Three mutations in GJA8 and CRYAA were identified in 3 families.
Methods
Patients
Nine families with congenital cataract and microcornea were collected at the Pediatric and Genetic Eye Clinic of the Zhongshan Ophthalmic Center, Guangzhou, China. Written informed consent conforming to the tenets of the Declaration of Helsinki and following the Guidance of Sample Collection of Human Genetic Diseases (863-plan) by the Ministry of Public Health of China were obtained from the participating individuals or their guardians before the study. Congenital cataract represents cataract presented at birth or noticed in the first few months after birth. Microcornea represents a cornea with horizontal diameter of less than 10 mm. Genomic DNA was prepared from leukocytes of peripheral venous blood using the standard phenol/chloroform method [22].
Mutation detection
Genomic bioinformation of the 6 genes was obtained from the National Center for Biotechnology Information (NCBI): CRYAA (NCBI human genome build 37.2, NC_000021.8 for gDNA, NM_000394.2 for mRNA and NP_000385.1 for protein), CRYBB1 (NCBI human genome build 37.2, NC_000022.10 for gDNA, NM_001887.3 for mRNA and NP_001878.1 for protein), CRYBA4 (NCBI human genome build 37.2, NC_000022.10 for gDNA, NM_001886.2 for mRNA and NP_001877.1 for protein), CRYGC (NCBI human genome build 37.2, NC_000002.11 for gDNA, NM_020989.3 for mRNA and NP_066269.1 for protein), CRYGD (NCBI human genome build 37.2, NC_000002.11 for gDNA, NM_006891.3 for mRNA and NP_008822.2 for protein), and GJA8 (NCBI human genome build 37.2, NC_000001.10 for gDNA, NM_005267.4 for mRNA and NP_005258.2 for protein). Primers used to amplify the coding exons and adjacent intronic regions of the 6 genes were referred to a previous publication [23] with modification for a few primers (Table 1). Individual exon was amplified by polymerase chain reaction (PCR). The sequence of the amplicons was determined with the ABI BigDye Terminator cycle sequencing kit v3.1 on a genetic analyzer (ABI Applied Biosystems, Foster City, CA). Sequencing results from patients were aligned with consensus sequences to identify variations by using the SeqManII program of the Lasergene package (DNAStar Inc., Madison, WI). A variant detected in patient was further evaluated in controls by sequencing 96 normal individuals.
Table 1. Primers used to amplify the coding and adjacent regions of the 6 genes.
| Gene | Primer name | Primer sequence (5′→3′) | Product length (bp) | Annealing temperature (°C) |
|---|---|---|---|---|
|
CRYAA |
1F |
GCTGGGGGCGGGCACTTG |
552 |
68 |
| |
1R |
TGGGGACACAGGCTCTCG |
|
|
| |
2F |
GGTGACCGAAGCATCTCTGT |
295 |
68 |
| |
2R |
CGTGACCCCCTTGTCCTC |
|
|
| |
3F |
ACCCGGCCCCTGTGAGAG |
438 |
59 |
| |
3R |
AAAGGGAAGCAAAGGAAGACA |
|
|
|
CRYGC |
1–2F |
CCAAATAAAAGCAACACAGAGC |
671 |
63.8 |
| |
1–2R |
AAACCTCCCTCCCTGTAACC |
|
|
| |
3F |
CGCAGCAACCACAGTAATCT |
579 |
59.2 |
| |
3R |
CCCACCCCATTCACTTCTTA |
|
|
|
CRYGD |
1–2F |
GGGCCCCTTTTGTGCGGTTCT |
643 |
65 |
| |
1–2R |
GTGGGGAGCAAACTCTATTGA |
|
|
| |
3F |
TGCTCGGTAATGAGGAGTTT |
506 |
63 |
| |
3R |
AAATCAGTGCCAGGAACACA |
|
|
|
GJA8 |
1aF |
CAGATATTGACTCAGGGTTGC |
475 |
60 |
| |
1aR |
CCGCTGCTCTTCTTGACG |
|
|
| |
1bF |
ATTCGCCTCTGGGTGCTG |
571 |
58 |
| |
1bR |
CCTTGGCTTTCTGGATGG |
|
|
| |
1cF |
GCAGCAAAGGCACTAAGAA |
578 |
60 |
| |
1cR |
CACCTGAGCGTAGGAAGG |
|
|
| |
1dF |
ATCGTTTCCCACTATTTCC |
559 |
56 |
| |
1dR |
GATCATGTTGGCACCTTTT |
|
|
|
CRYBB1 |
1F |
GGTAATGGAGGGTGGAAC |
|
|
| |
1R |
GAGAATAGGGACAGAGGATAAG |
672 |
62 |
| |
2F |
GGAGGACAGGATCATTTCA |
|
|
| |
2R |
ATAATGTATGTGCCAGGAGTA |
387 |
62 |
| |
3F |
CCTTTGGACTTTCCTACTG |
|
|
| |
3R |
GCTTTTGTGCTTATCATTT |
483 |
58 |
| |
4F |
TAGACAGCAGTGGTCCCT |
|
|
| |
4R |
TTGATTACTCCTTCAACCC |
571 |
60 |
| |
5F |
TAGCCAGGACAGAAGTGAGA |
|
|
| |
5R |
ATTGAACATGAAGAAGGGTT |
362 |
60 |
|
CRYBA4 |
1F |
CCCTAGCCCAGTCACTCCT |
|
|
| |
1R |
TGAGCCTTGATTGCACCTCT |
289 |
60 |
| |
2F |
GGCACCTGTGCTGTCTAGTG |
|
|
| |
2R |
GCCTAGGGAGAGGGGACCTA |
396 |
62 |
| |
3F |
CTCCCCTAGTCGTGACAACC |
|
|
| |
3R |
TTTCAACTCTGGAACCTTTGA |
394 |
62 |
| |
4F |
TTATTGCCCTTCCAAAAGGTT |
|
|
| |
4R |
TGTTCTCCTCTGGAATGTGG |
397 |
62 |
| |
5F |
AAAAGAAAGGCTGGGATGGT |
|
|
| 5R | AAAACCGGTTCTTTGAAAAGATTA | 584 | 62 |
Variations analysis through online tools
The effects of alterations were evaluated by Polymorphism Phenotyping (PolyPhen-2) [24,25] and Sorting Intolerant From Tolerant (SIFT) [26] at the protein level.
Results
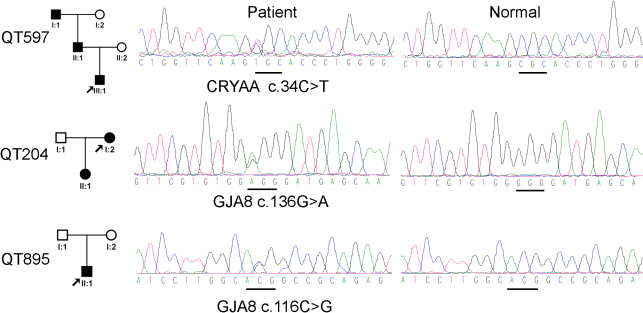
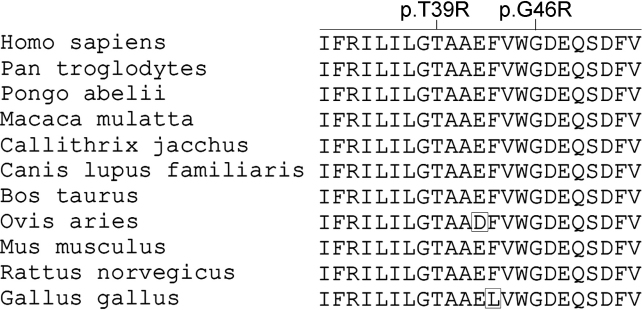
Upon complete analysis of the 6 genes, three heterozygous mutations in 2 genes were detected in 3 families (Figure 1), including c.34C>T (p.Arg12Cys) mutation in CRYAA, and c.116C>G (p.Thr39Arg) and c.136G>A (p.Gly46Arg) mutations in GJA8, where the last two mutations are novel. Both of the c.116C>G and c.136G>A mutations in GJA8 are predicted to be “probably damaging” by PolyPhen-2 and “intolerant” by SIFT. The p.Thr39Arg would change the Blosum62 score from 4 to −1 whereas the p.Gly46Arg would change the Blosum62 score from 6 to −2. The p.Thr39Arg and p.Gly46Arg variants involved residues that are conserved across different species (Figure 2).
Figure 1.
Mutations identified in 3 unrelated families with congenital cataract and microcornea. Pedigrees are shown in the left column. Sequence chromatography with mutation in each family is shown in the middle and the sequences from normal controls are aligned on the right column. Mutations in the 3 families were described under each sequence followed the nomenclature recommended by Human Genome Variation Society (HGVS).
Figure 2.
Protein sequence alignment of eleven GJA8 orthologs. The regions with the novel p.T39R and p.G46R mutations are highly conserved in the eleven species.
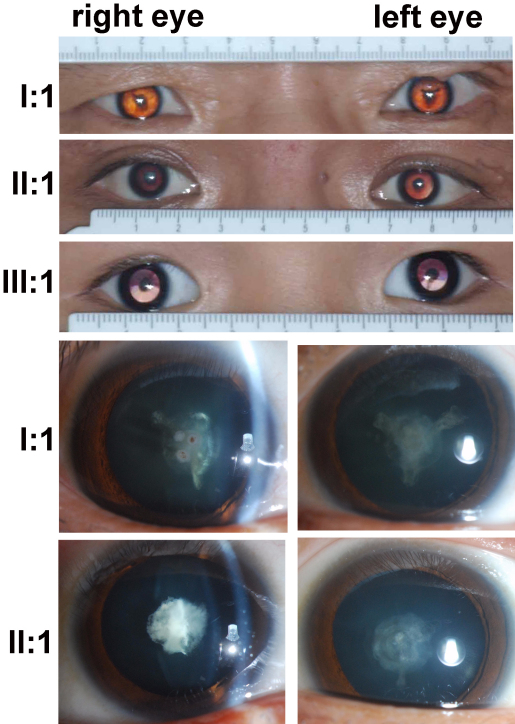
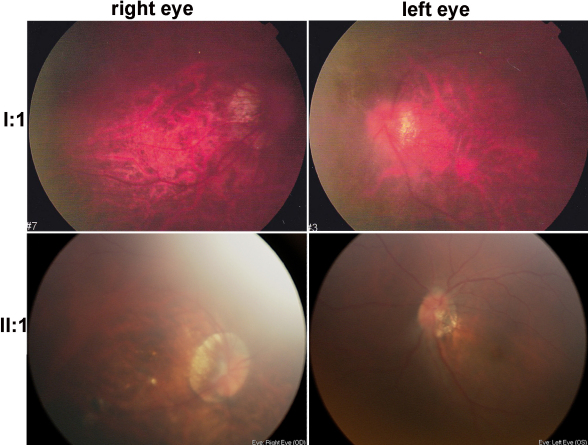
The heterozygous c.34C>T mutation in CRYAA was identified in all three patients in a three-generation family (QT597), where all patients had congenital nuclear cataract and microcornea (Figure 3, Table 2). Myopic fundus change in both eyes were observed in the affected father (II:1) and affected grandfather (I:1; Figure 4). Ocular ultrasound recorded axial lengths of 24.47 mm for the right eye and 24.61 mm for the left eye of II:1 and that of 27.82 mm for the right eye and 26.35 mm for the left eye of I:1. The proband had −3.00D for both eyes at the age of 4 years old.
Figure 3.
Photos shows the microcornea and nuclear cataracts of the three affected patients with a c.34C>T mutation in CRYAA in Family QT597. I:1, II:1, and III:1 on the left is the individual identification numbers that are the same as in the pedigree for QT597 in Figure 1. The top three photos demonstrated bilateral microcornea and bilateral nuclear cataracts in the three patients. The bottom two rows show the nuclear cataracts with suture opacity in I:1 and shell-like opacity in II:1.
Table 2. Listed below is the clinical information of the patients with mutations.
|
ID |
Mutation |
Gender |
Age (years) at |
Inheritance |
Visual acuity (right;left) |
Cataract types |
Cornea size (right;left; mm) |
Axial length (mm) (right;left) |
|
|---|---|---|---|---|---|---|---|---|---|
| exam | onset | ||||||||
| QT597I:1 |
c.34C>T; CRYAA |
male |
47 |
at birth |
AD |
0.04; 0.04 |
nuclear |
10; 10 |
27.82; 26.35 |
| QT597II:1 |
c.34C>T; CRYAA |
male |
24 |
at birth |
AD |
0.04; 0.08 |
nuclear |
10; 10 |
24.47; 24.16 |
| QT597III:1 |
c.34C>T; CRYAA |
male |
4 |
at birth |
AD |
N/A |
nuclear |
9.5; 9.5 |
N/A |
| QT204I:2 |
c.136G>A; GJA8 |
female |
34 |
at birth |
AD |
NLP; 0.03 |
total |
9; 9 |
N/A |
| QT204II:1 |
c.136G>A; GJA8 |
female |
5 |
at birth |
AD |
0.2; 0.25 |
total |
7; 7 |
N/A |
| QT895 | c.116C>G; GJA8 | male | 7 | at birth | Sporadic | 0.05; 0.1 | total | 6; 6 | N/A |
Figure 4.
Fundus photos demonstrate obvious crescent choroidal defects in the temporal region of the optic disc. Tigroid retinal changes are present in posterior fundus.
The c.136G>A mutation in GJA8 was identified in a two-generation family (QT204) with complete opacity of the lens and microcornea (Table 2). Horizontal cornea diameter was 9 mm for both eyes of the affected mother and 7 mm for both eyes of the affected daughter at the age of 5 years old.
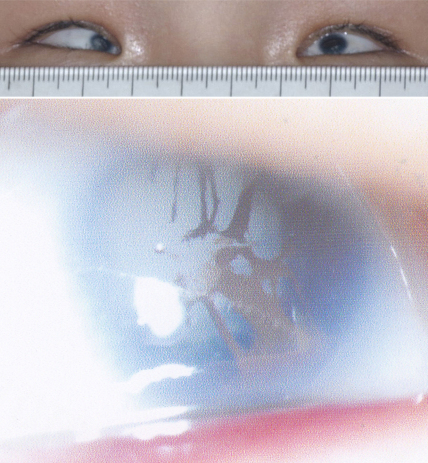
The c.116C>G mutation in GJA8 was identified in a sporadic patient (QT895) of 7 years old with microcornea, complete opacity of lenses, and iris hypoplasia (Figure 5, Table 2). Horizontal corneal diameter was about 6 mm for both eyes.
Figure 5.
Microcornea and congenital cataracts observed in patient QT895. The top photo shows the small corneas and the bottom photo shows complete opacity of the lens and iris hypoplasia.
Discussion
In this study, we screened 6 genes for mutations in 9 Chinese families with congenital cataract and microcornea. Three mutations were identified in 3 of the 9 (30%) families, including a c.34C>T (p.Arg12Cys) in CRYAA, and a c.136G>A (p.Gly46Arg) and a c.116C>G (p.Thr39Arg) in GJA8, respectively.
CRYAA is located in 21q22.3 and encodes the α-A-crystallin in lens epithelial cells and fiber cells. α-A-crystallin is a member of small heat shock proteins with the chaperone activity which contributes to keeping lens transparent [6,10,27]. Up to now, there were eight mutations of CRYAA found in sixteen families most of which involved substitutions from or to arginine [5]. And the corresponding phenotypes of the mutations were related with congenital cataract with or without microcornea, microphthalmia, or iris coloboma.
We found a known c.34C>T (p.Arg12Cys) mutation in CRYAA of three patients from a family with congenital nuclear cataract and microcornea. Previously, this mutation has been identified in four families with nuclear or lamellar cataracts, and some patients accompanied with microcornea or microphthalmia [6,10,28,29]. Elongation of axial length or myopia has not been observed in previous studies.
GJA8 is located in chromosome 1q21.1 and encodes the gap junction proteins, connexin50. GJA8 is one of the most common genes causing congenital cataract with or without other ocular abnormalities. Previous studies showed that GJA8-knockout mice developed nuclear cataract and microphthalmia, from which it is considered that GJA8 plays a role not only in keeping lens transparent but in ocular growth [30,31]. Up to now, about twenty mutations in GJA8 have been associated with congenital cataracts in at least 21 families. Of these mutations, five were identified in five families with microcornea and two families accompanied with microphthamia [32,33].
In this study, we found two novel missense mutations c.136G>A and c.116C>G in GJA8 in two families with congenital cataract and microcornea. The c.136G>A mutation led to a substitution from glycine to arginine at the amino acid position 46, and the c.116C>G mutation led to a substitution from threonine to arginine at the amino acid position 39. Both the 46 and 39 positions are located in the first transmembrane domain. In a previous study, Minogue et al. [32] identified a c.137G>T (p.Gly46Val) mutation in GJA8 of a proband with early-onset total cataract accompanied with small eyes and pupils. Therefore, the three mutations may result in phenotype by the similar mechanism.
In summary, a known c.34C>T mutation in CRYAA and two novel mutation in GJA8 were identified in 3 of 9 families after comprehensive analysis of 6 genes known to cause cataract and microcornea. Our results expand the mutation spectrum of GJA8 and phenotypic variations associated with CRYAA mutations. Patients without mutation in the 6 genes are potential candidate for future study of additional causative genes for cataracts and microcornea.
Acknowledgments
The authors thank the family members for their participation. This study was supported by the National Science Fund for Distinguished Young Scholars (30725044 to Q.Z.) and National 973 Plan of China (2010CB529904 to Q.Z.).
References
- 1.Wilson ME, Pandey SK, Thakur J. Paediatric cataract blindness in the developing world: surgical techniques and intraocular lenses in the new millennium. Br J Ophthalmol. 2003;87:14–9. doi: 10.1136/bjo.87.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reddy MA, Francis PJ, Berry V, Bhattacharya SS, Moore AT. Molecular genetic basis of inherited cataract and associated phenotypes. Surv Ophthalmol. 2004;49:300–15. doi: 10.1016/j.survophthal.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Bermejo E, Martinez-Frias ML. Congenital eye malformations: clinical-epidemiological analysis of 1,124,654 consecutive births in Spain. Am J Med Genet. 1998;75:497–504. [PubMed] [Google Scholar]
- 4.Hejtmancik JF. Congenital cataracts and their molecular genetics. Semin Cell Dev Biol. 2008;19:134–49. doi: 10.1016/j.semcdb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shiels A, Bennett TM, Hejtmancik JF. Cat-Map: putting cataract on the map. Mol Vis. 2010;16:2007–15. [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen L, Yao W, Eiberg H, Kjaer KW, Baggesen K, Hejtmancik JF, Rosenberg T. Genetic heterogeneity in microcornea-cataract: five novel mutations in CRYAA, CRYGD, and GJA8. Invest Ophthalmol Vis Sci. 2007;48:3937–44. doi: 10.1167/iovs.07-0013. [DOI] [PubMed] [Google Scholar]
- 7.Litt M, Kramer P, LaMorticella DM, Murphey W, Lovrien EW, Weleber RG. Autosomal dominant congenital cataract associated with a missense mutation in the human alpha crystallin gene CRYAA. Hum Mol Genet. 1998;7:471–4. doi: 10.1093/hmg/7.3.471. [DOI] [PubMed] [Google Scholar]
- 8.Richter L, Flodman P, Barria von-Bischhoffshausen F, Burch D, Brown S, Nguyen L, Turner J, Spence MA, Bateman JB. Clinical variability of autosomal dominant cataract, microcornea and corneal opacity and novel mutation in the alpha A crystallin gene (CRYAA). Am J Med Genet A. 2008;146:833–42. doi: 10.1002/ajmg.a.32236. [DOI] [PubMed] [Google Scholar]
- 9.Vanita V, Singh JR, Hejtmancik JF, Nuernberg P, Hennies HC, Singh D, Sperling K. A novel fan-shaped cataract-microcornea syndrome caused by a mutation of CRYAA in an Indian family. Mol Vis. 2006;12:518–22. [PubMed] [Google Scholar]
- 10.Zhang LY, Yam GH, Tam PO, Lai RY, Lam DS, Pang CP, Fan DS. An alphaA-crystallin gene mutation, Arg12Cys, causing inherited cataract-microcornea exhibits an altered heat-shock response. Mol Vis. 2009;15:1127–38. [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou G, Zhou N, Hu S, Zhao L, Zhang C, Qi Y. A missense mutation in CRYBA4 associated with congenital cataract and microcornea. Mol Vis. 2010;16:1019–24. [PMC free article] [PubMed] [Google Scholar]
- 12.Willoughby CE, Shafiq A, Ferrini W, Chan LL, Billingsley G, Priston M, Mok C, Chandna A, Kaye S, Heon E. CRYBB1 mutation associated with congenital cataract and microcornea. Mol Vis. 2005;11:587–93. [PubMed] [Google Scholar]
- 13.Zhang L, Fu S, Ou Y, Zhao T, Su Y, Liu P. A novel nonsense mutation in CRYGC is associated with autosomal dominant congenital nuclear cataracts and microcornea. Mol Vis. 2009;15:276–82. [PMC free article] [PubMed] [Google Scholar]
- 14.Wang KJ, Wang BB, Zhang F, Zhao Y, Ma X, Zhu SQ. Novel beta-crystallin gene mutations in Chinese families with nuclear cataracts. Arch Ophthalmol. 2011;129:337–43. doi: 10.1001/archophthalmol.2011.11. [DOI] [PubMed] [Google Scholar]
- 15.Devi RR, Vijayalakshmi P. Novel mutations in GJA8 associated with autosomal dominant congenital cataract and microcornea. Mol Vis. 2006;12:190–5. [PubMed] [Google Scholar]
- 16.Hansen L, Eiberg H, Rosenberg T. Novel MAF mutation in a family with congenital cataract-microcornea syndrome. Mol Vis. 2007;13:2019–22. [PubMed] [Google Scholar]
- 17.Vanita V, Singh D, Robinson PN, Sperling K, Singh JR. A novel mutation in the DNA-binding domain of MAF at 16q23.1 associated with autosomal dominant “cerulean cataract” in an Indian family. Am J Med Genet A. 2006;140:558–66. doi: 10.1002/ajmg.a.31126. [DOI] [PubMed] [Google Scholar]
- 18.Kloeckener-Gruissem B, Vandekerckhove K, Nurnberg G, Neidhardt J, Zeitz C, Nurnberg P, Schipper I, Berger W. Mutation of solute carrier SLC16A12 associates with a syndrome combining juvenile cataract with microcornea and renal glucosuria. Am J Hum Genet. 2008;82:772–9. doi: 10.1016/j.ajhg.2007.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu S, Wang B, Zhou Z, Zhou G, Wang J, Ma X, Qi Y. A novel mutation in GJA8 causing congenital cataract-microcornea syndrome in a Chinese pedigree. Mol Vis. 2010;16:1585–92. [PMC free article] [PubMed] [Google Scholar]
- 20.Jamieson RV, Munier F, Balmer A, Farrar N, Perveen R, Black GC. Pulverulent cataract with variably associated microcornea and iris coloboma in a MAF mutation family. Br J Ophthalmol. 2003;87:411–2. doi: 10.1136/bjo.87.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan AO, Aldahmesh MA, Meyer B. Recessive congenital total cataract with microcornea and heterozygote carrier signs caused by a novel missense CRYAA mutation (R54C). Am J Ophthalmol. 2007;144:949–52. doi: 10.1016/j.ajo.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Wang Q, Wang P, Li S, Xiao X, Jia X, Guo X, Kong QP, Yao YG, Zhang Q. Mitochondrial DNA haplogroup distribution in Chaoshanese with and without myopia. Mol Vis. 2010;16:303–9. [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen L, Mikkelsen A, Nurnberg P, Nurnberg G, Anjum I, Eiberg H, Rosenberg T. Comprehensive mutational screening in a cohort of Danish families with hereditary congenital cataract. Invest Ophthalmol Vis Sci. 2009;50:3291–303. doi: 10.1167/iovs.08-3149. [DOI] [PubMed] [Google Scholar]
- 24.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hicks S, Wheeler DA, Plon SE, Kimmel M. Prediction of missense mutation functionality depends on both the algorithm and sequence alignment employed. Hum Mutat. 2011;32:661–8. doi: 10.1002/humu.21490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andley UP, Mathur S, Griest TA, Petrash JM. Cloning, expression, and chaperone-like activity of human alphaA-crystallin. J Biol Chem. 1996;271:31973–80. doi: 10.1074/jbc.271.50.31973. [DOI] [PubMed] [Google Scholar]
- 28.Santana A, Waiswol M, Arcieri ES, Cabral de Vasconcellos JP, Barbosa de Melo M. Mutation analysis of CRYAA, CRYGC, and CRYGD associated with autosomal dominant congenital cataract in Brazilian families. Mol Vis. 2009;15:793–800. [PMC free article] [PubMed] [Google Scholar]
- 29.Devi RR, Yao W, Vijayalakshmi P, Sergeev YV, Sundaresan P, Hejtmancik JF. Crystallin gene mutations in Indian families with inherited pediatric cataract. Mol Vis. 2008;14:1157–70. [PMC free article] [PubMed] [Google Scholar]
- 30.White TW, Goodenough DA, Paul DL. Targeted ablation of connexin50 in mice results in microphthalmia and zonular pulverulent cataracts. J Cell Biol. 1998;143:815–25. doi: 10.1083/jcb.143.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rong P, Wang X, Niesman I, Wu Y, Benedetti LE, Dunia I, Levy E, Gong X. Disruption of Gja8 (alpha8 connexin) in mice leads to microphthalmia associated with retardation of lens growth and lens fiber maturation. Development. 2002;129:167–74. doi: 10.1242/dev.129.1.167. [DOI] [PubMed] [Google Scholar]
- 32.Minogue PJ, Tong JJ, Arora A, Russell-Eggitt I, Hunt DM, Moore AT, Ebihara L, Beyer EC, Berthoud VM. A mutant connexin50 with enhanced hemichannel function leads to cell death. Invest Ophthalmol Vis Sci. 2009;50:5837–45. doi: 10.1167/iovs.09-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ponnam SP, Ramesha K, Tejwani S, Ramamurthy B, Kannabiran C. Mutation of the gap junction protein alpha 8 (GJA8) gene causes autosomal recessive cataract. J Med Genet. 2007;44:e85. doi: 10.1136/jmg.2007.050138. [DOI] [PMC free article] [PubMed] [Google Scholar]