Abstract
Tail-anchored (TA) proteins are post-translationally targeted to and inserted into the endoplasmic reticulum (ER) membrane through their single C-terminal transmembrane domain. Membrane insertion of TA proteins in mammalian cells is mediated by the ATPase TRC40/Asna1 (Get3 in yeast) and a receptor in the ER membrane. We have identified tryptophan-rich basic protein (WRB), also known as congenital heart disease protein 5 (CHD5), as the ER membrane receptor for TRC40/Asna1. WRB shows sequence similarity to Get1, a subunit of the membrane receptor complex for yeast Get3. Using biochemical and cell imaging approaches, we demonstrate that WRB is an ER-resident membrane protein that interacts with TRC40/Asna1 and recruits it to the ER membrane. We identify the coiled-coil domain of WRB as the binding site for TRC40/Asna1 and show that a soluble form of the coiled-coil domain interferes with TRC40/Asna1-mediated membrane insertion of TA proteins. The identification of WRB as a component of the TRC (Get) pathway for membrane insertion of TA proteins raises new questions concerning the proposed roles of WRB (CHD5) in congenital heart disease, and heart and eye development.
Key words: WRB, Get1, TRC40/Asna1, Endoplasmic reticulum, Tail-anchored proteins
Introduction
Tail-anchored (TA) proteins span the lipid bilayer with their C-terminal transmembrane domain (TMD) and expose the N-terminal domain to the cytoplasm (Kutay et al., 1995; Kutay et al., 1993). They are post-translationally inserted into the membrane of the endoplasmic reticulum (ER) or the outer membrane of mitochondria and chloroplasts (Borgese et al., 2007; Borgese and Fasana, 2010). Post-translational targeting of TA proteins usually involves chaperones, targeting factors and receptors in the destination membrane. Recently, a distinct targeting pathway for the insertion of TA proteins into the membrane of the ER was discovered in mammalian cells that exploits as a central component the targeting factor TRC40 (Stefanovic and Hegde, 2007), also called Asna1 (Favaloro et al., 2008). TRC40 is a conserved cytosolic ATPase that recognizes the TMD of TA proteins and delivers them to the ER membrane for insertion (Favaloro et al., 2008; Stefanovic and Hegde, 2007). Among the TA proteins targeted by the TRC40 pathway are components of the Sec61 translocation channel (Sec61β), ribosome-associated membrane protein 4 (RAMP4) and SNAREs involved in vesicular trafficking
TRC40-mediated insertion of TA proteins requires a receptor at the ER membrane that is currently unknown (Favaloro et al., 2010; Schuldiner et al., 2008; Stefanovic and Hegde, 2007). In yeast, the structural and functional homolog of TRC40 is called Get3 (guided entry of TA proteins). Crystal structures of this protein reveal how a Get3 dimer might interact with TA proteins and deliver them in an ATP-dependent manner to the ER membrane (Bozkurt et al., 2009; Mateja et al., 2009; Suloway et al., 2009; Yamagata et al., 2010). Using genetics and biochemical approaches, additional components of the yeast Get pathway have been identified and partially characterized (Jonikas et al., 2009; Schuldiner et al., 2008). Get4 and Get5 have been proposed to function together with the tetratricopeptide-repeat-containing protein Sgt2 in the efficient capture of newly synthesized TA proteins at the ribosome and their delivery to Get3 (Chang et al., 2010; Leznicki et al., 2010; Simpson et al., 2010). Get1 and Get2 are transmembrane proteins that associate in a hetero-oligomeric complex and recruit Get3 to the ER membrane (Jonikas et al., 2009; Schuldiner et al., 2008). On the basis of sequence similarity, WRB (tryptophan-rich basic protein), also called CHD5 (congenital heart disease 5 protein), has been suggested as the possible mammalian homologue of Get1 (Borgese and Fasana, 2010; Rabu et al., 2009; Schuldiner et al., 2008), but no experimental evidence has yet been provided. Furthermore, a previous study has suggested a nuclear localization of WRB (Egeo et al., 1998). Here, we provide several lines of evidence that WRB is the receptor, or part of the receptor complex, for TRC40-mediated membrane insertion of TA proteins at the ER and that its coiled-coil domain functions as docking site for TRC40.
Results
WRB shows sequence similarity to yeast Get1
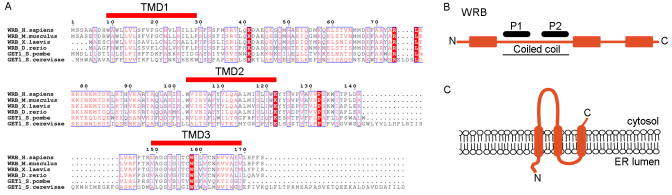
TRC40 (Get3)-mediated targeting and membrane insertion of TA proteins involves a receptor at the ER membrane (Favaloro et al., 2010; Schuldiner et al., 2008; Stefanovic and Hegde, 2007). Although such a receptor has been identified for yeast Get3 as the heterodimeric Get1–Get2 complex, orthologs in higher eukaryotes are still unknown. Based on sequence similarity (Fig. 1A), tryptophan-rich basic protein (WRB) has been suggested as a candidate for the mammalian ortholog of Get1 (Borgese and Fasana, 2010; Rabu et al., 2009; Schuldiner et al., 2008). WRB has three predicted TMDs and a cytosolically exposed coiled-coil domain (UniProtKB/Swiss-Prot O00258) (Fig. 1B,C). The coiled-coil domain shows the highest degree of conservation (23% identity, 49% similarity) between Homo sapiens WRB and Saccharomyces pombe Get1.
Fig. 1.
WRB shows sequence similarity to yeast Get1. (A) Multiple sequence alignment of Get1 homologs of eukaryotic organisms. Identical residues are indicated in white on a red background and similar residues in red. Alignment was performed by ESPript software (Gouet et al., 1999). (B) Linear outline of WRB. The predicted TMDs (red bars), the coiled-coil (cc) domain and the peptides (P1 and P2) used to raise anti-WRB antibodies are indicated. (C) Predicted membrane topology of WRB.
WRB is an ER-resident membrane protein
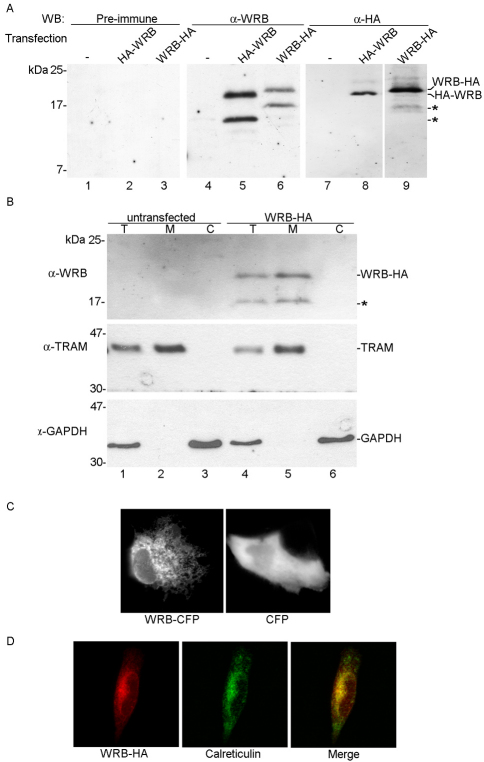
To further characterize WRB and find out whether it functions as a receptor for TRC40 at the ER membrane, we raised antibodies in rabbits against two peptides of the coiled-coil domain of WRB (WRBcc) (Fig. 1B). These anti-WRB antibodies specifically recognized on western blots of cell lysates 19 kDa HA-tagged forms of WRB, which were also recognized by an anti-HA antibody (Fig. 2A, lanes 5–6 and 8–9). Besides the 19 kDa protein, smaller molecular weight forms of HA–WRB and WRB–HA were recognized by the WRB antibodies (Fig. 2A, lanes 5–6). As the calculated molecular weight of WRB is 19 kDa, we conclude that the 19 kDa forms are the full-length WRB proteins and suggest that the faster migrating versions represent incomplete forms or degradation intermediates of WRB.
Fig. 2.
WRB is a 19 kDa membrane protein of the ER. (A) Identification of WRB–HA and HA–WRB. N-terminally or C-terminally HA-tagged forms of WRB (HA–WRB or WRB–HA) were expressed in HeLa cells. Proteins were separated by SDS-PAGE and analyzed by immunoblotting using the pre-immune serum (lanes 1–3), the anti-WRB serum (lanes 4–6) or anti-HA antibody (lanes 7–9). The asterisks indicate incomplete WRB. (B) Membrane association of WRB–HA. Untransfected HeLa cells (lanes 1–3) or HeLa cells transfected with HA–WRB (lanes 4–6) were analyzed by SDS-PAGE and immunoblotting using anti-WRB, anti-TRAM and anti-GAPDH antibodies. HeLa cells were permeabilized with digitonin, and cell constituents separated into a cytosolic and membrane fractions by centrifugation. Proteins of whole cell lysate (lanes 1 and 4), membrane (lanes 2 and 5) and cytosolic fractions (lanes 3 and 6) were analyzed. (C) Localization of WRB–CFP. CFP-tagged WRB (WRB–CFP) or CFP were expressed in RPE-1 cells and visualized by fluorescence microscopy. (D) ER localization of WRB–HA. WRB–HA was expressed in RPE-1 cells, immunostained with an anti-HA antibody and visualized with a fluorescent secondary antibody (red) by fluorescence microscopy. As an ER marker, calreticulin was visualized similarly using an anti-calreticulin antibody and a fluorescent secondary antibody (green).
To see whether WRB localizes to the membrane fraction of mammalian cells, we permeabilized HeLa cells transfected with WRB–HA using digitonin to release the cytosolic content, and solubilized membranes with Triton X-100. WRB–HA was detected in the total cell lysate and the membrane fraction (Fig. 2B, lanes 4–5). The anti-WRB antibodies were unable to detect endogenous WRB in the membranes of untransfected HeLa cells (Fig. 2B, lanes 1 to 3) and purified dog pancreas rough microsomes (data not shown). We tested the sensitivity of the anti-WRB antibodies in western blot analysis and applied decreasing amounts of recombinant WRBcc to an SDS gel. The antibodies could not detect WRBcc protein below 50 ng (supplementary material Fig. S1). This suggests that the amount of WRB within the membrane fraction of loaded HeLa cells or rough microsomes is less than 50 ng (5 pmol).
To determine the cellular localization of WRB, we expressed CFP- or HA-tagged forms of WRB (WRB–CFP or WRB–HA) in RPE-1 cells and located these proteins using fluorescence microscopy. Cells expressing WRB–CFP displayed a reticular network characteristic of the ER (Fig. 2C). The ER localization was confirmed by immunofluorescence analysis, in which WRB–HA was found to largely colocalize with the ER lumenal protein calreticulin (Fig. 2D).
WRB interacts with TRC40 in a cell lysate and at the ER membrane
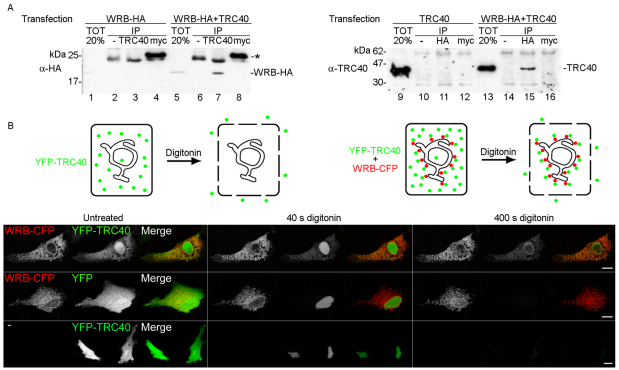
It has been shown previously that TRC40-mediated membrane insertion of TA proteins requires a receptor at the ER membrane (Favaloro et al., 2010; Schuldiner et al., 2008; Stefanovic and Hegde, 2007). To see whether WRB functions as this receptor and interacts with TRC40, we expressed TRC40 and WRB–HA either alone or together in HeLa cells, lysed the cells, and used an anti-TRC40, an anti-HA or an unrelated antibody (anti-myc) in co-immunoprecipitation experiments. The anti-TRC40 antibody immunoprecipitated TRC40 and in addition WRB–HA when both proteins were expressed (Fig. 3A, lane 7). Conversely, immunoprecipitation with the anti-HA antibody resulted in co-immunoprecipitation of TRC40 (Fig. 3A, lane 15). This suggests that WRB–HA and TRC40 associate either directly or indirectly with each other in cell lysates.
Fig. 3.
TRC40 associates with WRB–HA at the ER. (A) Co-immunoprecipitation of WRB–HA with TRC40 (left panel) and TRC40 with WRB–HA (right panel). HeLa cells were transfected with WRB–HA alone and with WRB–HA and TRC40 (left panel), or with TRC40 alone and with WRB–HA and TRC40 (right panel). 20% of the cell lysates were directly loaded onto the gel (lanes 1, 5 and 9, 13) and the rest were used for immunoprecipitation with the anti-TRC40 antibody (lanes 3 and 7), the anti-HA antibody (lanes 11, 15), an unrelated antibody (anti-myc) (lanes 4, 8 and 12, 16) or without an antibody (lanes 2, 6 and 10, 14). Lysates (TOT) and immunoprecipitates (IP) were analyzed by SDS-PAGE and immunoblotting using the anti-HA (left panel) or anti-TRC40 (right panel) antibodies. The asterisk indicates light chains of anti-myc antibody. (B) Colocalization of YFP–TRC40 with WRB–CFP at the ER. WRB–CFP was co-expressed with YFP–TRC40 (upper panel) or with YFP (middle panel), or YFP–TRC40 was expressed alone (lower panel). Cells were permeabilized with digitonin to release cytosolic content, and the CFP (red), YFP (green) signals or both signals were recorded over time using confocal microscopy. Untreated cells or cells treated with digitonin for 40 seconds and 400 seconds are shown. Scale bars: 10 μm. A graphical interpretation of the colocalization of YFP–TRC40 (green) with WRB–CFP (red) is shown on top of the figure.
As WRB is a protein of the ER membrane and TRC40 a soluble cytoplasmic protein, we asked whether TRC40 interacts with WRB at the ER membrane. To address this, we expressed YFP–TRC40 alone or in combination with WRB–CFP in RPE-1 cells, permeabilized the plasma membrane with digitonin, and followed the release of YFP–TRC40 over time using confocal microscopy. When WRB–CFP and YFP–TRC40 were co-expressed, most of YFP–TRC40 was detectable as a diffusely distributed pool in the cytoplasm and the nucleus. Upon digitonin application, this population disappeared by diffusion out of the cell, revealing a previously hidden population of YFP–TRC40, which was distinctively overlapping the WRB–CFP signal (Fig. 3B, upper panel; supplementary material Movie 1). As a control, YFP co-expressed with WRB–CFP completely diffused out of the cells after digitonin permeabilization (Fig. 3B, middle panel; supplementary material Movie 1). When TRC40 alone was expressed, it was initially diffusely distributed in the cells, was still seen in the nuclei 40 seconds after digitonin permeabilization and essentially disappeared from cells after 400 seconds (Fig. 3B, lower panel; supplementary material Movie 1). This demonstrates that WRB–CFP can efficiently recruit YFP–TRC40 to the ER membrane, and suggests that the endogenous level of WRB is very low and insufficient to recruit detectable amounts of TRC40 to the ER membrane.
The coiled-coil domain of WRB is the docking site for TRC40
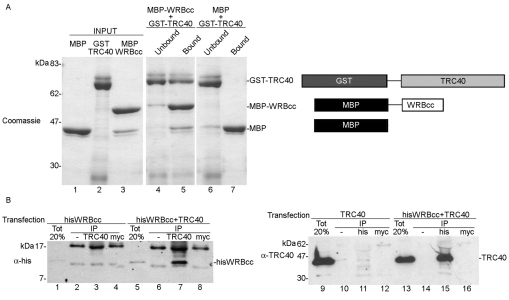
The coiled-coil domain of WRB (WRBcc) is predicted to be exposed on the cytoplasmic side of the ER [HMMTOP software (Tusnady and Simon, 2001)] and thus represents a good candidate for a docking site for TRC40 at the ER membrane. To test whether WRBcc alone can interact with TRC40, we expressed tagged forms of WRBcc [maltose-binding protein (MBP)–WRBcc] and TRC40 (GST–TRC40), and MBP in Escherichia coli (supplementary material Fig. S2), and allowed binding of MBP–WRBcc or MBP to an amylose resin. When GST–TRC40 was then passed over the resin, it was exclusively retained by the resin with bound MBP–WRBcc (Fig. 4A, lane 5). This suggests that the coiled-coil domain of WRB can directly associate with TRC40.
Fig. 4.
WRBcc interacts with TRC40. (A) Purified GST–TRC40 and MBP–WRBcc interact with each other. MBP (lane 1) and MBP–WRBcc (lane 3) were bound to amylose resin. GST–TRC40 (lane 2) was then incubated with the resins, and the unbound and bound proteins characterized by SDS-PAGE and Coomassie Blue staining. (B) HisWRBcc co-immunoprecipitates with TRC40 and TRC40 with HisWRBcc. HisWRBcc (lanes 1–4) and TRC40 (lanes 9–12) were expressed alone or together (lanes 5–8 and 13–16) in HeLa cells, and either TRC40 (left panel) or hisWRBcc (right panel) was immunoprecipitated by their respective antibodies (anti-TRC40 or anti-his). Co-immunoprecipitated proteins were then characterized by western blot analysis using the anti-his (left panel) or anti-TRC40 (right panel) antibody.
To see whether WRBcc can also associate with TRC40 in the cytosol of a eukaryotic cell, we co-expressed histidine-tagged WRBcc (HisWRBcc) with TRC40 in HeLa cells. We then asked whether HisWRBcc can be co-immunoprecipitated with TRC40 and vice versa. WRBcc efficiently co-immunoprecipitated with TRC40 (Fig. 4B, lane 7) and TRC40 with HisWRBcc (Fig. 4B, lane 15). This shows that WRBcc can also tightly interact with TRC40 in the cytosol. Taken together, these data suggest that the coiled-coil domain of WRB is the docking site for TRC40 at the ER membrane.
A recombinant coiled-coil domain of WRB interferes with TRC40-mediated insertion of TA proteins into the ER membrane in vitro
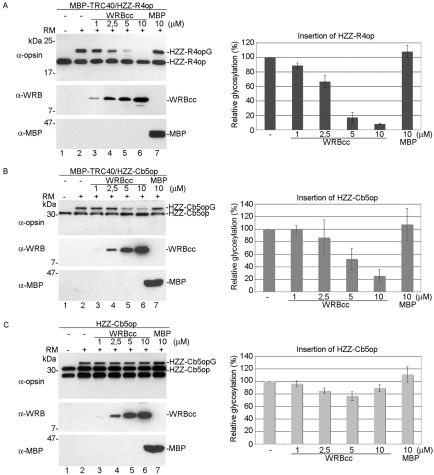
As the coiled-coil domain of WRB is able to interact with TRC40, it might compete with ER-resident WRB for binding to TRC40 and thereby interfere with membrane insertion of TA proteins. To test the effect of WRBcc on membrane insertion of TA proteins, we expressed WRBcc in E. coli and purified it (supplementary material Fig. S2). We then tested whether WRBcc affects TRC40-mediated TA protein insertion into the membrane of rough microsomes (RMs) derived from the pancreatic ER. A recombinant soluble complex formed by MBP–TRC40 and the cargo TA protein HZZ-RAMP4op was incubated in presence of RMs and ATP with increasing amounts of WRBcc. Membrane insertion was monitored after SDS-PAGE by a shift in molecular mass due to glycosylation (membrane insertion) of the C-terminal opsin tag of RAMP4op, as described previously (Favaloro et al., 2010) (Fig. 5A, lanes 1 and 2). WRBcc interfered with HZZ-RAMP4op insertion in a dose-dependent manner (Fig. 5A, lanes 3 to 6). The amount of glycosylated (membrane-inserted) HZZ-RAMP4op was reduced by up to 92% (Fig. 5A) when a 100-fold molar excess (10 μM) of WRBcc over MBP–TRC40–HZZ-RAMP4op was added. Adding similar amounts of MBP did not have any effect on membrane insertion of HZZ-RAMP4op (Fig. 5A, lane 7).
Fig. 5.
Recombinant WRBcc specifically prevents TRC40-dependent membrane insertion of TA proteins. (A) WRBcc was expressed in E. coli and tested for its ability to affect TRC40-mediated membrane insertion (glycosylation) of the TA protein RAMP4op. HZZ-RAMP4op–MBP–TRC40 complexes were produced in E. coli, purified and incubated in the absence (lane 1) or presence (lanes 2–7) of RMs, and increasing amounts of purified WRBcc (lanes 3–6) or MBP (lane 7). Proteins were analyzed by SDS-PAGE and immunoblotting using anti-opsin, anti-MBP or anti-WRB antibodies. Quantification of relative amounts of glycosylated (membrane-inserted) HZZ-RAMP4op is shown in the graph. Error bars ± s.d. (B) Effect of WRBcc on membrane insertion of cytochrome b5 (Cb5) co-purified with MBP–TRC40. HZZ-Cb5op–MBP–TRC40 complexes were incubated in the absence (lane 1) or presence (lanes 2–7) of RMs, and increasing amounts of purified WRBcc (lanes 3–6) or MBP (lane 7). Proteins were analyzed by SDS-PAGE and immunoblotting as described above. (C) Effect of WRBcc on membrane insertion of cytochrome b5 (Cb5). HZZ-Cb5op was incubated in the absence (lane 1) or presence (lanes 2–7) of RMs, and increasing amounts of purified WRBcc (lanes 3–6) or MBP (lane 7). Proteins were analyzed by SDS-PAGE and immunoblotting as described above.
We then asked whether WRBcc has an effect on membrane insertion of a TA protein that can insert unassisted in vitro, but can also associate with TRC40 (Favaloro et al., 2010). WRBcc showed dose-dependent inhibition of insertion of the TA protein cytochrome b5 (HZZ-Cb5op) when it was co-purified with TRC40 (Fig. 5B, lanes 3 to 6), whereas no significant effect on membrane insertion of HZZ-Cb5op alone could be observed (Fig. 5C, lanes 3 to 6). WRBcc also had no effect on membrane insertion of the type II membrane protein invariant chain synthesized in vitro in the presence of RMs (supplementary material Fig. S3).
These results suggest that WRBcc can efficiently compete with WRB at the ER membrane and specifically prevent TRC40-mediated membrane insertion of TA proteins.
Discussion
TRC40-mediated targeting and membrane insertion of TA proteins involves a receptor at the ER membrane (Favaloro et al., 2010; Schuldiner et al., 2008; Stefanovic and Hegde, 2007). Here, we provide several lines of evidence that WRB is a component of the postulated mammalian receptor complex for the TRC40-mediated delivery of TA proteins to the ER membrane. WRB shares similar sequence and topology with yeast Get1, a subunit of the receptor for Get3. Both Get1 and WRB have three predicted TMDs, and a cytosolically exposed coiled-coil domain between the first and the second TMDs. The coiled-coil domain shows the highest degree of conservation between H. sapiens WRB and S. pombe Get1 (23% identity, 49% similarity). Evolutionary conservation of Get1 function between lower and higher eukaryotes is also suggested by the previous finding that Get3 of C. thermophilum is able to mediate insertion of TA proteins into the membrane of canine RMs (Bozkurt et al., 2009).
We show that WRB is a membrane protein of the ER and that it interacts with TRC40 there. In a previous study, when the subcellular localization of the WRB protein was characterized by immunofluorescence analysis with polyclonal antibodies (anti-peptide 33–46 of WRB), a positive signal was predominantly observed over the cell nucleus (Egeo et al., 1998). We have not been able to detect WRB in untransfected RPE-1 or HeLa cells using our anti-WRB antibodies in immunofluorescence analysis or western blotting.
We demonstrate an interaction between WRB and TRC40 by co-immunoprecipitation of the two proteins and also by colocalization of WRB–CFP with YFP–TRC40 at the ER membrane of RPE-1 cells. WRB–CFP recruits YFP–TRC40 to the ER membrane only when WRB–CFP is overexpressed. As YFP–TRC40 was not detectably recruited to the ER of untransfected HeLa cells, this also suggests that the endogenous level of WRB is rather low.
We show that a purified coiled-coil domain of WRB can efficiently interact with TRC40. This strongly suggests that this domain of WRB functions as the ER membrane docking site for TRC40 and the TRC40–TA protein complex. Moreover, WRBcc can interfere with TRC40-mediated membrane insertion of RAMP4 and also cytochrome b5 (Cb5), but not with TRC40-independent membrane insertion of Cb5 or with signal recognition particle (SRP)-dependent membrane insertion of the type II membrane protein invariant chain. This suggests that WRB is part of the ER membrane receptor complex for TRC40-mediated membrane insertion of TA proteins. Whereas WRB appears to be a well-conserved homolog of Get1, we could not identify by sequence similarity a candidate Get2 homolog in higher eukaryotes, although it is conserved among several yeast species. Whether WRB alone is sufficient to drive TRC40-dependent membrane insertion of TA proteins or a Get2 homolog is required remains to be investigated.
WRB has been described as a tryptophan-rich basic protein because of its overall basic character and its tryptophan-rich C-terminal region (Egeo et al., 1998; Murata et al., 2009). An alternative name for WRB is congenital heart disease 5 protein (CDH5) based on the finding that the human gene encoding WRB has been mapped to Down syndrome (DS) region-2 of chromosome 21 (21q22.3), within the congenital heart disease region (Egeo et al., 1998; Korenberg et al., 1992; Vidal-Taboada et al., 1998). Human WRB (CDH5) was found to be widely expressed in adult and fetal tissues, with higher expression levels in the heart, brain, liver, skeletal muscle and pancreas (Egeo et al., 1998). Knocking down CHD5 (WRB) function in medaka fish (Oryzias latipes) caused severe cardiac disorder, including abnormal chamber differentiation, abnormal looping and eye abnormalities such as cyclops (Murata et al., 2009). It was suggested that medaka CHD5 (WRB) is directly involved in the molecular mechanism of heart chamber differentiation and looping, and might be associated with brain and eye development (Murata et al., 2009). In zebrafish, a WRB mutant (pwi) showed photoreceptor-specific retinal degeneration (Gross et al., 2005). Our data suggest that WRB (CDH5) is involved in TA protein targeting and insertion into the ER membrane. From the ER, TA proteins are distributed to all membranes of the endomembrane system and play crucial roles in nearly all aspects of cell biology (Borgese et al., 2003). Examples of TA proteins are components of the translocation site (Sec61β, RAMP4), SNAREs involved in vesicular trafficking, components of the membrane-bound degradation machinery, structural proteins of the Golgi apparatus and enzymes whose activities are spatially restricted in the cell (Borgese et al., 2003). Deletion or mutation of WRB, the receptor for TRC40-mediated ER membrane insertion of TA proteins, is expected to have very pleiotropic effects related to irregular biosynthesis of the various TA proteins. The identification of WRB as a component of the membrane receptor complex for the TRC40-mediated insertion of TA proteins opens up the possibility to more directly investigate the role of the TRC (Get) system in, for example, early developmental processes, as observed in the WRB mutants (Gross et al., 2005; Murata et al., 2009).
Materials and Methods
Plasmids
Constructs used in this study were obtained by standard methods and verified by sequencing. For pRK5rs-HA-WRB, WRB was amplified from pCMV-Sport6-WRB (Image Consortium, Berlin, Germany) using the primers 5′-GACCATGAGCTCAGCCGCGGCCGACCACTG-3′ and 5′-CTCAAGCGTAATCTGGAACATCGTATGGGTAGCTGAACGGATGAAG-3′. The fragment was cloned into pCR-II-TopoTA (Invitrogen) and then subcloned into the pRK5rs backbone.
For pRK5rs-WRB-HA, the sequence encoding WRB was amplified using the primers 5′-GACCATGAGCTCAGCCGCGGCCGACCACTG-3 and 5′-CTCAAGCGTAATCTGGAACATCGTATGGGTAGCTGAACGGATGAAG-3′. The fragment was cloned into pCR-II-TopoTA (Invitrogen) and then subcloned into the pRK5rs backbone.
For pECFP-WRB, the sequence encoding WRB was amplified from pCMV-Sport6-WRB using the primers 5′-TATCTACTCGAGACCATGAGCTCAGCCGCG-3′ and 5′-TAGATAGGTACCAAGCTGAACGGATGAAG-3′, and the fragment was cloned into the pmCerulean-N1 vector (Rizzo et al., 2004).
For pEYFP-TRC40, a BamHI-XbaI fragment originated from pCDNA3-Asna1 (a kind gift from Blanche Schwappach, University of Göttingen, Germany) was cloned into pmVenus-C1vector (Nagai et al., 2002).
For pRK5rs-hisWRBcyt, a BamHI-HindIII fragment from pQE80-WRBcc was cloned into the pRK5rs backbone.
For pQE80MBP-WRBcc, the sequence encoding the coiled-coil domain of WRB was amplified from pCMV-Sport6-WRB using the primers 5′-TATCTAGGATCCTCCTTCATGTCCAGGGTG-3′ and 5′-ATAGATGAATTCTTATTTTATCTTGGCTAA-3′. The fragment was cloned into pQE80 backbone (Qiagen) and then subcloned into pQE80-MBPtev (Favaloro et al., 2010).
For pGEX-TRC40, the sequence encoding TRC40 was amplified from pCDNA3-Asna1 using the primers 5′-GAATTCTCCACCATGGCGGCAGGGGTG-3′ and 5′-GTATATCCCAAGCTTCTACTGGGCACTGGG-3′. The fragment was cloned into pCR-II-TopoTA (Invitrogen) and then subcloned into the pGEX-5X-1 backbone (Amersham).
Vectors for bacterial expression of the MBP–TRC40–TA protein complexes and HZZ-Cb5op (pT5L/T7-MBP-Asna1/HZZ-R4op, pT5L/T7-MBP-Asna1/HZZ-Cb5op and pQET328-HZZ-Cb5op) were previously described (Favaloro et al., 2010).
pGEM4Z-MNCb-RAMP4 and pGem4Ii for in vitro transcription of RAMP4op and invariant chain mRNAs were previously described (Favaloro et al., 2008).
Antibodies
Mouse monoclonal anti-HA is from Covance, mouse monoclonal anti-polyhistidine was from Sigma, rabbit polyclonal anti-calreticulin from ABR, mouse monoclonal anti-MBP from NEB, rabbit monoclonal anti-GAPDH from Cell Signaling. Mouse monoclonal anti-opsin, mouse monoclonal anti-myc, rabbit polyclonal anti-TRAM and rabbit polyclonal anti-TRC40 were previously described (Favaloro et al., 2010). The anti-WRB antibodies were raised in rabbits against two ovalbumin-conjugated synthetic peptides (LQKDAEQESQMRAEIQDMKQELS and ARLERKINKMTDKLKTHVKART) by Peptide Speciality Laboratories, Heidelberg, Germany. Rabbit anti-mouse IgG and goat anti-rabbit IgG secondary antibodies were from Sigma. Fluorescent secondary antibodies Alexa-Fluor-546 goat anti-rabbit IgG and Alexa-Fluor-568 goat anti-mouse IgG were from Invitrogen.
Cell culture and transfection
HeLa cells (ATCC, CCL-2) were grown in DMEM containing 4.5 g/l glucose, 10% FBS and 2 mm L-glutamine. For western blot analysis, HeLa cells were seeded in six-well slots. 18 hours after calcium phosphate transfection (Chen and Okayama, 1987), cells were supplied with fresh medium.
RPE-1 cells were grown in DMEM/F12 supplemented with 10% FBS and 2 mm L-glutamine. Cells were transiently transfected in 12-well slots with Fugene HD (Roche), as suggested by the manufacturer.
Cell lysis and western blot
48 hours after transfection, HeLa cells were lysed in Triton X-100 lysis buffer (50 mM Hepes-NaOH pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 10% glycerol, 1 mM EGTA, 1 mM phenylmethylsulfonyl fluoride and 1% Triton X-100). Non-solubilized material was removed by centrifugation at 25,000 g for 10 minutes at 4°C.
For fractionation, cells were first permeabilized for 10 minutes with 0.02% digitonin (50 mM Hepes-NaOH pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 10% glycerol, 0.02% digitonin, 1 mM EGTA, 1 mM phenylmethylsulfonyl fluoride) to release cytosolic proteins. The permeabilized cells were pelleted at 15,000 g for 10 minutes at 4°C, and lysed in Triton X-100 lysis buffer to solubilize membrane proteins. Non-solubilized material was removed by centrifugation at 25,000 g for 10 minutes at 4°C. Protein lysates were separated by 16.5% Tris-Tricine SDS-PAGE. For western blots, blocking and antibody incubation were done with 7% w/v skimmed milk (Carl Roth) in 1× PBS-0.02% Tween.
For densitometric analysis of the amount of glycosylated protein, ImageJ software was used (NIH, http://www.rsbweb.nih.gov/ij/).
Immunofluorescence
RPE-1 cells were fixed with 2% paraformaldehyde in PBS supplemented with 125 mM sucrose for 20 minutes followed by 10 minutes incubation in 1% paraformaldehyde in PBS. Cells were then permeabilized with 0.3% Triton X-100 and 0.05% SDS, and incubated for 45 minutes at room temperature with primary antibodies (1:300 dilution). Secondary antibody incubation was performed for 45 minutes at room temperature (1:500 dilution) with Alexa-Fluor-546 goat anti-rabbit IgG or Alexa-Fluor-568 goat anti-mouse IgG. Cells were mounted with Mowiol (Sigma).
Microscopy
Wide-field fluorescence images (as shown in Fig. 2) were acquired with a CellR IX81 microscope system (Olympus) using an UPLSAPO 60× 1.35 numerical aperture (NA) oil objective lens and appropriate filter settings. Confocal microscopy (as shown in Fig. 3) was performed on a TCS SP2 laser-scanning system (Leica). All images were taken with a 63× 1.4 NA oil HCX Plan Apochromat objective (Leica) and identical pinhole settings of 2 airy units. For multi-channel imaging, each fluorophore was imaged sequentially in the frame-interlace mode to eliminate cross-talk between the channels. The methodology and a detailed protocol for the permeabilization of cells with digitonin for imaging purposes have been described (Lorenz et al., 2006). Z-stacks of images were recorded over time and displayed as maximum intensity projections.
Image processing was performed using ImageJ (NIH, http://www.rsbweb.nih.gov/ij/).
Immunoprecipitation
Lysates were diluted in a 1:4 ratio with IP buffer (20 mM Hepes-NaOH pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% Triton X-100). Antibodies were added at 1:150 final dilution and incubated for 2 hours. Protein A–sepharose beads (Amersham Biosciences) were added and incubated for one further hour. Beads were washed five times with IP buffer and immunoprecipitated proteins were eluted with 2× SDS sample buffer.
In vitro post-translational membrane insertion
In vitro reconstitution of post-translational membrane insertion of TA proteins from recombinant TRC40–TA protein complexes and RM was performed as previously described (Favaloro et al., 2010). Where indicated, recombinant WRBcc was added to the reaction mixture.
Protein expression and purification
Expression of MBP, MBP–WRBcc and GST–TRC40 was induced in BL21AI E. coli strain with 1 mM IPTG for 2 hours at 30°C. Cells were then harvested and resuspended in ice-cold LS buffer (50 mM HEPES, 150 mM KOAc, 10 mM MgOAc2, 10% glycerol, 1 mM PMSF, pH 7.0) containing 10 μg/ml DNase I. After lysis with Avestin Emulsiflex-C5, aggregates were removed by centrifugation at 100,000 g for 30 minutes at 4°C. For MBP and MBP–WRBcc purification, bacterial lysates was loaded onto an amylose resin (NEB) column. The resin was washed with 10 volumes LS buffer containing 5 mM ATP, 10 volumes HS buffer (50 mM HEPES, 500 mM KOAc, 10 mM MgOAc2, 10% glycerol, 1 mM PMSF, pH 7.0) and 10 volumes LS buffer. Proteins were eluted with LS buffer containing 20 mM maltose.
To remove the N-terminal tag from MBP–WRBcc, the resin was incubated with histidine-tagged TEV protease in a 1:30 w/w ratio for 2 hours at 4°C. TEV protease was removed by incubation with Ni-NTA resin (Qiagen). GST–TRC40 was purified using GST trap HP columns (GE Healthcare), as suggested by the manufacturer, and proteins were eluted in LS buffer containing 10 mM reduced glutathione.
Purification of MBP–TRC40–TA protein complexes and HZZ-Cb5op has previously been described (Favaloro et al., 2010).
Pull down
2 μM MBP or MBP–WRBcc were bound to amylose resin (NEB) together with 2 μM GST–TRC40, and incubated for 1 hour in the cold with gentle shaking. Bound and unbound proteins were analyzed by SDS-PAGE and Coomassie Blue staining.
In vitro transcription and translation
mRNAs were synthesized from the SP6 promoter using linearized plasmid DNA. Proteins were synthesized in rabbit reticulocyte lysate (RRL) according to the manufacturer's instructions (Promega) in the presence of 35S-labelled methionine and cysteine (7.5 μCi per 10 μl reaction) and in the absence or presence of RMs. Translation and membrane insertion of the TA protein RAMP4op and the invariant chain of MHC class II molecules were done as described previously (Favaloro et al., 2008), with the exception that increasing amounts of purified recombinant WRBcc were added as indicated in supplementary material Fig. S3.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB 638 A2). We thank Klaus Meese for excellent technical assistance, Irmgard Sinning, Günes Bozkurt and Matthias Seedorf for critically reading the manuscript and helpful suggestions, and members of our group for stimulating discussions. The data included in this manuscript have never been either submitted for publication or published elsewhere. The authors declare no conflict of interest.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/124/8/1301/DC1
References
- Borgese N., Fasana E. (2010). Targeting pathways of C-tail-anchored proteins. Biochim. Biophys. Acta 1808, 937-946 [DOI] [PubMed] [Google Scholar]
- Borgese N., Colombo S., Pedrazzini E. (2003). The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J. Cell Biol. 161, 1013-1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N., Brambillasca S., Colombo S. (2007). How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol. 19, 368-375 [DOI] [PubMed] [Google Scholar]
- Bozkurt G., Stjepanovic G., Vilardi F., Amlacher S., Wild K., Bange G., Favaloro V., Rippe K., Hurt E., Dobberstein B., et al. (2009). Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc. Natl. Acad. Sci. USA 106, 21131-21136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. W., Chuang Y. C., Ho Y. C., Cheng M. Y., Sun Y. J., Hsiao C. D., Wang C. (2010). Crystal structure of Get4-Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J. Biol. Chem. 285, 9962-9970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Okayama H. (1987). High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7, 2745-2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeo A., Mazzocco M., Sotgia F., Arrigo P., Oliva R., Bergonon S., Nizetic D., Rasore-Quartino A., Scartezzini P. (1998). Identification and characterization of a new human cDNA from chromosome 21q22.3 encoding a basic nuclear protein. Hum. Genet. 102, 289-293 [DOI] [PubMed] [Google Scholar]
- Favaloro V., Spasic M., Schwappach B., Dobberstein B. (2008). Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J. Cell Sci. 121, 1832-1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro V., Vilardi F., Schlecht R., Mayer M. P., Dobberstein B. (2010). Asna1/TRC40-mediated membrane insertion of tail-anchored proteins. J. Cell Sci. 123, 1522-1530 [DOI] [PubMed] [Google Scholar]
- Gouet P., Courcelle E., Stuart D. I., Metoz F. (1999). ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305-308 [DOI] [PubMed] [Google Scholar]
- Gross J. M., Perkins B. D., Amsterdam A., Egana A., Darland T., Matsui J. I., Sciascia S., Hopkins N., Dowling J. E. (2005). Identification of zebrafish insertional mutants with defects in visual system development and function. Genetics 170, 245-261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas M. C., Collins S. R., Denic V., Oh E., Quan E. M., Schmid V., Weibezahn J., Schwappach B., Walter P., Weissman J. S., et al. (2009). Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323, 1693-1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenberg J. R., Bradley C., Disteche C. M. (1992). Down syndrome: molecular mapping of the congenital heart disease and duodenal stenosis. Am. J. Hum. Genet. 50, 294-302 [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Hartmann E., Rapoport T. A. (1993). A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 3, 72-75 [DOI] [PubMed] [Google Scholar]
- Kutay U., Ahnert-Hilger G., Hartmann E., Wiedenmann B., Rapoport T. A. (1995). Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 14, 217-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leznicki P., Clancy A., Schwappach B., High S. (2010). Bat3 promotes the membrane integration of tail-anchored proteins. J. Cell Sci. 123, 2170-2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz H., Hailey D. W., Lippincott-Schwartz J. (2006). Fluorescence protease protection of GFP chimeras to reveal protein topology and subcellular localization. Nat. Methods 3, 205-210 [DOI] [PubMed] [Google Scholar]
- Mateja A., Szlachcic A., Downing M. E., Dobosz M., Mariappan M., Hegde R. S., Keenan R. J. (2009). The structural basis of tail-anchored membrane protein recognition by Get3. Nature 461, 361-366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K., Degmetich S., Kinoshita M., Shimada E. (2009). Expression of the congenital heart disease 5/tryptophan rich basic protein homologue gene during heart development in medaka fish, Oryzias latipes. Dev. Growth Differ. 51, 95-107 [DOI] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A. (2002). A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87-90 [DOI] [PubMed] [Google Scholar]
- Rabu C., Schmid V., Schwappach B., High S. (2009). Biogenesis of tail-anchored proteins: the beginning for the end? J. Cell Sci. 122, 3605-3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo M. A., Springer G. H., Granada B., Piston D. W. (2004). An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 22, 445-449 [DOI] [PubMed] [Google Scholar]
- Schuldiner M., Metz J., Schmid V., Denic V., Rakwalska M., Schmitt H. D., Schwappach B., Weissman J. S. (2008). The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134, 634-645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson P. J., Schwappach B., Dohlman H. G., Isaacson R. L. (2010). Structures of Get3, Get4, and Get5 provide new models for TA membrane protein targeting. Structure 18, 897-902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S., Hegde R. S. (2007). Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 128, 1147-1159 [DOI] [PubMed] [Google Scholar]
- Suloway C. J., Chartron J. W., Zaslaver M., Clemons W. M., Jr (2009). Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc. Natl. Acad. Sci. USA 106, 14849-14854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady G. E., Simon I. (2001). The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849-850 [DOI] [PubMed] [Google Scholar]
- Vidal-Taboada J. M., Bergonon S., Sanchez M., Lopez-Acedo C., Groet J., Nizetic D., Egeo A., Scartezzini P., Katsanis N., Fisher E. M., et al. (1998). High resolution physical mapping and identification of transcribed sequences in the Down syndrome region-2. Biochem. Biophys. Res. Commun. 243, 572-578 [DOI] [PubMed] [Google Scholar]
- Yamagata A., Mimura H., Sato Y., Yamashita M., Yoshikawa A., Fukai S. (2010). Structural insight into the membrane insertion of tail-anchored proteins by Get3. Genes Cells 15, 29-41 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.