Abstract
We have undertaken an extensive screen to identify Saccharomyces cerevisiae genes whose products are involved in cell cycle progression. We report the identification of 113 genes, including 19 hypothetical ORFs, which confer arrest or delay in specific compartments of the cell cycle when overexpressed. The collection of genes identified by this screen overlaps with those identified in loss-of-function cdc screens but also includes genes whose products have not previously been implicated in cell cycle control. Through analysis of strains lacking these hypothetical ORFs, we have identified a variety of new CDC and checkpoint genes.
Cell cycle studies performed with Saccharomyces cerevisiae have served as a guideline for understanding eukaryotic cell cycle progression. In particular, the now classic screens by Hartwell and colleagues (1–6) to identify temperature-sensitive mutants with specific arrest points throughout the cell division cycle (cdc) have shed light on numerous aspects of cell cycle, including progression through START (e.g., CDC28), regulation of DNA replication (e.g., CDC6), and control of mitosis (e.g., CDC14).
Although the cumulative cdc screens performed to date have been extensive, Hartwell and colleagues proposed that there may be additional CDC genes that are difficult or impossible to uncover by using a recessive loss-of-function approach (6). Reasons for this difficulty were speculated to include: (i) a lack of susceptibility of the gene product to generation of a temperature-sensitive allele; (ii) the presence in the genome of a closely related gene with similar or redundant function (e.g., cyclins); (iii) the presence of CDC genes that are helpful but not absolutely required for cell cycle progression; and (iv) the assumption of a morphologically homogeneous terminal phenotype for all cdc mutants (6). In addition, it has since become apparent that protein degradation during the cell cycle is a highly regulated process that contributes to many stages of cell cycle progression. A change in turnover of some encoded gene products can lead to cell cycle arrest (7–9). Such genes would likely escape detection in the cdc screens.
In an effort to complement the original cdc screens and in the interest of uncovering new CDC genes from within the categories described above, we undertook an extensive overexpression screen to identify genes that cause an alteration in cell cycle progression. By using this approach, we expected to uncover genes whose overexpression leads to hypermorphic, antimorphic, or neomorphic effects. Many of these genes should be involved at some level in the yeast cell cycle. To further delineate these putative cell cycle genes, we analyzed all of the initial positives by a variety of other assays to establish their roles in cell cycle control.
We used a conditional overexpression approach with a S. cerevisiae cDNA library under control of the GAL1 promoter (10). A second smaller screen was performed by using a sheared genomic library under GAL1 control (11). Several previous studies have been reported that used GAL1 or GAL10 promoter-driven yeast libraries to identify genes whose overexpression is lethal (10–13). A moderate number of genes have been identified in these screens, and a few of them have been demonstrated to cause a cell cycle specific arrest (12–13). However, a large-scale analysis of cell cycle effects has not previously been undertaken. We hypothesized that in some, if not many, instances, effects on the cell cycle might be apparent in the absence of complete lethality. For this reason, we devised a protocol that would uncover not only those genes whose overproduction is lethal, but also those where overproduction causes impaired growth. We also reasoned that moderate overproduction of proteins might be more physiologically relevant than dramatic overproduction, therefore we used GAL promoter-driven libraries expressed from ARS-CEN vectors to control levels of gene expression.
Materials and Methods
Screening of Libraries.
Yeast strain K699 (W303 background) was transformed as previously described (14) with the cDNA library or the sheared genomic library, and transformants were allowed to grow for 2 days at 30°C on glucose-ura plates. These plates were then replica-plated to both raffinose-ura and galactose-ura plates. After 1 day at 30°C, the galactose plates were replica-plated to a second set of galactose plates and grown for 1 day. A second round of replica-platings on galactose ensured adequate induction by galactose. Comparison of the raffinose and galactose plates to the original glucose plates allowed the identification of clones that were completely unable to grow on galactose, as well as those that showed reduced growth. These clones were then retested for their glucose, raffinose, and galactose phenotypes. Those that recapitulated the slow-growth/loss-of-growth phenotype on galactose were subjected to plasmid loss by growing on 0.15% 5-fluoroorotic acid to demonstrate that the phenotype was plasmid-dependent. Plasmids were recovered from these clones and retransformed into K699, and the phenotypes were checked once again. In several instances, clones were also retransformed into strain BY4741 (S288C background) to ensure that the phenotype could be recapitulated in both strain backgrounds. In all cases, the phenotypes were identical in both strains.
For the cDNA library, approximately 150,000 colonies were screened, and 562 plasmid-dependent clones were identified that grew poorly on galactose. These 562 clones were partially sequenced from both ends and found to represent 179 different genes or hypothetical ORFs, all of which were full-length. For the sheared genomic library, approximately 30,000 colonies were screened, identifying 7 gene fragments that caused compromised growth on galactose. All genomic clones are N-terminally truncated.
Induction of Expression and Flow Cytometry Analysis of Clones.
Clones were patched onto raffinose plates and grown at 30°C for 1–2 days. Cells were then removed from the plate and placed in liquid medium containing 2% galactose for 6–8 h at 30°C. After induction, cells were fixed in 70% EtOH overnight at 4°C. Cells were then digested with 1 mg/ml RNase A (37°C for 4 h) and 5 mg/ml pepsin (37°C for 30 min) and stained with 50 μg/ml propidium iodide (4°C overnight). DNA content was analyzed by flow cytometry (FACSCalibur, Becton Dickinson). Results represent data from two to seven independent experiments.
4′,6-Diamidino-2-Phenylindole (DAPI) Staining.
Clones expressing hypothetical ORFs were induced as described above. Cells were fixed for 20 min in 70% EtOH and stained with 0.1 μg/ml DAPI for 15 min at room temperature. Fixed and stained cells were attached to polylysine-coated slides and examined by fluorescence microscopy. A minimum of 200 cell nuclei were counted for each sample to determine whether the increase in 2C DNA content was because of a specific increase in G2 (pre-M), early, or late M phase. Results shown are the mean of two or more independent experiments.
Construction of Deletion Strains.
Strains deleted for the hypothetical ORFs were purchased (Research Genetics, Huntsville, AL) or were generated in strain BY4741 by PCR-mediated disruption as previously described (15). For construction of double deletion strains, diploid strains possessing a homozygous deletion of one of each pair of ORFs were purchased (Research Genetics), and the second ORF in each pair was targeted for disruption by PCR. The resulting strains, homozygous null at one locus and heterozygous at the second locus, were sporulated, and tetrads were dissected to allow analysis of haploid double deletion strains.
Phenotype Assays.
For each phenotype test, wild-type control strain, BY4741, and each deletion strain were grown in yeast extract–peptone–dextrose (YPD) medium to the same density. Five 10-fold serial dilutions were made, and 15 μl of each was spotted onto YPD plates containing the appropriate drug or treated as indicated. Unless otherwise indicated, all plates were grown at 30°C. Plates were photographed after 2–3 days. Conditions for each test were as follows: temperature sensitivity, 37°C; cold sensitivity, 18°C; benomyl sensitivity and resistance, 10–20 μg/ml; methyl methanesulfonate (MMS) sensitivity, 0.005%, UV sensitivity, 100 J/m2.
Results
Numerous Genes Cause Impaired Growth When Overexpressed.
Of ≈180,000 total clones screened, we identified 569 clones, representing 185 genes, that caused impaired growth when overexpressed. Thirty-six of these genes are hypothetical ORFs [Table 1; see also Table 5, which is published as supplemental data on the PNAS web site (www.pnas.org)]. Of the 185 genes that caused growth arrest when overexpressed, 113 (19 of them hypothetical ORFs) were found to alter cell cycle profiles when induced for 6–8 h in galactose-containing medium and analyzed by flow cytometry (Table 1 and supplemental data). The remaining genes and ORFs showed either no change in cell cycle profile after induction when compared with control vector or had effects that could not be reproduced reliably (supplemental data). Because it was anticipated that overexpression might confer less dramatic effects than traditional loss-of-function screens, we included genes whose elevated expression caused even very slight, albeit reproducible, effects on the cell cycle.
Table 1.
Effect on cell cycle distribution as determined by flow cytometry
| Shift | Shift
toward 1C DNA content
|
Shift toward 2C DNA content
|
||||
|---|---|---|---|---|---|---|
| ORF | Description | Notes | ORF | Description | Notes | |
| >20% | CDC14 | Phosphatase with function in late cell cycle | NE*† | TUB2 | β-Tubulin | E†§ |
| GCN4 | Transcription factor regulating amino acid biosynthesis | NE‡ | ||||
| MGA1 | Null diploids show increased random budding | NE‡ | ||||
| PPZ1 | Phosphatase that inhibits G1/S transition | NE‡ | ||||
| STE4 | β-Subunit of G protein coupled to mating factor receptor | NE†‡§ | ||||
| TPK1 | Catalytic subunit of PKA; inhibits filamentous growth | NE*†§ | ||||
| TPK2 | Catalytic subunit of PKA; activates filamentous growth | NE†‡ | ||||
| YFL010C | Unknown | NE†‖ | ||||
| >10% | ALO1 | d-arabino-1,4-lactone oxidase | NE* | ACS2 | Acetyl coA synthetase | NE |
| CCC1 | Potential role in calcium regulation and meiosis | NE‡ | ACT1 | Actin | E*§ | |
| DAT1 | DNA-binding protein | NE* | ARF1 | Transport to or within Golgi; “CDC One Suppressor” | NE*‡‡ | |
| FUN14 | “Function Unknown Now” | NE‡ | BFA1 | Component of Bub2-dependent checkpoint pathway | NE*††† | |
| MGE1 | Homologue of E. coli GrpE | E‡ | BNI4 | Involved in cell wall maintenance and cytokinesis | NE*‡‡†† | |
| MKS1 | Proper levels required for formation of pseudohyphae | NE* | CDH1 | Involved in Clb2 proteolysis | NE†‡‡‡ | |
| MRT4 | Involved in mRNA turnover | NE‡ | CLB3 | B-type cyclin | NE*§§ | |
| MSS11 | Required for nitrogen starvation-induced diploid filamentous growth | NE* | ERV14 | Required for axial budding pattern in haploids | NE* | |
| NAM8 | Involved in meiotic recombination; induced in S, G2 | NE* | KES1 | Implicated in ergosterol biosynthesis | NE* | |
| NOP2 | Nucleolar protein | E* | NHP6B | Affects CLN1 transcription | NE*§ | |
| PHO87 | Member of phosphate permease family | NE* | PDS1 | Anaphase inhibitor | NE*§§ | |
| RPG1 | “Required for Progression through G1”; translation initiation factor eIF3 | E*† | PPH21 | Ser/Thr phosphatase 2A; ts mutants arrest in G2 | NE* | |
| RPL4A | Ribosomal protein L4 | NE‡ | SDS22 | Glc7p-Sds22p holoenzyme function required for M | E‡ | |
| SLG1 | Involved in cell wall integrity | NE‡ | SED5 | Required in ER to Golgi transport | E‡ | |
| YRB1 | CST20 = “Chromosomal Stability” | E* | SPC42 | Spindle pole component | E‡§§ | |
| SPC98 | Spindle pole component | E*††† | ||||
| SSU81 | Required for normal pseudohyphal development | NE*‡‡ | ||||
| ZDS1 | Regulates SWE1 and CLN2 transcription | NE*‡‡†† | ||||
| YIL041W | Unknown | NE‖ | ||||
| YLR057W | Unknown | NE‡ | ||||
| >5% | AAC3 | Member of mitochondrial carrier family | NE*¶ | ABF1 | Activation of DNA replication | E‡ |
| AAH1 | Adenine deaminase | NE* | ARF2 | GTP-binding protein of ARF family | NE*‡‡ | |
| ABF2 | Mitochondrial HMG-1 homologue | NE* | ARP1 | Actin-related protein required for spindle orientation | NE* | |
| ATP4 | Subunit 4 of F0-ATP synthase | NE* | BIM1 | Required for a cell cycle checkpoint | NEत | |
| GBP2 | Potential telomere-associated protein | NE*¶ | CAJ1 | Homologue of E. coli DnaJ | NE‡‡‡ | |
| MCM1 | Exerts control over G1/S transition by regulating genes affecting CLN1 and CLN2 expression | E*§ | CDC20 | Required for microtubule function and exit from anaphase | E* | |
| MRPL31 | Mitochondrial ribosomal protein | NE‡ | CDC55 | Protein phosphatase 2A regulatory subunit | NE*§§ | |
| PDB1 | β-Subunit of pyruvate dehydrogenase | NE‡ | CLB2 | B-type cyclin | NE‡§§ | |
| PSE1 | β-Karyopherin involved in nuclear import | NE* | FTI1 | Rad52 inhibitor | NE‖ | |
| RFX1 | Repressor of DNA damage inducible genes | NE‡ | HHO1 | Histone H1; induced in late G1 | NE‡ | |
| RPL3 | Ribosomal protein L3 | E‡ | HSC826 | Chaperonin homologous to mammalian Hsp90 | NE‡‡‡ | |
| RPL13B | Ribosomal protein L13 | NE‡ | MYO26 | Vesicle transport along actin cables to the bud site | E‡‡‡ | |
| RPS31 | Ribosomal protein S31 | NE* | PAP1 | Poly(A)polymerase | E‡ | |
| SDH1 | Succinate dehydrogenase flavoprotein (Fp) subunit | NE*¶ | SAC3 | Role in cytoskeletal functions and mitosis | NE‡ | |
| SUB2 | Member of DEAD box family of RNA helicases | E* | SEC4 | Transport between Golgi and plasma membrane | E* | |
| SUP45 | Translational release factor eRF1 | E* | SLI15 | Mitotic spindle protein | E* | |
| TOM20 | Mitochondrial import receptor of the outer membrane | NE* | TIF46316 | mRNA cap-binding protein eIF4F | NE*‡‡ | |
| YRA1 | Protein with RNA:RNA annealing activity | E‡ | ULP1 | Required for progression through G2/M | E* | |
| YGR235C | Unknown | NE‡ | YCL028W | RNQ1; function unknown | NE‡ | |
| YIL036W | CST6 = “Chromosomal Stability” | NE¶‖** | YGR284C | Unknown | NE‖ | |
| YIL157C | Unknown | NE‖ | ||||
| YKL195W | Unknown | E‖ | ||||
| YML068W | Unknown | NE‖ | ||||
| YPL137C | Unknown | NE‖ | ||||
Bold, previously implicated in cell cycle; NE, nonessential gene; E, essential gene; *, published;
, overexpression results in accumulation of cells with homogeneous morphology;
, Yeast Deletion Project;
, reported in other overexpression screens;
, variable results upon overexpression;
, this study; **, deletion is slow-growing;
, overexpression also causes some accumulation of cells with 4C DNA content;
, identified in the genomic library screen.
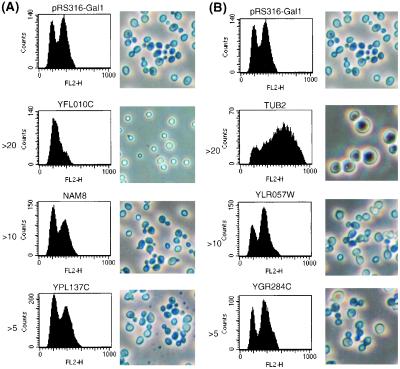
Genes were broken down into groups based on the extent to which they affected cell cycle, ranging from no effect to >20% shift in cell cycle profile (Table 1 and supplemental data, www.pnas.org). Genes with the most robust effects, such as YFL010C and TUB2, resulted in the accumulation of >90% of the cells at a specific cell cycle stage, whereas those with the weakest effects showed movement of 3–5% of the population (Table 1, supplemental data, and Fig. 1). Genes known to be involved in cell cycle progression can be found throughout the entire distribution, confirming that even genes causing subtle changes should not be overlooked. Overall, 43% of the known genes identified here have been previously implicated in cell cycle control (Table 1 and supplemental data). Conversely, in a control experiment, 18 random clones from the cDNA library were sequenced, revealing 14 known genes and 4 hypothetical ORFs. Only 2 (14%) of the 14 known genes have previously characterized cell cycle functions, indicating that the screen has produced significant enrichment (data not shown). As further validation, a number of the genes identified are known to arrest cells in specific phases of the cycle when overexpressed. Examples include: TUB2, overexpression of which causes an imbalance that results in loss of microtubule structure and arrest at G2 with large-budded cells (16); the redundant pair of cyclins, CLB2 and CLB3 (17); and CDC14, a classical CDC gene (3). From the genomic screen, an N-terminally truncated form of PDS1, which lacks its cyclin destruction box, was isolated. Such alleles of PDS1 prevent anaphase initiation when overexpressed (7).
Figure 1.
Representative examples of genes/ORFs whose overexpression causes >5->20% shift toward 1C (A) or 2C (B). DNA content. Cells carrying control plasmid (pRS316-GAL1) or plasmids encoding genes/ORFs of interest were induced in galactose containing medium for 6–8 h, and DNA content was assayed by flow cytometry. Photographs of induced cultures are included to demonstrate presence or absence of a homogeneous terminal morphology.
During our studies, we decided not to exclude genes from further analysis that had subtle overexpression effects. In some cases, likely due to differences in expression levels, genes identified in our screen had more modest effects than previously reported. For example, in a study that used extreme overexpression methods (12), MCM1 was found to cause nearly 100% accumulation at G1, whereas in this study, we see a smaller shift (5–10%) toward G1 with MCM1. We also noted that in some instances, genes causing only small to modest shifts in cell cycle profile on overexpression cause dramatic effects when deleted. ULP1 and SLI15 represent examples of this phenomenon. When overexpressed, ULP1 and SLI15 were found to shift FACS profiles toward 2C DNA content by 5–9%. However, in studies recently published, it was demonstrated that both Δulp1 and Δsli15 strains are inviable and that these genes have authentic roles in cell cycle progression (18–19). These two examples show situations in which the overexpression screens, even when yielding only minor changes in cell cycle profile, identified critical cell cycle regulators. Therefore, all new genes/ORFs were studied in detail for a role in cell division control.
Characterization of the Hypothetical ORFs.
One of the main objectives of this study was to uncover genes involved in cell cycle control. Numerous hypothetical ORFs were identified in the screen, and we therefore speculated that some of these genes might be involved in aspects of cell cycle control (Table 1, supplemental data, and Fig. 1). The hypothetical ORFs were further characterized in several ways. As an initial test, we microscopically examined 4′,6-diamidino-2-phenylindole-stained cells from the seven hypothetical ORFs whose overexpression increased 2C DNA content to determine more definitively where in G2/M the cells were arrested or delayed. Analysis of TUB2 (Table 2) and other known genes (not shown) served as a positive control. Of the seven ORFs characterized, six caused specific increases in either early or late M phase. Hence, their gene products may antagonize passage through these specific cell cycle compartments.
Table 2.
DAPI staining of ORFs that cause an increase in 2C DNA content
| Gene/ORF | Phase increased | % change from control |
|---|---|---|
| TUB2 | Pre-M | 33.7 +/− 2.05 |
| YCL028W | Early M | 20.0 +/− 2.0 |
| YGR284C | Late M | 15.0 +/− 1.0 |
| YIL041W | Late M | 11.7 +/− 4.6 |
| YLL023C | Late M | 14.0 +/− 4.0 |
| YLR057W | Late M | 13.5 +/− 0.5 |
| YMR067C | None | |
| YNL224C | Late M | 12.5 +/− 2.5 |
Large-budded cells were counted and determined to be pre-M phase (one nucleus in the mother cell), early-M phase (a single nucleus within the bud neck), or late-M phase (two clearly separated nuclei). TUB2 was used as a control. Data represent two to three independent experiments. DAPI, 4′6-diamidino-2-phenylindole.
Deletion Analysis of Hypothetical ORFs.
To identify additional CDC genes, hypothetical ORFs were systematically deleted, and their phenotypes were characterized. Of 36 total hypothetical ORFs, 33 were not required for viability, although two of these were slow growing (Δydr470c retained a larger proportion of cells in G1 compared with the wild-type control strain, whereas Δyil036w showed an increase in cells with 2C DNA content) (Table 3 and data not shown). The three ORFs that proved to be required for viability were examined for any apparent cdc phenotype. Heterozygous diploids of each deletion were sporulated, and the resulting inviable microcolonies were microdissected to determine whether the cells in the colony had accumulated with a homogeneous morphology. Indicative of a cdc phenotype, deletion of each of the three ORFs, YGL068W, YKL195W, and YPL063W resulted in inviable microcolonies that were comprised of 90, 84, and 100% unbudded cells, respectively, whereas the population of unbudded cells in matched wild-type colonies was 52% (Table 3).
Table 3.
Phenotypic analysis of deletion strains
| ORF | Tetrad analysis | ts/cs | MMS | UV | Ben |
|---|---|---|---|---|---|
| YCL036W | 4:0 | wt | wt | wt | wt |
| YDR470C | 2:2 (wt:slow) | Weak cs | S | S | wt |
| YGL068W | 2:2 93% Unbudded* | ||||
| YHR192W | 4:0 | wt | wt | wt | wt |
| YIL124W | 4:0 | wt | wt | wt | wt |
| YKR100C | 4:0 | wt | wt | wt | wt |
| YOR271C | 4:0 | wt | wt | wt | wt |
| YPL063W | 2:2 100% Unbudded* | ||||
| YGR235C | 4:0 | wt | wt | wt | wt |
| YHR181W | 4:0 | wt | wt | wt | wt |
| YIL157C | 4:0 | wt | wt | wt | wt |
| YKL195W | 2:2 84% Unbudded* | ||||
| YOR227W | 4:0 | wt | wt | wt | wt |
| YPL137W | 4:0 | wt | wt | wt | wt |
| YDR514C | 4:0 | wt | wt | wt | wt |
| YML068W | 4:0 | wt | wt | wt | wt |
| YMR067C | 4:0 | wt | wt | wt | wt |
| YIL036W | 2:2 (wt:slow) | Weak cs | wt | wt | R |
| YNL224C | 4:0 | Weak ts | wt | wt | S |
| YCL028W | 4:0 | wt | wt | wt | wt |
| YGR284C | 4:0 | wt | wt | wt | wt |
| YLR057W | 4:0 | wt | wt | wt | wt |
| YPL020C (ULP1) | 2:2 100% Budded* | ||||
| YFL010C | 4:0 | wt | wt | wt | wt |
| YIL041W | 4:0 | wt | wt | wt | wt |
Tetrad analysis of heterozygous strains was performed to determine viability of the deletion strain. Viable haploid strains were examined for temperature sensitivity (37°C), cold sensitivity (18°C), sensitivity to MMS (0.005%), sensitivity to UV (100 J/m2), and sensitivity or resistance to benomyl (10 μg/ml and 20 μg/ml, respectively).
Inviable microcolonies were dissected to determine the ratio of budded to unbudded cells. wt, wild-type; cs, cold-sensitive; ts, temperature-sensitive; R, resistant; S, sensitive; slow, slow-growing at all temperatures.
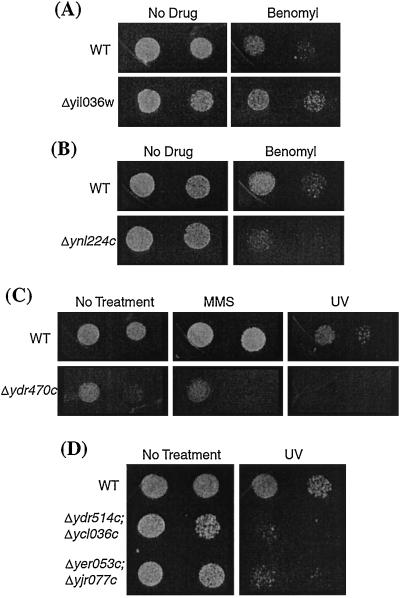
Because strains with inactive checkpoint genes are often healthy under normal growth conditions but show increased lethality when a checkpoint is activated (20), we tested whether any of the viable deletions strains were sensitive to checkpoint induction. All of the viable deletion strains were tested for their sensitivity to UV, benomyl, and MMS (Tables 3 and 4 and Fig. 2). Three of the deletion strains displayed altered sensitivity to these agents. Δynl224c was found to be sensitive to benomyl, whereas Δydr470c was found to be sensitive to both MMS and UV (Table 3 and Fig. 2). It may be that these ORFs act in part to arrest cell cycle progression when the appropriate checkpoint pathway is induced. Δyil036w was found to be benomyl resistant, a phenotype often associated with a direct or indirect role in microtubule function and cell cycle control (21, 22).
Table 4.
Phenotypic analysis of deletions of ORF pairs with high similarity
| ORF pair | P value | Tetrad analysis | ts/cs | MMS | UV | Ben |
|---|---|---|---|---|---|---|
| YPL137C | 10−175 | 2:2 95% | ||||
| YOR227W | budded* | |||||
| YDR514C | 10−88 | 2:2 | wt | wt | S | wt |
| YCL036W | (wt:slow) | |||||
| YBR177C | 10−162 | 4:0 | wt | wt | wt | wt |
| YPL095C | ||||||
| YGL224C | 10−60 | 2:2 84% | ||||
| YER037W | unbudded* | |||||
| YHR162W | 10−44 | 2:2 | wt | wt | wt | wt |
| YGR243W | (wt:slow) | |||||
| YPR125W | 10−70 | 4:0 | wt | wt | wt | wt |
| YOL027C | ||||||
| YER053C | 10−52 | 2:2 | wt | wt | S | wt |
| YJR077C | (wt:slow) |
The ORF listed first in each pair was recovered from the screen, whereas the second ORF was identified by database search. Double deletion strains were generated by PCR-mediated disruption of the second ORF in a diploid strain homozygously deleted for the first ORF. Tetrad analysis of the resulting heterozygotes was performed to determine viability of the double deletion strains. Viable haploid strains were further examined for temperature sensitivity (37°C) or cold sensitivity (18°C), sensitivity to MMS (0.005%), sensitivity to UV (100 J/m2), and sensitivity or resistance to benomyl (10 μg/ml and 20 μg/ml, respectively).
Inviable microcolonies were dissected to determine the ratio of budded to unbudded cells. Ben, benomyl; wt, wild-type growth; cs, cold-sensitive; ts, temperature-sensitive; S, sensitive; slow, slow-growing at all temperatures.
Figure 2.
Checkpoint sensitivity of deletion strains. Cultures of wild-type (BY4741) and deletion strains were grown to the same density. Ten-fold serial dilutions were made and spotted onto YPD plates without (no drug/no treatment) or with 20 μg/ml benomyl (A and B) or 0.005% MMS (C). Additional plates were subjected to 100 J/m2 UV (C and D). Photographs were taken after 2 days at 30°C.
Search for ORFs with Redundant Function.
We had anticipated that K cell cycle genes with functionally redundant partners would be identified in the screen. These genes represent a category of cell division cycle genes that would have been missed in loss-of-function screens. Comparison of the sequences of all of the hypothetical ORFs against the S. cerevisiae database identified several instances where a highly similar ORF was present in the yeast genome (Table 4). These ORFs were cloned and overexpressed. In four of seven cases, overexpression of the putative partner did not significantly impair growth on galactose. However, in the remaining cases, a phenotype virtually identical to the partner was observed on overexpression (Table 4). To see whether deletion of both partners affected viability, diploid strains entirely lacking one ORF and heterozygous for the other were generated. Upon sporulation and tetrad analysis, it was found that in two cases the double deletion resulted in loss of viability, whereas single deletions of each individual ORF were viable (Table 4 and data not shown). Microdissection of inviable colonies showed >90% large-budded cells for a Δypl137c; Δyor227w strain and 84% unbudded cells for a Δygl224c; Δyer037w strain. It is likely that these gene pairs have redundant or overlapping functions, and the homogeneous morphology seen in the inviable colonies suggests a cdc phenotype. The precise cell cycle role of these pairs is currently under investigation.
Viable double deletion strains, three of which were slow-growing, were tested for sensitivity to temperature, UV, MMS, and benomyl (Table 4 and Fig. 2). Δydr514c;Δycl036w and Δyer053c;Δyjr077c strains were both found to be sensitive to UV (Table 4 and Fig. 2), while strains bearing single deletions of these ORFs were not (data not shown), again suggesting some level of redundant function. The deletion pairs for which no defects were found may not be functionally redundant, or it may be that their redundant roles were simply not uncovered in these assays. They may also be members of larger families of related proteins, and in some instances, deletion of additional family members may ultimately reveal a cell cycle phenotype.
Discussion
We have undertaken an extensive overexpression screen to identify genes involved in cell cycle progression in the hopes of complementing the large body of information generated by the cdc screens (1–6). To that end, we have identified a large number of genes representative of categories that may have been difficult or impossible to uncover using a recessive loss-of-function approach. Categories of dominant effects that we expected to uncover in our screen include: (i) dominant negative effects caused by overexpression of a gene fragment; (ii) dominant positive effects due to elevated protein activity that is able to escape normal regulatory controls; and (iii) imbalances in critical protein complexes caused by excess of one component. Cell cycle genes identified in this screen can be found in all of these categories. Overexpression of N-terminally truncated PDS1 acts in a dominant-negative fashion to arrest cells at anaphase (7). Excess levels of Cdc14p cannot be appropriately regulated and result in arrest at G1 (9), while checkpoint pathways are often triggered by inappropriate levels of key proteins such as Bfa1p (23). In addition, overexpression of the β-tubulin gene, TUB2, results in the disruption of microtubules and arrest at G2 (16). Lastly, overexpression of cell cycle regulatory genes, such as the redundant cyclins, CLB2 and CLB3, can also cause specific arrest (24). Since our screen uncovered all of these genes, as well as other known genes from within these categories, we expected that many of the unknown ORFs would also fall into these classes. Indeed, from our additional analyses of strains deleted for these ORFs, this appears to be the case.
Identification of New CDC Genes.
Although the new classical cdc screens were extensive (6), we were able to identify CDC genes in our overexpression screen. Of the 36 hypothetical ORFs identified in our screen, deletion of three of them (YGL068W, YPL063W, YKL195W) resulted in arrest in a specific phase of the cell cycle, reflecting a role for these gene products in cell cycle progression. It may be that these genes are not amenable to the generation of temperature-sensitive alleles, which would account for their failure to be detected in the original cdc screens. It is likely that there are additional CDC genes awaiting identification. Further overexpression studies could help to uncover them.
Overexpression Identifies Genes That Are Helpful for Cell Cycle Progression.
Although the identification of (to our knowledge) new CDC genes was fortuitous, we originally designed our screen with the aim of recovering alternative categories of cell cycle genes. For example, while genes whose products are helpful but are not absolutely required for cell cycle progression are inevitably missed in a recessive loss-of-function approach, overexpression has the potential to detect them. Checkpoint genes such as BFA1, which was recovered from the genomic library screen, would fall into such a category. Under normal growth conditions, strains deleted for BFA1 grow at a wild-type rate and do not exhibit any apparent cell division defects (23); however, when stressed by microtubule inhibitors (benomyl or nocodazole), these strains are unable to arrest appropriately and fail to grow (23, 25). We have identified two ORFs, YIL036W and YNL224C, whose disruption results in strains with altered sensitivity to benomyl. Additionally, strains disrupted for a third ORF, YDR47°C, are sensitive to the DNA damaging agents MMS and UV. It is therefore suggested that these gene products, while not absolutely required for normal cell cycle divisions, may play an important role under circumstances of stress.
An additional subcategory of cell cycle genes that play helpful or auxiliary roles in cell cycle progression might include those where deletion results in strains with slow-growth phenotypes. Within this category of genes are likely to be many important cell cycle regulators, an example of which is the mitotic exit network component, LTE1 (26, 27). Lte1p is a guanidine exchange factor for the Tem1p GTPase. Activation of Tem1p is believed to be necessary for release of the Cdc14p phosphatase from the nucleolus, allowing it to dephosphorylate its substrates and promote mitotic exit (9, 28). Although TEM1 is an essential gene, LTE1 is not (29). Strains deleted for LTE1 arrest only at low temperatures (29), whereas at 25°C they merely show a transient delay at telophase (26). Thus, although Lte1p is not strictly required for cell cycle progression, it is required for efficient progression. Consequently, it is possible that the slow-growing strains, Δyil036w and Δydr470c, both of which are also cold-sensitive, represent further examples of such a class of cell cycle genes. Notably, overexpression of LTE1 has only a small effect on cell cycle progression (26). The same is true for YDR470C and YIL036W. The clear cell cycle role demonstrated for LTE1 further emphasizes the importance of examining genes that exhibit subtle cell cycle effects when overexpressed.
Identification of Redundant CDC Genes.
One of the major strengths of overexpression lies in the ability to recover genes with redundant function. Roles for cell cycle genes with redundant function have long been recognized and are best exemplified by the cyclins (17). We set out to recover more of these redundant CDCs by determining which of the genes pulled out of our screen have closely related sequence partners in the S. cerevisiae genome and then creating double-deletion strains of each pair. Because our screen had successfully recovered gene pairs with known redundant functions, CLB2 and CLB3 (17), NHP6A and NHP6B (30), and TPK1 and TPK2 (31), it was not unlikely that others existed from within our panel of hypothetical ORFs. We were able to identify two pairs of genes, YPL137C;YOR227W and YGL224C;YER037W, where double deletion resulted in the accumulation of cells with homogeneous morphology (budded for Δypl137c;Δyor227w and unbudded for Δygl224c; Δyer037w). These pairs represent examples of redundant CDCs. In three other cases, double-deletion strains were slow-growing and two of these displayed sensitivity to UV, suggesting that these two gene pairs may have some redundant role in checkpoint function. Further studies will be necessary to determine in greater detail the specific roles of these genes in cell cycle progression.
Future Overexpression Analyses.
Despite examining a large number of colonies, these screens are clearly not saturated. One hundred four of 179 genes recovered in the cDNA library screen and all seven genes recovered in the genomic screen were identified only once. However, the availability of the complete S. cerevisiae genome sequence and the ability to systematically clone all genes into an overexpression vector may allow analysis of the entire genome. These types of resources are now available for yeast (32) and are presently being constructed for other organisms (ref. 33 and J. LaBaer, personal communication). The results of the screens described here indicate that such an undertaking would be extremely informative and would provide a valuable complement to these screens and the growing number of genome-wide analyses of the yeast cell cycle (34–36). Furthermore, we anticipate modifying these yeast screens to be able to analyze the genomes of other organisms and, more immediately, we might expect to extrapolate our present yeast data to other organisms. In particular, mammalian systems have long been most efficiently studied in culture by overexpression. Establishing a gene's role by overexpression in yeast will make the experimental leap for its mammalian ortholog easier. In this way, we can accelerate our understanding of cell cycle in several arenas.
Supplementary Material
Acknowledgments
We thank Jeff Flick for providing the cDNA library and Steve Elledge for the genomic library. We are grateful to Leonard Guarante and David McNabb for advice and technical assistance. We are also indebted to Sofie Salama for expert advice on flow cytometry analysis, Angelika Amon for helpful discussion, and Marc Vidal and Iswar Hariharan for comments on the manuscript.
Abbreviations
- cdc
cell division cycle
- MMS
methyl methanesulfonate
- YPD
yeast extract–peptone–dextrose
References
- 1.Hartwell L H, Culotti J, Reid B. Proc Natl Acad Sci USA. 1970;66:352–359. doi: 10.1073/pnas.66.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartwell L H. J Mol Biol. 1971;59:183–194. doi: 10.1016/0022-2836(71)90420-7. [DOI] [PubMed] [Google Scholar]
- 3.Culotti J, Hartwell L H. Exp Cell Res. 1971;67:389–401. doi: 10.1016/0014-4827(71)90424-1. [DOI] [PubMed] [Google Scholar]
- 4.Hartwell L H. Exp Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- 5.Hartwell L H, Culotti J, Pringle J R, Reid B J. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- 6.Pringle J R, Hartwell L H. In: The Molecular Biology of the Yeast Saccharomyces. Strathern J N, Jones E W, Broach J R, editors. Vol. 1. Plainview, NY: Cold Spring Harbor Lab. Press; 1981. pp. 97–142. [Google Scholar]
- 7.Cohen-Fix O, Peters J M, Kirschner M W, Koshland D. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 8.Schwab M, Lutum A S, Seufert W. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 9.Visintin R, Prinz S, Amon A. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Krizek J, Bretscher A. Genetics. 1992;132:665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramer S W, Elledge S J, Davis R W. Proc Natl Acad Sci USA. 1992;89:11589–11593. doi: 10.1073/pnas.89.23.11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espinet C, de la Torre M, Adea M, Herrero E. Yeast. 1995;11:25–32. doi: 10.1002/yea.320110104. [DOI] [PubMed] [Google Scholar]
- 13.Akada R, Yamamoto J, Yamashita I. Mol Gen Genet. 1997;254:267–274. doi: 10.1007/s004380050415. [DOI] [PubMed] [Google Scholar]
- 14.Becker D M, Guarante L. In: Guide to Yeast Genetics and Molecular Biology. Guthrie C, Fink G R, editors. San Diego: Academic; 1991. pp. 182–187. [Google Scholar]
- 15.Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Hieter P, Boeke J D. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 16.Burke D, Gasdaska P, Hartwell L. Mol Cell Biol. 1989;9:1049–1059. doi: 10.1128/mcb.9.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fitch I, Dahmann C, Surana U, Amon A, Nasmyth K, Goetsch L, Byers B, Futcher B. Mol Biol Cell. 1992;3:805–818. doi: 10.1091/mbc.3.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S-J, Hochstrasser M. Nature (London) 1999;398:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 19.Kim J, Kang J, Chan C S M. J Cell Biol. 1999;145:1381–1394. doi: 10.1083/jcb.145.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartwell L H, Weinert T A. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 21.Bauer A, Kolling R. J Cell Sci. 1996;109:1575–1583. doi: 10.1242/jcs.109.6.1575. [DOI] [PubMed] [Google Scholar]
- 22.Foreman P K, Davis R W. Genetics. 1996;144:1387–1397. doi: 10.1093/genetics/144.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li R. Proc Natl Acad Sci USA. 1999;96:4989–4994. doi: 10.1073/pnas.96.9.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stueland C S, Lew D J, Cismowski M J, Reed S I. Mol Cell Biol. 1993;13:3744–3755. doi: 10.1128/mcb.13.6.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexandru G, Zachariae W, Schleiffer A, Nasmyth K. EMBO J. 1999;18:2707–2721. doi: 10.1093/emboj/18.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bardin A J, Visintin R, Amon A. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- 27.Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E. Mol Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- 28.Shou W, Seol J H, Shevchenko A, Baskerville C, Moazed D, Chen Z W, Jang J, Charbonneau H, Deshaies R. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 29.Shirayama M, Matsui Y, Toh-E A. Mol Cell Biol. 1994;14:7476–7482. doi: 10.1128/mcb.14.11.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costigan C, Kolodrubetz D, Snyder M. Mol Cell Biol. 1994;14:2391–2403. doi: 10.1128/mcb.14.4.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toda T, Cameron S, Sass P, Zoller M, Wigler M. Cell. 1987;50:277–287. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]
- 32.Hudson J R, Dawson E P, Rushing K L, Jackson C H, Lockshon D, Conover D, Lanciault C, Harris J R, Simmons S J, Rothstein R, Fields S. Genome Res. 1997;7:1169–1173. doi: 10.1101/gr.7.12.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walhout AJ, Temple G F, Brasch M A, Hartley J L, Lorson M A, van den Heuvel S, Vidal Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- 34.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futzher B. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winzeler E A, Shoemaker D D, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke J D, Bussey H. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 36.Uetz P, Giot L, Cagney G, Mansfield T A, Judson R S, Knight J R, Lockshon D, Narayan V, Srinivasan M, Pochart P. Nature (London) 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.