Abstract
Current technology to isolate viable cytokine-producing antigen-specific primary human T cells is limited to bi-specific antibody capture systems, which suffer from limited sensitivity and high background. Here, we describe a novel procedure for isolating antigen-specific human T cells based on their ability to produce tumor necrosis factor (TNF)-α. Unlike many cytokines, TNF-α is initially produced in a biologically active membrane-bound form that is subsequently cleaved by TNF-α converting enzyme (TACE) to release the soluble form of TNF-α. By preventing this cleavage event, we show that TNF-α can be ‘trapped’ on the surface of the T cells from which it originates and directly labeled for viable isolation of these antigen-specific T cells. Together with other existing sorting procedures to isolate activated T cells, this new technique should permit the direct isolation of multi-functional T lymphocytes for further protein and gene expression analyses, as well as a detailed functional assessment of the potential role that TNF-α producing T cells play in the adaptive immune system.
Keywords: Cell Sorting, T cells, TNF-α, Viable, Functional
1. Introduction
CD8+ T cells are essential for the effective control and clearance of intracellular pathogens, and play a central role in tumor surveillance. In response to antigenic stimuli, CD8+ T cells can deploy an extensive array of effector functions, including perforin-mediated cytotoxicity, the induction of apoptosis via Fas and TNF-α (TNF from hereon), and the production of numerous immunomodulatory cytokines and chemokines (Harty et al., 2000). Procedures to identify and isolate CD8+ T cells with these particular properties remain critically important for our understanding of CD8+ T cell function and the development of CD8+ T cell-based immunotherapeutic strategies.
Although basic analysis of CD8+ T cell function and phenotype is relatively straightforward through the use of flow cytometry, there are few procedures that permit the isolation of viable antigen-specific CD8+ T cells for further study. Staining with fluorescent MHC class I tetramers, or multimers with higher order valencies, remains the gold standard for the precise identification and isolation of antigen-specific CD8+ T cells (Altman et al., 1996; Wooldridge et al., 2009). However, this approach is limited to defined specificities, requires unique and costly reagents, and provides no assessment of functional capacity. Furthermore, it is clear that not all tetramer+CD8+ T cells are functional, at least as assessed by current techniques, and that not all functional cells can be identified with MHC class I tetramers due to threshold differences that distinguish activation from physical staining (Goepfert et al., 2000; Chattopadhyay et al., 2008). Currently, there are few available methods for the isolation of viable antigen-specific CD8+ T cells based on functional responses. As we have shown previously, exposure of CD107a on the CD8+ T cell surface after activation can be used to isolate viable antigen-specific CD8+ T cells capable of degranulation (Betts et al., 2003; Rubio et al., 2003). The only current techniques that enable the isolation of CD8+ T cells based on their ability to produce cytokines, however, are capture assays (Brosterhus et al., 1999; Douek et al., 2002b; Lichterfeld et al., 2004). These assays use a bi-specific antibody that recognizes the cytokine of interest, such as IFNγ, together with lineage specific markers, such as the pan-lymphocyte receptor CD45. Although useful, this procedure requires proprietary reagents and alternate procedural steps that can lead to specificity and sensitivity concerns.
Here, we sought to expand upon this repertoire of sorting methods by developing an assay to sort functional CD8+ T cells based on TNF production. TNF is an important cytokine produced by a variety of cell types, including CD8+ T cells, that has pleiotropic effects including apoptosis induction and complex immunoregulatory and systemic functions (Kull, 1988; Lazdins et al., 1997; Badovinac et al., 2000; Aggarwal, 2003). Compared to other cytokines, TNF production and release is somewhat unusual, in that TNF is produced initially as a membrane-bound protein that requires subsequent cleavage by the TNF-α converting enzyme (TACE) to be released (Bjornberg et al., 1994; Black et al., 1997). Both forms of TNF are biologically active, with potentially different functions. Thus, membrane-bound TNF binds preferentially to TNFRII to induce apoptosis, whereas soluble TNF binds primarily to TNFRI and exerts immunomodulatory properties (Chen and Goeddel, 2002; Wajant et al., 2003). Importantly, the TNF-α Processing Inhibitor 0 (TAPI-0) can directly prevent TNF release from the cell surface (Crowe et al., 1995; Pagan et al., 2003; Huse et al., 2006), effectively trapping TNF on the surface of the producing cell. Through the use of TAPI-0, we now demonstrate that human antigen-specific CD8+ T cells can be live-sorted based upon their ability to produce TNF. We show that this assay is highly specific and compatible with the isolation of mRNA from TNF-producing CD8+ T cells, as well as culture and in vitro expansion of antigen-specific CD8+ T cells.
2. Materials and Methods
2.1 Cells
Peripheral blood mononuclear cells (PBMC) were obtained from the University of Pennsylvania’s Center for AIDS Research Human Immunology Core Facility in compliance with the guidelines set by the respective institutional internal review boards. PBMC were isolated by standard Hypaque-Ficoll separation and cryopreserved in fetal bovine serum (FBS; ICS Hyclone, Logan, Utah) containing 10% dimethyl sulfoxide (DMSO; Fisher Scientific, Pittsburgh, Pennsylvania). Individual peptide stimuli were determined by prior epitope mapping using standard IFN-γ ELISpot assays and confirmed by intracellular cytokine flow cytometry.
2.2 ELISA
Cryopreserved human PBMC were thawed and stimulated with the Epstein-Barr virus (EBV) EBNA3A-derived RAKFKQLL (RL8) peptide (2 µM) (New England Peptide, Gardner,MA) restricted by HLA-B*08 (HLA-B8 from hereon unless specified), or Staphylococcus Enterotoxin B (SEB; 1µg/ml; Sigma-Aldrich, St. Louis, Missouri) for periods of 4, 2, and 1 hours in the presence or absence of 10 µM TAPI-0 (Calbiochem, La Jolla, California). Supernatants were then assayed using the TNF Quantikine ELISA Kit (R&D Systems, Minneapolis, Minnesota).
2.3 Antibodies and peptide/MHC class I tetramers
Monoclonal antibodies (mAbs) for surface staining included: (i) anti-CD4 PECy5-5, anti-CD19 Alexa-750, anti-CD19 Pac Blue and anti-CD8 TRPE (Invitrogen, Carlsbad, California); (ii) anti-CD107a FITC, anti-CD3 APC-H7, anti-CD14 Pac Blue, anti-CD16 Pac Blue, anti-TNF-α PECy7, anti-TNF-α PE and anti-TNF-α APC (BD Biosciences, San Jose, California); (iii) anti-CD27 PECy5 and anti-CD45RO ECD (Beckman Coulter, Fullerton, California); and, (iv) anti-CD4 Qdot 585, anti-CD8 Qdot 655, anti-CD57 Qdot 565 and anti-CD45RO Qdot 705 (custom). For intracellular staining, mAbs included anti-CD3 Qdot 585 (custom) and anti-IFN-γ Alexa-700 (BD Pharmingen, San Diego, California). Custom conjugations to quantum dot (Qdot) nanocrystals were performed in our laboratory as described previously (Chattopadhyay et al., 2006) with reagents purchased from Invitrogen. Peptide/MHC class I tetramers were produced as described previously.(Price et al., 2005)
2.4 TNF surface staining assay
Cryopreserved human PBMC were washed and resuspended at 106 cells/ml in RPMI (Mediatech Inc., Manassas, Virginia) supplemented with 10% FBS, 1% L-glutamine (Mediatech Inc.) and 1% penicillin-streptomycin (Lonza, Walkersville, Maryland); complete RPMI (cRPMI) medium was sterile filtered prior to use. Cells were then stimulated with SEB (1 µg/ml; Sigma-Aldrich) or peptide (2 µM) as indicated in the presence of 15 µl of anti-TNF-α PE or anti-TNF-α PECy7 (BD Biosciences) and 10µM of TAPI-0 (Calbiochem) for 4 hours at 37 °C, 5% CO2. Note that following the 4-hour incubation period with anti-TNF-α mAb, cells were not re-stained with anti-TNF-α in any subsequent steps. Following incubation, cells were stained with surface markers and any other relevant markers as described in section 2.5.
2.5 Flow cytometry staining assay
Cryopreserved PBMC were thawed and rested overnight at 37°C, 5% CO2 in cRPMI medium at a concentration of 2×106 cells/ml. Subsequently, the cells were washed with cRPMI and resuspended at a concentration of 1×106 cells/ml with costimulatory mAbs (anti-CD28 and anti-CD48d; 1 µg/ml final concentration; BD Biosciences). Peptide stimulations were performed at a final concentration of 2 µM; SEB (1 µg/ml; Sigma-Aldrich) was used as a positive control. Stimulation tubes were incubated at 37°C, 5% CO2 for 4.5 hours. The cells were then washed once with PBS and stained with Aqua Blue amine-reactive viability dye (Invitrogen) for 10 minutes in the dark at room temperature; a mAb cocktail was then used to stain surface markers for an additional 20 minutes. The cells were then washed again with PBS containing 1% bovine serum albumin (BSA; Fisher Scientific) and 0.1% sodium azide (Fisher Scientific) and either fixed in paraformaldehyde (Sigma-Aldrich) or fixed/permeabilized using the Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer's instructions. A cocktail of mAbs against intracellular markers was added to the fixed/permeabilized cells, which were then incubated further for 1 hour in the dark at room temperature. These cells were then washed once with Perm Wash buffer (BD Biosciences) and fixed in PBS containing 1% paraformaldehyde (Sigma-Aldrich). Fixed cells were stored in the dark at 4°C until the time of collection.
2.6 Flow cytometric analysis
For each specimen, between 0.5 and 1×106 total events were acquired on a modified LSRII flow cytometer (BD Immunocytometry Systems, San Jose, California) equipped for the detection of 18 fluorescent parameters. Anti-mouse Igκ antibody capture beads (BD Biosciences) were used to prepare individual compensation tubes for each mAb used in the experiment. Data analysis was performed using FlowJo version 9.1 (TreeStar, Ashland, Oregon).
2.7 Cell sorting
A 30µM final concentration of TAPI-0 and 100 µl of anti-TNF-α were added to a final cell concentration of 70×106 PBMC/ml; 15 µg/ml of mouse anti-human CD3 mAb (AbD Serotech, Oxford, UK) was added to the stimulated condition. Cells were incubated for 4 hours, washed with PBS and stained with Aqua Blue. Subsequently, cells were surface stained for CD4, CD8, CD14, CD16 and CD19, then sorted using a modified FACSAria flow cytometer (BD Immunocytometry Systems) at 70 lb/in2. Alternatively, PBMC were stained with cognate APC-conjugated RL8/HLA-B*0801 tetramer at 37°C for 15 minutes, washed twice and then surface stained for the markers listed above. Cells from each experimental condition were either sorted directly into microfuge tubes containing 150 µl RNAlater (Applied Biosystems, Foster City, California) and stored at −80°C until RNA extraction or sorted into 1.7 ml Eppendorf tubes for proliferation studies and incubated at 37°C, 5% C02. The sorting scheme is described in Supplemental Figure 1.
2.8 Amnis Image Stream
Cells were stimulated with SEB (1 µg/ml; Sigma-Aldrich) in the presence of TAPI-0 (30µM) and 100µl of anti-TNF-α for 4.5 hours at a final concentration of 70×106 PBMC/ml. After washing with PBS and staining to exclude non-viable events (Aqua Blue), the cells were surface stained for CD8 (FITC) and CD14, CD16, CD19 (Pacific Blue). Samples were run on an ImageStream C28 equipped with three lasers (405nm, 488nm and 658nm). All samples were collected with 100 mW 405, 200 mW 488, and 80 mW 658 laser powers. Parameters were set to prevent the collection of small debris and large clumps. From 10,000 to 17,000 events were collected for each experimental sample. Single color controls were used to create a compensation matrix that was applied to all files to correct for spectral overlap. The data were analyzed using IDEAs software version 4.0.53(Amnis Corporation, Seattle, Washington).
2.9 RNA extraction and clonotypic analysis
Total mRNA was extracted using the Oligotex Direct mRNA Mini Kit (Qiagen). Unbiased amplification of all expressed TRB gene products was conducted using a template-switch anchored RT-PCR as described previously (Douek et al., 2002a; Price et al., 2004). Amplicons were purified, cloned and sequenced as described previously(Price et al., 2005). The international ImMunoGeneTics (IMGT) nomenclature is used throughout.(Lefranc et al., 1999)
2.10 CFSE staining
Carboxyfluorescein succinimidyl ester (CFSE) staining was performed as described previously (Brenchley et al., 2002).
2.11 Figures and data analysis
Canvas software, version 10.4.9 (ACD Systems, Miami, Florida), and Prism software, version 5.0 (Graphpad, La Jolla, California), were used to create the figures. Labels and boxes were added to raw data images in Canvas.
3. Results
3.1 Prevention of TNF release from activated PBMC directly ex vivo
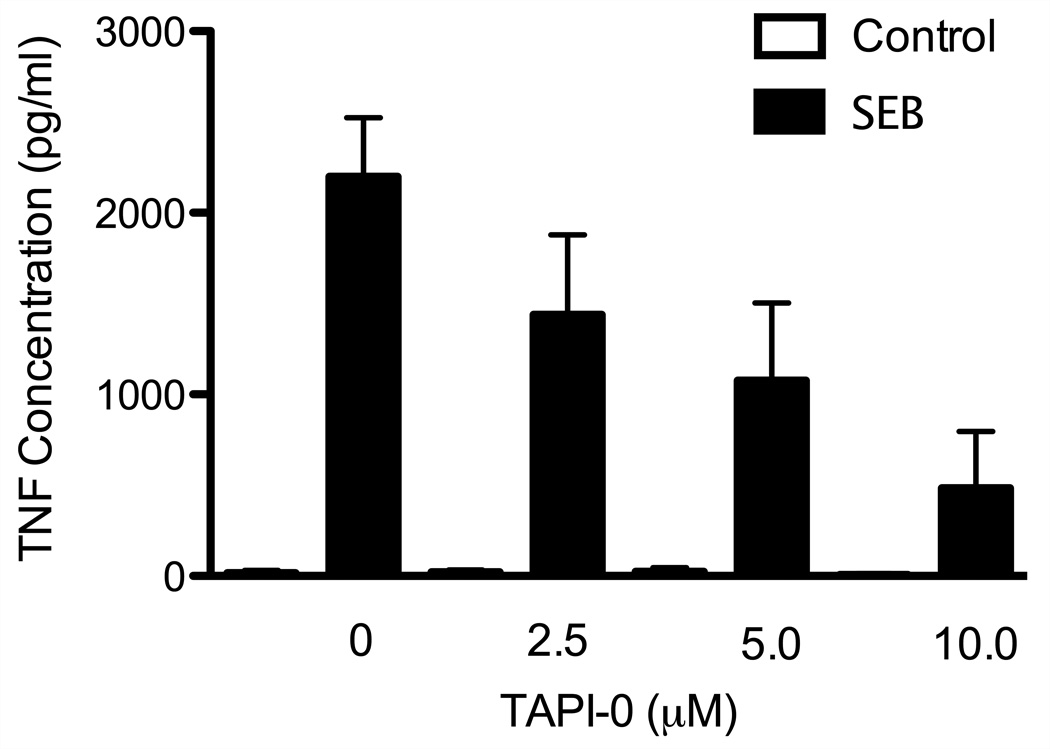
TNF is initially produced as a membrane bound protein that is subsequently cleaved by TACE to release soluble TNF (Black et al., 1997). To prevent this cleavage event, and thereby trap TNF on the producing cells, we used the TACE protease inhibitor TAPI-0. Initially, we used TNF ELISA assays to determine the optimal dosage of TAPI-0 necessary to reduce the cleavage of TNF (Figure 1) and prevent its release from stimulated cells. We stimulated human PBMC directly ex vivo with SEB in the presence of 0, 2.5, 5, and 10 µM of TAPI-0 for 4 hours. Unstimulated PBMC incubated with same dosages of TAPI-0 were included as an additional control (Figure 1). As expected, SEB stimulation potently induced the production of TNF, which was readily detectable in the culture supernatant. TAPI-0 directly inhibited TNF release from SEB-stimulated human PBMC, reaching a maximal effect at 10 µM; at this concentration, TNF content in the culture supernatant was reduced by nearly 4-fold compared to untreated cultures. Together, these data indicate that the TACE inhibitor TAPI-0 can directly inhibit TNF release from activated human PBMC.
Figure 1. TAPI-0 prevents TNF secretion.
Human PBMC were stimulated directly ex vivo with SEB in the presence of TAPI-0 at the indicated concentrations. TNF concentration in the supernatant was determined by ELISA. Open bars depict unstimulated conditions; black bars depict stimulated conditions.
3.2 TNF can be detected by flow cytometry on unfixed/unpermeabilized cells
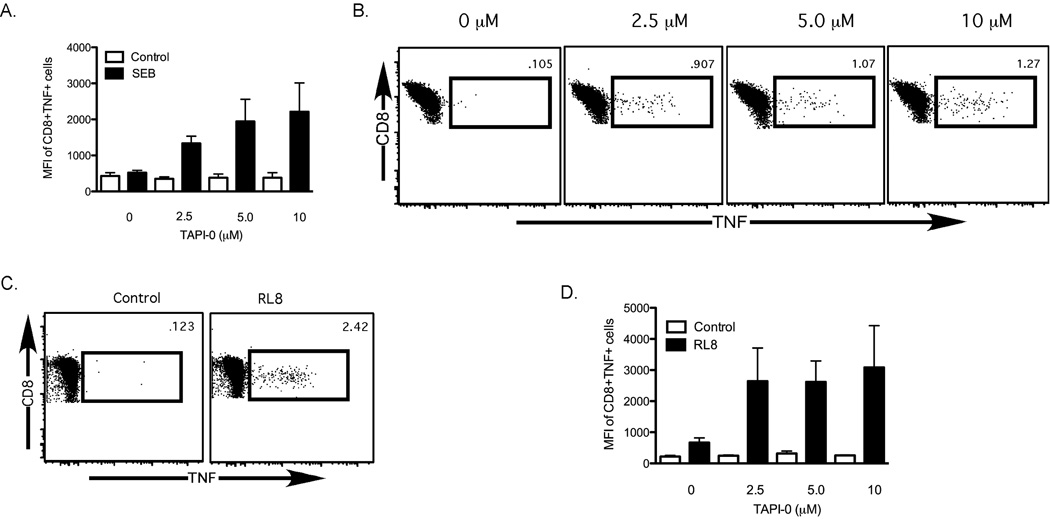
We next examined whether prevention of TNF release from activated cells by TAPI-0 could be measured by flow cytometry. As before, we stimulated human PBMC with SEB in the presence of increasing concentrations of TAPI-0. To maximize the potential staining of surface-trapped TNF, we included a fluorophore-labeled anti-TNF-α mAb during the incubation period (Supplemental Figure 2). This technique can enhance the staining of surface-exposed markers that may be internalized during T cell activation, as has been shown for other activation-exposed proteins on T cells such as CD107a and CD40L (Betts et al., 2003; Chattopadhyay et al., 2005). Unlike a standard TNF intracellular cytokine stain, no secretion inhibitors were included, and the cells were not permeabilized, fixed or re-stained for TNF following the incubation period. Using this procedure, we could directly visualize TNF-expressing CD8+ T cells after activation with SEB. Similar to our findings by ELISA, we found that increasing concentrations of TAPI-0 prevented the release of TNF from activated CD8+ T cells (a representative flow plot is shown in Figure 2B; n=3). This effect could be detected both by an increase in the median fluorescence intensity of TNF+CD8+ T cells (Figure 2A), and by an increase in the total number of TNF+CD8+ T cells (Figure 2B).
Figure 2. Expression of TNF on the surface of human PBMC can be quantified by flow cytometry.
(A) Increasing the concentration of TAPI-0 results in an increase in the amount of TNF trapped on the surface of CD8+ T cells. Open bars depict unstimulated controls; black bars depict SEB-stimulated conditions (n=3). Whiskers represent standard error. (B&C) Flow cytometric representation of the dose-dependent increase in TNF surface expression on SEB-stimulated (B) and EBNA3A-RL8 peptide-stimulated (C) CD8+ T cells. (D) Median fluorescence intensity (MFI) of surface-stained TNF in EBNA3A-RL8-specific CD8+ T cells from 3 different human donors with increasing concentrations of TAPI-0. Open bars depict unstimulated conditions; black bars depict stimulated conditions. Whiskers represent standard error.
Although SEB provides a strong stimulus to CD8+ T cells from nearly any healthy human donor, we wanted to determine whether TNF production could be detected after an antigen-specific stimulus. For this purpose, we obtained PBMC from a human donor with a previously characterized CD8+ T cell response to the HLA-B8-restricted EBNA-3A-derived EBV epitope RAKFKQLL (EBNA3A-RL8; data not shown). We stimulated donor PBMC with EBNA3A-RL8 in the presence of increasing TAPI-0 concentrations and anti-TNF-α mAb. As shown in Figure 2C, TNF was readily detectable on EBNA3A-RL8 specific CD8+ T cells. Similar data were obtained with other donors and different antigenic peptides, thereby confirming the general applicability of this approach (Supplemental Figure 3 and data not shown). Furthermore, unlike SEB stimulation, the same dose-response effect was not observed using peptide stimulation (Figure 2D), suggesting that lower concentrations of TAPI-0 may be sufficient to trap TNF on the surface of CD8+ T cells after peptide-specific stimulation.
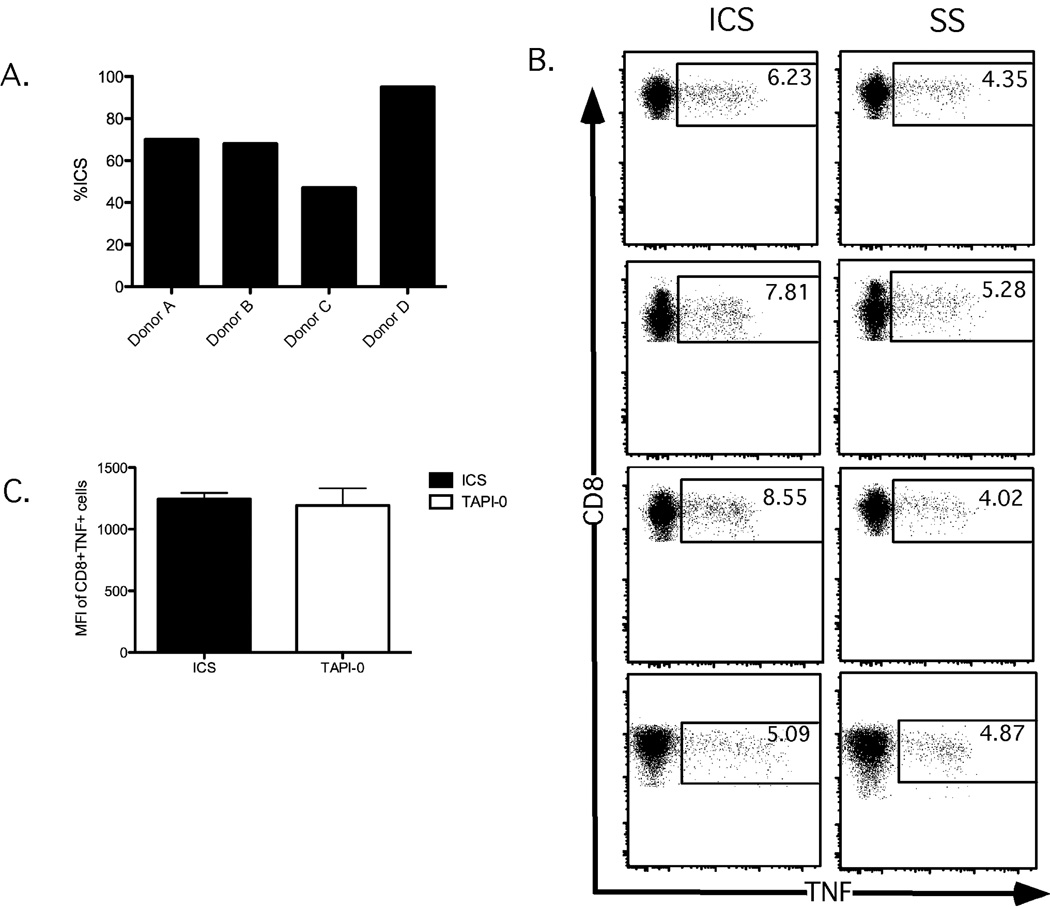
Typically, intracellular cytokine staining (ICS) is considered the ‘gold standard’ for the detection of TNF production by T cells. We therefore compared TNF responses in the CD8+ T cell population as detected by ICS or TNF-surface staining (SS) after stimulation with the SEB superantigen across 4 different donors. As shown in Figure 3A, the detected SEB-responding CD8+ T cells that produced TNF in SS conditions varied between 45%–95% of the traditional ICS methods. Two caveats are important to note from this comparison. First, background levels of TNF responses tend to be slightly higher using the TNF-SS procedure compared to TNF-ICS (data not shown). Furthermore, background staining of T cells by TNF-ICS is often higher than that for other cytokines such as IFNγ and IL-2 (Horton et al., 2007). Thus, it is not entirely surprising that the TNF background may be detected using the active labeling procedure for surface staining. Second, TNF expression visualized by SS does not result in a more uniform MFI for TNF compared to ICS(Figure 3B). This is likely due to the fact that the label only detects TNF that reaches the cell surface in the SS procedure, whereas standard ICS conditions trap TNF within the cell, allowing it to accumulate for staining. Despite these minor caveats, the data indicate that the TNF-SS procedure achieves a similar efficiency of TNF staining compared to TNF-ICS.
Figure 3. Cell surface staining for TNF recapitulates intracellular TNF staining.
Human PBMC were stimulated with SEB in the presence of TAPI-0 for 4.5 hours and then either fixed (for surface TNF staining; left panels) or permeabilized (for intracellular TNF staining; right panels) before analysis. Values represent percent TNF+CD8+ T cells. Unstimulated control CD8+ T cells are shown in the top panels; SEB-stimulated CD8+ T cells are shown in the bottom panels.
3.3 TNF can be detected on the surface of natural killer cells and CD4+ T cells
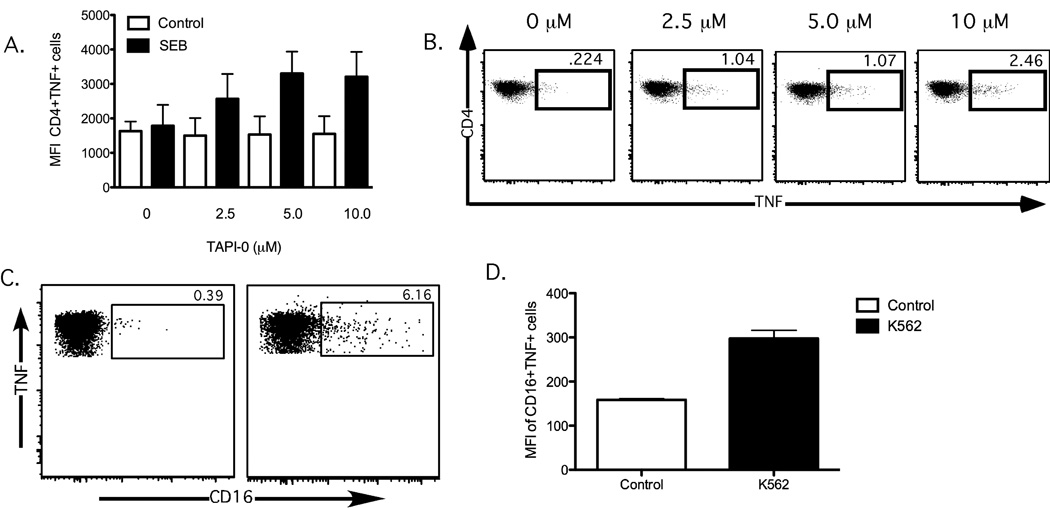
Many different lymphocytes are able to produce TNF upon activation, including CD4+ and CD8+ T cells, NK cells, B cells, monocytes, macrophages and dendritic cells (Wajant et al., 2003). We therefore examined whether TAPI-0 could similarly allow surface labeling of some of these other cell types. Similar to our findings with CD8+ T cells, SEB stimulation of CD4+ T cells induced appreciable amounts of TNF that could be directly surface stained in the presence of TAPI-0 (Figure 4A–B). Notably, CD4+ T cells often show an appreciable background level of TNF production in ICS assays (substantially higher than CD8+ T cells); this was also apparent with the TNF-SS method. Background expression of TNF was not due to the inclusion of the TAPI-0 inhibitor, as unstimulated CD4+ T cells both with and without TAPI-0 exhibited similar TNF levels. We also assessed the ability of CD16+ NK cells to produce TNF upon recognition of K562 target cells. As shown in Figure 4C, TNF produced by responding NK cells could also be surface labeled in the presence of TAPI-0.
Figure 4. Cell surface staining of TNF on activated CD4+ T cells and natural killer cells.
(A) TAPI-0 prevents TNF release from activated CD4+ T cells (n=3). Open bars depict MFI unstimulated controls; black bars depict MFI SEB-stimulated conditions; Whiskers represent standard error. (B) Flow cytometric representation of the dose-dependent increase in TNF surface expression on SEB-stimulated CD4+ T cells (C) TNF produced by activated NK cells can be surface stained after addition of TAPI-0. PBMC from a healthy human donor were co-cultured at a 1:1 ratio with K562 cells for 4 hours and then stained for the NK cell marker CD16. The frequency of responding NK cells is shown for unstimulated PBMC (left panel) and K562-stimulated PBMC (right panel)(D) MFI of CD16+TNF+ cells(n=3); open bars depict MFI of unstimulated controls; black bars depict MFI of k562 stimulated conditions
3.4 Direct visualization of TNF on the surface of CD8+ T cells
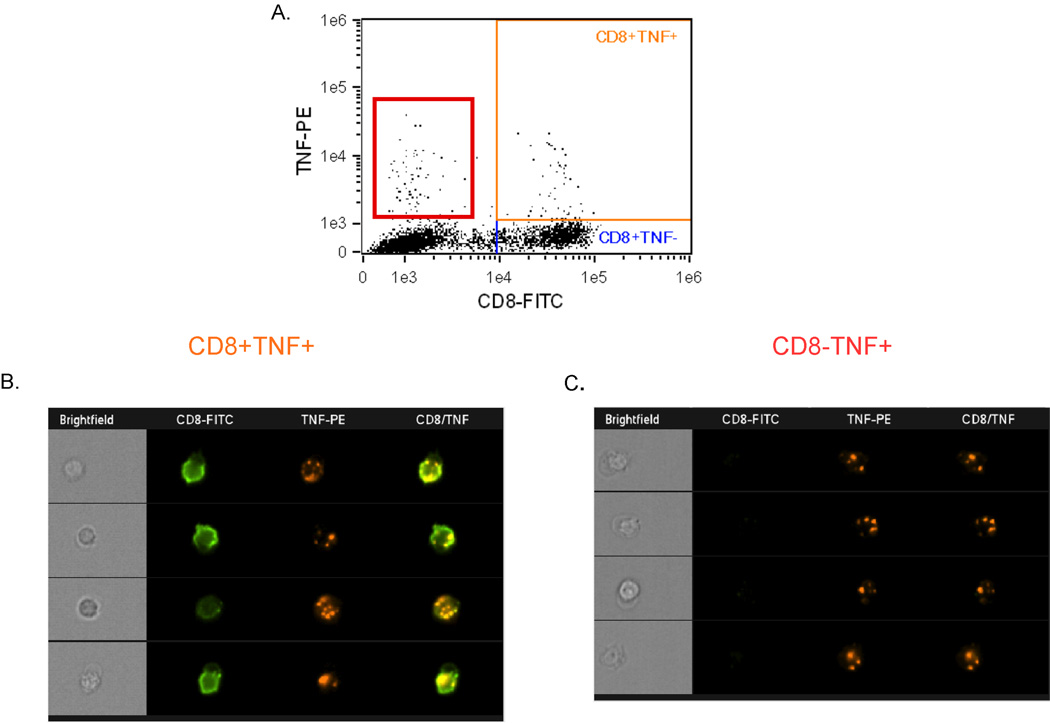
From the flow-based analysis, it was unclear whether TNF stained on the surface of CD8+ T cells remained exposed or was subsequently internalized. We therefore used an Amnis Image Stream cytometer to perform a population-based assessment of TNF-producing CD8+ T cells to determine TNF localization to the cell surface. We stimulated donor PBMC with SEB in the presence of TAPI-0 and anti-TNF-α conjugated to phycoerythrin for 4 hours. Approximately 4% of CD8+ cells produced TNF, as shown in Figure 5A (orange box). Examination of the individual responding TNF+CD8+ T cells revealed a punctate staining pattern for TNF (Figure 5B, column “TNF-PE”), which often clustered with CD8, suggesting co-localization (Figure 5B, column "CD8/TNF”). As evidenced by Figure 5A (red box) and Figure 5C, TNF can also be visualized on non-CD8+ T cells. We next used the Amnis internalization analysis feature to determine the surface localization of TNF. As shown in Supplemental Figure 4, 42.6% of the responding TNF+CD8+ cells expressed TNF on the surface, whereas 57.4% of the cells internalized the fluorophore-labeled TNF.
Figure 5. Direct visualization of TNF staining on the surface of activated CD8+ T cells.
Human donor PBMC were stimulated with SEB and stained for TNF production in the presence of TAPI-0 for 4 hours, then visualized by Image Stream technology. (A) Representative flow plot of TNF production after SEB stimulation. The orange box represents TNF+CD8+ events, whereas the red box represents TNF+CD8− events. (B&C) Representative images of TNF+CD8+ (B) and TNF+CD8− (C) cells display the cellular localization of TNF.
3.5 TNF+CD8+ T cells are a subset of MHC class I tetramer-defined clonotypes
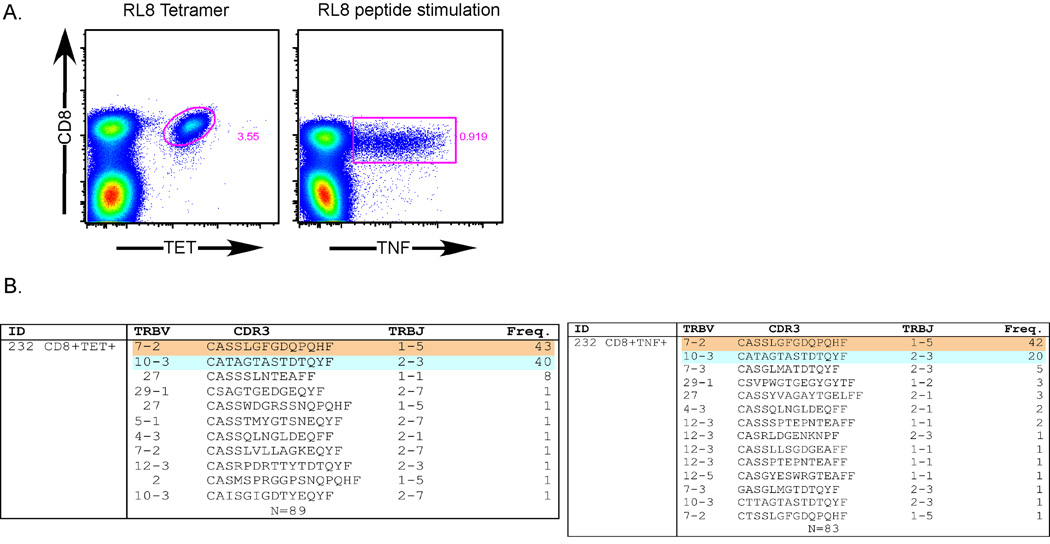
To definitively show that the TNF-SS assay is antigen-specific, we performed a clonotypic analysis of ENBA3A-RL8-specific CD8+ T cells from donor 232. CD8+ T cells were labeled with the RL8/HLA-B*0801 tetramer and, in parallel, stimulated with the EBNA3A-RL8 peptide in the presence of TAPI-0 and anti-TNF-α mAb. Tetramer+CD8+ or TNF+CD8+ T cells were then sorted viably using a modified FACS Aria flow cytometer (representative data are shown in Figure 6). We then isolated total mRNA and performed a quantitative, unbiased clonotypic analysis of all expressed TRB gene products. As shown in Figure 6, the tetramer+CD8+ T cells were composed primarily of two major clonotypes, TRBV7-2/TRBJ1-5 (CASSLGFGDQPQHF) and TRBV10-3/TRBJ2-3 (CATAGTASTDTQYF), which were equally represented and accounted for ~83% of all tetramer+CD8+ T cells. These same major clonotypes were also identified after sorting based on TNF surface expression. Interestingly, the two clonotypes were not equally represented in the TNF+CD8+ T cell population, indicating that the EBNA3A-RL8-specific tetramer+CD8+ T cell clonotypes possessed differential TNF production capacities.
Figure 6. Confirmation of TNF-SS assay specificity through TCR clonotypic analysis.
(A) Representative flow cytometry plots obtained from resting human PBMC stained with the RL8/HLA-B*0801 tetramer (upper panel) or stimulated with RL8 peptide for 4.5 hours in the presence TAPI-0 (50µM) and anti-TNF-α mAb (lower panel). The tetramer+CD8+ and the TNF+CD8+ populations were then sorted for clonotypic analysis (n=2). (B) TRBV and TRBJ usage, CDR3 amino acid sequence and clonotype frequency are shown for each distinct sequence. Tetramer+CD8+ T cell clonotypes are shown in the top panel; TNF+CD8+ T cell clonotypes are shown in the bottom panel. The number (N) of sequences obtained from the respective sorted populations is shown at the bottom of each panel.
3.6 CD8+ T cells identified by TNF-SS can be sorted viably for cell culture and expansion
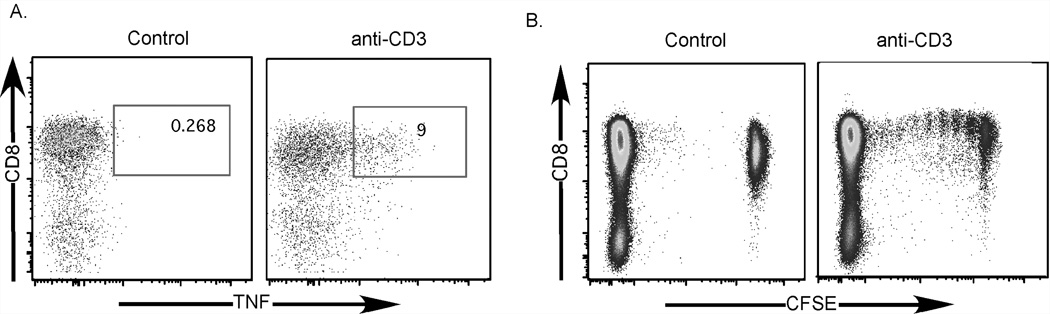
Thus far, we had established that our TNF-SS assay worked well for flow cytometry, was reproducible in other cell types, and was applicable for assays requiring isolation of intact mRNA. We next addressed whether cells sorted on the basis of TNF production could survive in culture and proliferate. To this end, we stimulated CFSE-stained human PBMC with anti-CD3 mAb in the presence of TAPI-0 and anti-TNF-α mAb, then sorted CD8+ T cells using a FACSVantage flow cytometer based on their ability to produce TNF (Figure 7A). Unstimulated CD8+CFSE+ cells were collected as a control. We recovered approximately 370,000 TNF+CD8+ T cells and placed them into culture for 5 days. The TNF+CD8+CFSE+ cells underwent 6 rounds of cell division and 34.5% of the cells divided. An appreciable population did not undergo cell division, which likely represents a lack of co-stimulation during culture (Figure 7B). Together, these data show that the TNF-SS assay can be used to isolate viable cells for further culture, as well as for mRNA extraction directly from functional antigen-specific CD8+ T cells.
Figure 7. CD8+ T cells identified by TNF-SS can be sorted viably for cell culture and expansion.
(A) Human PBMC were labeled with CFSE, rested overnight and then stimulated (right panel) with anti-CD3 for 4 hours in the presence of TAPI-0(30µM) and anti-TNF-α mAb. Control cells were not stimulated (left panel). TNF+CD8+ T cells were then sorted using a FACS DiVa flow cytometer and placed into culture for 5 days with unstimulated CFSE-autologous PBMC. (B) CFSE proliferation plots of TNF−CD8+CFSE+ (unstimulated control, left plot) and TNF+CD8+CFSE+ (anti-CD3 stimulated, right plot) T cells. The CFSE− cells in each plot are unstimulated autologous feeder cells remaining in the culture after the incubation period.
4. Discussion
In this study, we have demonstrated a novel procedure by which to isolate activated TNF-producing lymphocytes. Through the use of a TACE inhibitor, we take advantage of the unique mechanism of TNF production in order to prevent TNF release from activated T cells, thereby allowing their direct identification and isolation. We find that this procedure is highly reproducible, yields similar results to intracellular cytokine staining, and is of sufficient sensitivity to identify rare populations of antigen-specific CD8+ T cells. We demonstrate that this assay can be used to isolate viable TNF-producing antigen-specific T cells, allowing for subsequent studies requiring cell culture or RNA-based analyses.
Many different cell types of both lymphoid and non-lymphoid origin produce TNF; we show here that our assay is amenable for the identification of TNF-producing CD4+ and CD8+ T lymphocytes, as well as NK cells. It is important to note in particular that CD4+ T cells appear to exhibit substantially higher background TNF staining in the absence of stimulation compared to both CD8+ T cells and NK cells, as has been reported previously (Horton et al., 2007). Although this would likely reduce the sensitivity of our assay for CD4+ T cell sorting, combining TNF labeling with another measure of CD4+ T cell activation, such as upregulation of CD40L (Chattopadhyay et al., 2005) or CD69 (Waldrop et al., 1998) will reduce sensitivity concerns and still allow CD4+ T cells to be sorted viably and specifically based on TNF production.
Viable isolation of functional antigen-specific T cells is of particular importance for both basic and translational immunological research. As we have shown, cell sorting via TACE-inhibition of TNF release permits the direct isolation of viable antigen-specific CD8+ T cells for clonotypic analysis of expressed TCRs. Our results demonstrate both the specificity and utility of the TNF-SS sorting procedure. Thus, compared to CD8+ T cells sorted on the basis of physical binding to cognate MHC class I tetramer, the TNF-SS sorting procedure identified the same dominant clonotypes at the molecular level. Interestingly, however, the TNF assay revealed that the dominant clonotypes identified by MHC class I tetramer binding were not equally capable of producing TNF, thereby indicating functional heterogeneity within the peptide-specific CD8+ T cell population at the clonotypic level.
From the standpoint of translational research, there is a need for procedures that permit the isolation and expansion of antigen-specific T cells for use in immunotherapeutic strategies. Commonly, MHC class I tetramers/multimers are used for such isolation procedures; however, as shown herein and elsewhere (Appay et al., 2000; Oxenius et al., 2002; Gu et al., 2007; Rehr et al., 2008), not all MHC class I tetramer-binding cells are fully functional. For instance, not all MHC class I tetramer-binding cells in a population produce IFN-γ after stimulation with cognate peptide (Goepfert et al., 2000). Alternatively, surface mobilized CD107a can be used as a means to isolate viable CD8+ T cells. Again, however, not all antigen-specific CD107a+CD8+ T cells produce IFN-γ(Makedonas et al.; Betts et al., 2004; Betts et al., 2006). In addition to this, as T cells mature and become terminally differentiated, TNF production is lost (La Gruta et al., 2004). Together, this demonstrates functional heterogeneity within the overall population of responding T cells to any given antigen. Thus, the ability to isolate cells based upon the ability to produce TNF is of particular use, as it will allow the isolation of CD8+ T cells with increased functional capacity (Gu et al., 2007) at an earlier stage of differentiation. This may be of great importance, as T cell-based therapies typically require in vitro expansion of the T cells to generate sufficient numbers for effective transfer.
In conclusion, we have demonstrated a novel procedure by which to identify and isolate antigen-specific T cells based upon their ability to produce TNF. This procedure is highly sensitive and specific, and can be used to isolate viable cells for mRNA-based analyses as well as subsequent cell culture and proliferation. Alone, or in combination with other functional sorting methods, this procedure will facilitate further investigations into the underlying biological properties of antigen-specific T cells. This procedure will also be valuable for T cell-based immunological therapies, as TNF-producing T cells can either be enriched or depleted from therapeutic products.
Supplementary Material
Cell were sorted as depicted in the schematic.
Human PBMC were stimulated with SEB in the presence of 10µM TAPI-0 with the indicated concentrations of anti-TNF-α mAb. Unstimulated (NS) controls are shown in the top panel; stimulated cells are shown in the bottom panel.
PBMC from 3 human donors were stimulated with previously mapped optimal EBV(SENDRLRLL, YLQQNWWTL)or CMV(TPRVTGGGA) (2 µM) peptides in the presence of anti-TNF-α mAb and TAPI-0 (10µM) for 4.5 hours. Experiments were performed as described in the Materials and Methods.
(A) Histogram plot depicts the internalization score of TNF obtained using Amnis Image Stream co-localization analysis. Internalization scores below 0 indicate cell surface TNF labeling, scores between 0 and 1 indicate a mix of cell surface and internalized label, and scores >1 indicate internalization of labeled TNF. R5 (red) and R6 (green) gates on the histogram plot denote internalization ranges for cells shown in (B) and (C). (B) Representative images of cells with internalization scores ≤1. (C) Representative images of cells with internalization scores >1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- Altfeld M, Addo MM, Eldridge RL, Yu XG, Thomas S, Khatri A, Strick D, Phillips MN, Cohen GB, Islam SA, Kalams SA, Brander C, Goulder PJ, Rosenberg ES, Walker BD. Vpr is preferentially targeted by CTL during HIV-1 infection. J Immunol. 2001;167:2743–2752. doi: 10.4049/jimmunol.167.5.2743. [DOI] [PubMed] [Google Scholar]
- Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- Appay V, Nixon DF, Donahoe SM, Gillespie GM, Dong T, King A, Ogg GS, Spiegel HM, Conlon C, Spina CA, Havlir DV, Richman DD, Waters A, Easterbrook P, McMichael AJ, Rowland-Jones SL. HIV-specific CD8(+) T cells produce antiviral cytokines but are impaired in cytolytic function. J Exp Med. 2000;192:63–75. doi: 10.1084/jem.192.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badovinac VP, Corbin GA, Harty JT. Cutting edge: OFF cycling of TNF production by antigen-specific CD8+ T cells is antigen independent. J Immunol. 2000;165:5387–5391. doi: 10.4049/jimmunol.165.10.5387. [DOI] [PubMed] [Google Scholar]
- Betts MR, Brenchley JM, Price DA, De Rosa SC, Douek DC, Roederer M, Koup RA. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J Immunol Methods. 2003;281:65–78. doi: 10.1016/s0022-1759(03)00265-5. [DOI] [PubMed] [Google Scholar]
- Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts MR, Price DA, Brenchley JM, Lore K, Guenaga FJ, Smed-Sorensen A, Ambrozak DR, Migueles SA, Connors M, Roederer M, Douek DC, Koup RA. The functional profile of primary human antiviral CD8+ T cell effector activity is dictated by cognate peptide concentration. J Immunol. 2004;172:6407–6417. doi: 10.4049/jimmunol.172.10.6407. [DOI] [PubMed] [Google Scholar]
- Bjornberg F, Lantz M, Olsson I, Gullberg U. Mechanisms involved in the processing of the p55 and the p75 tumor necrosis factor (TNF) receptors to soluble receptor forms. Lymphokine Cytokine Res. 1994;13:203–211. [PubMed] [Google Scholar]
- Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, Nelson N, Boiani N, Schooley KA, Gerhart M, Davis R, Fitzner JN, Johnson RS, Paxton RJ, March CJ, Cerretti DP. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- Brenchley JM, Douek DC, Ambrozak DR, Chatterji M, Betts MR, Davis LS, Koup RA. Expansion of activated human naive T-cells precedes effector function. Clin Exp Immunol. 2002;130:432–440. doi: 10.1046/j.1365-2249.2002.02015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosterhus H, Brings S, Leyendeckers H, Manz RA, Miltenyi S, Radbruch A, Assenmacher M, Schmitz J. Enrichment and detection of live antigen-specific CD4(+) and CD8(+) T cells based on cytokine secretion. Eur J Immunol. 1999;29:4053–4059. doi: 10.1002/(SICI)1521-4141(199912)29:12<4053::AID-IMMU4053>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay PK, Melenhorst JJ, Ladell K, Gostick E, Scheinberg P, Barrett AJ, Wooldridge L, Roederer M, Sewell AK, Price DA. Techniques to improve the direct ex vivo detection of low frequency antigen-specific CD8+ T cells with peptide-major histocompatibility complex class I tetramers. Cytometry A. 2008;73:1001–1009. doi: 10.1002/cyto.a.20642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay PK, Price DA, Harper TF, Betts MR, Yu J, Gostick E, Perfetto SP, Goepfert P, Koup RA, De Rosa SC, Bruchez MP, Roederer M. Quantum dot semiconductor nanocrystals for immunophenotyping by polychromatic flow cytometry. Nat Med. 2006;12:972–977. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay PK, Yu J, Roederer M. A live-cell assay to detect antigen-specific CD4+ T cells with diverse cytokine profiles. Nat Med. 2005;11:1113–1117. doi: 10.1038/nm1293. [DOI] [PubMed] [Google Scholar]
- Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science. 2002;296:1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- Crowe PD, Walter BN, Mohler KM, Otten-Evans C, Black RA, Ware CF. A metalloprotease inhibitor blocks shedding of the 80-kD TNF receptor and TNF processing in T lymphocytes. J Exp Med. 1995;181:1205–1210. doi: 10.1084/jem.181.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douek DC, Betts MR, Brenchley JM, Hill BJ, Ambrozak DR, Ngai KL, Karandikar NJ, Casazza JP, Koup RA. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J Immunol. 2002a;168:3099–3104. doi: 10.4049/jimmunol.168.6.3099. [DOI] [PubMed] [Google Scholar]
- Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002b;417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- Goepfert PA, Bansal A, Edwards BH, Ritter GD, Jr, Tellez I, McPherson SA, Sabbaj S, Mulligan MJ. A significant number of human immunodeficiency virus epitope-specific cytotoxic T lymphocytes detected by tetramer binding do not produce gamma interferon. J Virol. 2000;74:10249–10255. doi: 10.1128/jvi.74.21.10249-10255.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu XX, Yue FY, Kovacs CM, Ostrowski MA. The role of cytokines which signal through the common gamma chain cytokine receptor in the reversal of HIV specific CD4(+) and CD8(+) T cell anergy. PLoS One. 2007;2:e300. doi: 10.1371/journal.pone.0000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty JT, Tvinnereim AR, White DW. CD8+ T cell effector mechanisms in resistance to infection. Annu Rev Immunol. 2000;18:275–308. doi: 10.1146/annurev.immunol.18.1.275. [DOI] [PubMed] [Google Scholar]
- Horton H, Thomas EP, Stucky JA, Frank I, Moodie Z, Huang Y, Chiu YL, McElrath MJ, De Rosa SC. Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. J Immunol Methods. 2007;323:39–54. doi: 10.1016/j.jim.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse M, Lillemeier BF, Kuhns MS, Chen DS, Davis MM. T cells use two directionally distinct pathways for cytokine secretion. Nat Immunol. 2006;7:247–255. doi: 10.1038/ni1304. [DOI] [PubMed] [Google Scholar]
- Kull FC., Jr The TNF receptor in TNF-mediated cytotoxicity. Nat Immun Cell Growth Regul. 1988;7:254–265. [PubMed] [Google Scholar]
- La Gruta NL, Turner SJ, Doherty PC. Hierarchies in cytokine expression profiles for acute and resolving influenza virus-specific CD8+ T cell responses: correlation of cytokine profile and TCR avidity. J Immunol. 2004;172:5553–5560. doi: 10.4049/jimmunol.172.9.5553. [DOI] [PubMed] [Google Scholar]
- Lazdins JK, Grell M, Walker MR, Woods-Cook K, Scheurich P, Pfizenmaier K. Membrane tumor necrosis factor (TNF) induced cooperative signaling of TNFR60 and TNFR80 favors induction of cell death rather than virus production in HIV-infected T cells. J Exp Med. 1997;185:81–90. doi: 10.1084/jem.185.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP, Giudicelli V, Ginestoux C, Bodmer J, Muller W, Bontrop R, Lemaitre M, Malik A, Barbie V, Chaume D. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 1999;27:209–212. doi: 10.1093/nar/27.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichterfeld M, Yu XG, Waring MT, Mui SK, Johnston MN, Cohen D, Addo MM, Zaunders J, Alter G, Pae E, Strick D, Allen TM, Rosenberg ES, Walker BD, Altfeld M. HIV-1-specific cytotoxicity is preferentially mediated by a subset of CD8(+) T cells producing both interferon-gamma and tumor necrosis factor-alpha. Blood. 2004;104:487–494. doi: 10.1182/blood-2003-12-4341. [DOI] [PubMed] [Google Scholar]
- Makedonas G, Hutnick N, Haney D, Amick AC, Gardner J, Cosma G, Hersperger AR, Dolfi D, Wherry EJ, Ferrari G, Betts MR. Perforin and IL-2 upregulation define qualitative differences among highly functional virus-specific human CD8 T cells. PLoS Pathog. 6 doi: 10.1371/journal.ppat.1000798. e1000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxenius A, Sewell AK, Dawson SJ, Gunthard HF, Fischer M, Gillespie GM, Rowland-Jones SL, Fagard C, Hirschel B, Phillips RE, Price DA. Functional discrepancies in HIV-specific CD8+ T-lymphocyte populations are related to plasma virus load. J Clin Immunol. 2002;22:363–374. doi: 10.1023/a:1020656300027. [DOI] [PubMed] [Google Scholar]
- Pagan JK, Wylie FG, Joseph S, Widberg C, Bryant NJ, James DE, Stow JL. The t-SNARE syntaxin 4 is regulated during macrophage activation to function in membrane traffic and cytokine secretion. Curr Biol. 2003;13:156–160. doi: 10.1016/s0960-9822(03)00006-x. [DOI] [PubMed] [Google Scholar]
- Price DA, Brenchley JM, Ruff LE, Betts MR, Hill BJ, Roederer M, Koup RA, Migueles SA, Gostick E, Wooldridge L, Sewell AK, Connors M, Douek DC. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DA, West SM, Betts MR, Ruff LE, Brenchley JM, Ambrozak DR, Edghill-Smith Y, Kuroda MJ, Bogdan D, Kunstman K, Letvin NL, Franchini G, Wolinsky SM, Koup RA, Douek DC. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004;21:793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Rehr M, Cahenzli J, Haas A, Price DA, Gostick E, Huber M, Karrer U, Oxenius A. Emergence of polyfunctional CD8+ T cells after prolonged suppression of human immunodeficiency virus replication by antiretroviral therapy. J Virol. 2008;82:3391–3404. doi: 10.1128/JVI.02383-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Stuge TB, Singh N, Betts MR, Weber JS, Roederer M, Lee PP. Ex vivo identification, isolation and analysis of tumor-cytolytic T cells. Nat Med. 2003;9:1377–1382. doi: 10.1038/nm942. [DOI] [PubMed] [Google Scholar]
- Wajant H, Pfizenmaier K, Scheurich P. Tumor necrosis factor signaling. Cell Death Differ. 2003;10:45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- Waldrop SL, Davis KA, Maino VC, Picker LJ. Normal human CD4+ memory T cells display broad heterogeneity in their activation threshold for cytokine synthesis. J Immunol. 1998;161:5284–5295. [PubMed] [Google Scholar]
- Wooldridge L, Lissina A, Cole DK, van den Berg HA, Price DA, Sewell AK. Tricks with tetramers: how to get the most from multimeric peptide-MHC. Immunology. 2009;126:147–164. doi: 10.1111/j.1365-2567.2008.02848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cell were sorted as depicted in the schematic.
Human PBMC were stimulated with SEB in the presence of 10µM TAPI-0 with the indicated concentrations of anti-TNF-α mAb. Unstimulated (NS) controls are shown in the top panel; stimulated cells are shown in the bottom panel.
PBMC from 3 human donors were stimulated with previously mapped optimal EBV(SENDRLRLL, YLQQNWWTL)or CMV(TPRVTGGGA) (2 µM) peptides in the presence of anti-TNF-α mAb and TAPI-0 (10µM) for 4.5 hours. Experiments were performed as described in the Materials and Methods.
(A) Histogram plot depicts the internalization score of TNF obtained using Amnis Image Stream co-localization analysis. Internalization scores below 0 indicate cell surface TNF labeling, scores between 0 and 1 indicate a mix of cell surface and internalized label, and scores >1 indicate internalization of labeled TNF. R5 (red) and R6 (green) gates on the histogram plot denote internalization ranges for cells shown in (B) and (C). (B) Representative images of cells with internalization scores ≤1. (C) Representative images of cells with internalization scores >1.