Abstract
The Neolithic is a key period in the history of the European settlement. Although archaeological and present-day genetic data suggest several hypotheses regarding the human migration patterns at this period, validation of these hypotheses with the use of ancient genetic data has been limited. In this context, we studied DNA extracted from 53 individuals buried in a necropolis used by a French local community 5,000 y ago. The relatively good DNA preservation of the samples allowed us to obtain autosomal, Y-chromosomal, and/or mtDNA data for 29 of the 53 samples studied. From these datasets, we established close parental relationships within the necropolis and determined maternal and paternal lineages as well as the absence of an allele associated with lactase persistence, probably carried by Neolithic cultures of central Europe. Our study provides an integrative view of the genetic past in southern France at the end of the Neolithic period. Furthermore, the Y-haplotype lineages characterized and the study of their current repartition in European populations confirm a greater influence of the Mediterranean than the Central European route in the peopling of southern Europe during the Neolithic transition.
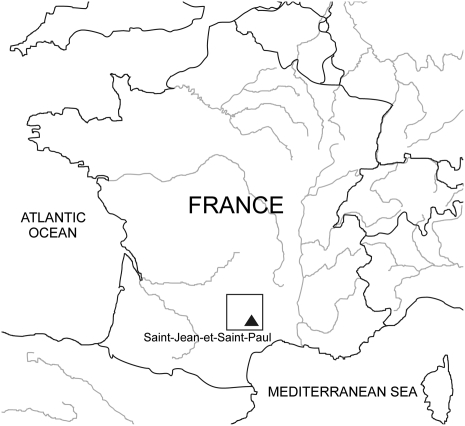
The Neolithic expansion was a major event in the European settlement and its impact on the European gene pool is still highly debated in terms of genetic flow and dispersal routes (i.e., Mediterranean vs. Central European) (1–4). In this context, molecular analyzes of ancient human populations of the end of the Neolithic are crucial to understand the origin and genetic structure of the European population. Because DNA is a very fragile molecule, rarely well preserved in ancient European specimens, only few molecular analyzes have been carried out on Neolithic remains, and they have often been limited to the study of mtDNA (4–9). The few published studies on nuclear DNA concern a small number of individuals (10–13). In the present work, the particularly good preservation of DNA in the samples excavated from a collective burial of the end of the Neolithic period (3000 B.C.) (14) allowed us to perform a study of short tandem repeats (STRs) and/or SNPs located on the nuclear DNA (Y-chromosome and autosomes) and mitochondrial DNA. Concretely, we analyzed DNA extracted from 53 individuals buried in Cave I of Treilles located in the Grands Causses region, at Saint-Jean-et-Saint-Paul, Aveyron, France (Fig. 1). The Treilles cultural group is a well identified archeological complex of the late Stone Age period, preserved of any major late Neolithic population movements as suggested by the absence of the Bell–Beaker culture influence in the second part of the third millennium B.C. The study of this cultural group should give a snapshot of the local genetic pool of the end of the Neolithic period in southern France before all recent migrations.
Fig. 1.
Location of the Grands Causses region (bounded by square) and of cave I of Treilles at Saint-Jean-et-Saint-Paul (France).
The two main objectives of this ancient DNA work were (i) to understand the structure of the Treilles community and its funeral practices by determining the sex of the individuals buried as well as putative close familial relationships; and (ii) to estimate the biogeographical origins of the specimens under study, and to infer the patterns of peopling of the region in this transitional period. To trace back the maternal and paternal lineages, we determined both mtDNA and Y-chromosomal haplogroups. We also typed a particular polymorphism associated with lactase persistence (i.e., ability to digest raw milk at adulthood) probably carried in western Europe with the Linearbandkeramic culture during the Neolithic (15).
Results
Necropolis Recruitment.
Partial autosomal profiles were obtained for 24 of the 53 specimens under study (Table S1). The amelogenin locus indicates that 22 individuals were male and two were female (subjects 573 and 614). For five samples (samples 571, 581, 603, 609, and 637), the molecular sex could not be determined. Autosomal STR kinship analyzes highlighted at least three close familial relationships within the necropolis: individuals 604 and 636 have a 99,9979% probability to have a father/son relationship [likelihood ratio (LR), 48,400]. Individuals 612 and 583 could be siblings (LR, 66,400), with a probability of 99.9985%, and subject 612 could also be the father of 616, with a probability of 99.9995% (LR, 22,4000).
Mitochondrial Results.
Reproducible HVI sequences were obtained for 29 of the 53 individuals tested. They were classified into 13 different haplotypes, which yielded a relatively high haplotype diversity (H) of 0.8966 ± 0.0354. All the haplogroups inferred by HVI sequencing were confirmed by typing of the mitochondrial coding region SNPs, for which the typing rate was as high as 98% (Table S2). Thanks to these coding region positions, the 13 haplotypes previously found could be classified in 11 different haplogroups or subhaplogroups: H1, H3, HV0, V, K1a, T2b, U, U5, U5b1c, X2, and J1.
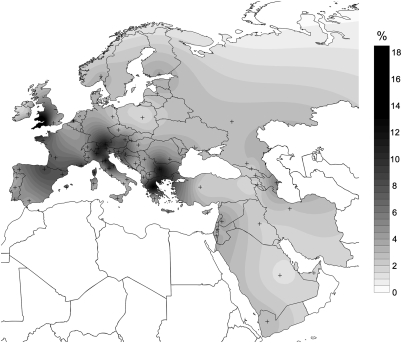
Analysis of the FST genetic distances based on HVI variation showed that the Treilles specimens were genetically close to all current European populations. Indeed, FST values were between 0 and 0.06 for all populations included in the database (Table S3 and Figs. S1 and S2). The study of shared lineages showed furthermore that the Treilles maternal lineages are found in all present-day European populations with percentages as high as nearly 18% (Fig. 2 and Table S4).
Fig. 2.
Map showing mitochondrial lineages shared between Treilles individuals and current European populations. Crosses denote the location of modern-day populations used in the analysis. The gray gradient indicates the percentage of shared lineages between modern local populations and ancient samples: the highest percentages are in black and the weakest are in gray.
Nonrecombining Region of Y-Chromosome Results.
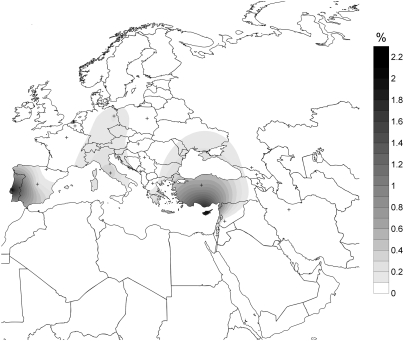
From the 22 ancient male specimens studied, three complete and 18 partial Y-STR-haplotypes were obtained (Table S5). Although all loci could not be clearly amplified for all specimens, most of the ancient individuals’ Y-STR haplotypes seem closely linked. This explains the very low average gene diversity over all loci obtained (H, 0.361664 ± 0.196576). Only individuals 577 and 596 seemed different from the other ones. Among the six nonrecombining region of Y-chromosome (NRY) SNPs typed to confirm the affiliation to haplogroups previously deduced from STR haplotypes, only three gave workable results (P15, M438, and P37.2). Nevertheless, the 22 male individuals were confirmed to belong to the Y-haplogroup previously inferred. As expected from Y-STR data, all samples were found to belong to Y-haplogroup G2a except samples 577 and 596, which belong to haplogroup I2a. Cross-population comparison tests showed a great or very great genetic differentiation between Treilles male samples and current western Eurasian populations (FST values >0.15 and as high as >0.45) except for Basque and Spanish populations, with FST values of 0.0014 and 0.007, respectively (Table S6 and Figs. S3 and S4). The analysis of shared lineages showed that the Treilles haplotypes are rarely observed in current western European populations: among the 4,791 haplotypes carried by the 10,488 European individuals included in the databases, the Treilles haplotypes were observed only 11 times (Table S7). The highest percentage of shared lineages were found mainly in Mediterranean populations: 2.06% in Cypriot, 1.98% in Portuguese, 0.7% in Turkish, 0.38% in Italian, and 0.35% in Lebanese populations (Fig. 3).
Fig. 3.
Map showing the Y-lineages shared between Treilles individuals and current European populations. Crosses denote the location of modern-day populations used in the analysis. The gray gradient indicates the percentage of shared lineages between modern local populations and ancient samples: the highest percentages are in black and the weakest are in gray.
To evaluate the molecular affinity between the G2a haplotypes from the Treilles samples and current G2a haplotypes found in European populations, we constructed a median-joining network of the G2a paternal haplogroup frequently observed in our ancient samples. The Treilles G2a haplotypes are located at the periphery of the network in a particular branch, suggesting that they are probably not the ancestral haplotypes (Fig. S5). Furthermore, they are located on a Mediterranean branch clearly differentiated from the Caucasian G2a, in which G2a is currently the most frequent in Europe, as high as approximately 30% (16).
Lactase Persistence Results.
The LP-13910-C/T SNP associated with lactase persistence was successfully typed for 26 of the 29 ancient samples tested. All were homozygous C/C for this marker, which suggests that the ancient Treilles individuals were probably not able to digest fresh milk.
Discussion
Authenticity of Results.
The main issues in ancient DNA studies is to avoid contamination by modern DNA templates and to produce authentic data. During all the steps of this study, extensive precautions were taken to avoid the amplification of contaminating contemporary DNA molecules (SI Materials and Methods). Despite the fact that not all of the classical authenticity criteria (17) could be satisfied, the following data support the authenticity of the results: (i) extraction controls, PCR blanks, and amplified products from animal remains were always negative; (ii) autosomal profiles were different from each other and different from those of researchers recently in contact with the samples; (iii) there was an inverse relationship between the amplification efficiency and length of the amplification products, especially with STR markers, which is characteristic of ancient degraded DNA; (iv) results of amplifications performed several times on various extractions were always concordant between each others; (v) results of SNP genotyping were also 100% concordant with mitochondrial and Y-chromosome haplotypes previously deduced from HVI sequencing and Y-STRs analysis; and (vi) results obtained are consistent with what can be expected on European ancient remains, as all samples were unambiguously affiliated to European haplogroups.
Social and Burial Implication.
According to molecular data, 22 individuals were male and two were female. Morphometric analysis on 30 well preserved hipbones (not included in the analysis) also showed an imbalance of sex ratio: 20 male and 10 female (14). Furthermore, in the Treilles samples, a very low gene diversity was calculated from Y-haplotypes (H, 0.361664 ± 0.196576), combined with a high gene diversity from HVI haplotypes (H, 0.8966 ± 0.0354). We can thus hypothesize that the necropolis was only dedicated to male specimens of the same paternal lineage (18).
In present-day populations, this particular sex-specific genetic structure often involves a limited gene flow within the male component of the populations and suggests that the communities are patrilocal (19). In our ancient samples, this genetic structure suggests that the community that used this burial cave was patrilocal, or that it reflects a particular funeral rite.
Maternal and Paternal Origins.
The results of the mitochondrial and Y-chromosome analyzes suggest that the maternal and paternal biogeographical origins of the Treilles samples might be substantially different.
Concerning the maternal origin, the gene pool of the Treilles samples seems to reflect a combination of the main events of the settlement of southwest Europe. Most of the mitochondrial haplogroups have an ancient ancestry consistent with the oldest episodes of settlement of western Europe from the Near East during the upper Paleolithic (52% of individuals are U, U5, HV0, X2, K1a, or T2b) (20–22) or from the Franco-Cantabrian region during the late glacial recolonization of the continent after the late glacial maximum (28% of individuals are H1, H3, V, or U5b1c) (23–25). A frequent haplogroup in Neolithic samples (4, 5, 8), haplogroup J1, found in six of 29 of the Treilles individuals, indicates also a Neolithic contribution of approximately 20% in the gene pool of our ancient samples (24, 26). The great haplotype diversity of the U5 cluster, one of the most ancient haplogroups found in Europe and very frequent in Neolithic and Mesolithic specimens (20), confirms moreover that part of the maternal lineage of Treilles samples is probably very ancient, originating from the upper Paleolithic. Similarly, the lack of haplotype diversity within haplogroup J1 confirms a probable recent origin of this haplogroup in the genetic pool of the Treilles samples.
On the contrary, similarly to southern Europe Neolithic specimens (4), there is no evidence in the Treilles samples of the N1a haplogroup, which was highly present in central Europe and Atlantic coast Neolithic cultures (6, 8). According to mitochondrial data, the Neolithic wave in the Treilles genetic pool is thus more likely to be Mediterranean than central European in origin.
The biogeographical origin of male samples appears less diverse. Treilles males belong to only two different haplogroups: I2a and G2a. In the Y phylogeny, haplogroup I is widespread over Europe but virtually absent elsewhere (27). Subclade I2a (formerly I1b1) probably originated in southern Europe during the Ice Age. Haplogroup G may represent a male contribution to a demic diffusion of farmers (1) from the Middle East to Europe (16, 28). G2a (formerly G2) is the major subclade of haplogroup G (29). Its origin in Europe is still unclear, but it could be a good marker for the Neolithic migrations of farmers into Europe (30).
The low percentage (<2%) of shared lineages between Treilles and current populations, and the fact that the ancestral and current G2a haplotypes do not seem related, imply that the G2a lineage of Treilles was probably lost between the end of the Neolithic and today. Few ancient data are currently available on Y-haplogroups to confirm this hypothesis, but G2a haplotypes have been found in other prehistoric remains; two ancient DNA studies revealed the presence of G2a in the Czech Republic during the seventh century (31) and in a German sample of a central European Neolithic culture (13), whereas this haplogroup is very rare in these places nowadays (32).
Anyway, even if the lineages shared between Treilles individuals and present-day populations are small, their location along the Mediterranean coast is consistent with an origin of part of the males’ gene pool in the Mediterranean Neolithic expansion (33).
Recent studies on modern samples link the geographical distribution of the R1b-M269 haplogroup to its spread from the Near East during the Neolithic (34). More specifically, subclade R1b-S116 has been linked with the early north-central European plain colonization (35). This haplogroup was not found in the Treilles samples. The Treilles group is strongly structured by paternal lineage, implying a low diversity among paternal lineages. The absence of the R1b haplogroup in the ancient samples could be linked to this particular genetic structure but it could also be caused by the absence of a Danubian route influence in the southwestern Mediterranean male gene pool. The latter hypothesis is highly compatible with shared lineages distribution.
In summary, even if the maternal lineages seem to have more diversified origins in time and space, both mitochondrial and NRY studies reveal a contribution of the Neolithic wave in the gene pool of the Treilles specimens. Furthermore, our results also show that, at least for the gene pool of the male samples, the Neolithic dispersals had to take place along the Mediterranean route.
Lactase Persistence in the Treilles Individuals.
The allele T located at position 13,910 bp upstream of the lactase gene is a polymorphism strongly associated with the ability to produce lactase, an intestinal enzyme that aids the digestion of untransformed milk. Largely widespread in northern and western current Europe, the 13910T allele is present in 43% of the present French population (36). This polymorphism is very rare or absent in Mesolithic Scandinavian samples and in early Neolithic Europeans (10, 12). According to a recent study, the T allele probably appeared in Europe in a region between the Balkans and central Europe and spread with the dissemination of the Linearbandkeramic culture over central Europe (15). This allele was not found in Treilles samples. This suggests that the Treilles individuals probably did not directly acquire the possibility to digest fresh milk from the farming communities of central Europe. This could also imply that the Treilles community was closer to the Mediterranean agropastoral cultures, which have an economy based on farming of sheep/goat and consumption of fermented milk (15) than to central European cultures, which practiced dairy farming. This finding also suggests that the peopling of southern France during the Neolithic expansion is more likely to have originated from the Mediterranean Sea than the central European plains.
Conclusion
All three systems used in this work to estimate the genetic origin of the Treilles samples (mtDNA, NRY, and lactase persistence SNP) are consistent with a substantial contribution of the Mediterranean Neolithic spread into the gene pool of ancient specimens. The absence of the mitochondrial haplogroup N1a and of the R1b Y-chromosomal haplogroup, both potentially associated with the spread of a Neolithic culture in Central Europe, confirms moreover the probable heterogeneity of Neolithic dispersals into Europe.
However, data obtained on the Y-chromosome suggest that the Treilles group was strongly structured by paternal lineage, and thus these data provide information on only a limited part of all of the existing lineages of southern European populations living nearby at the same period. New ancient Y-chromosomal studies from adjacent ancient populations will be needed in the future to give a complete overview on the Neolithic male diffusion through the Mediterranean route.
Materials and Methods
Samples.
The cave of Treilles is a collective burial site containing a minimum number of 149 individuals buried over a period of one or two centuries (14). Babies and young children were less represented than would be expected from the natural mortality of a community (63 children and subadults and 86 adults), and the adults’ bodies were partially disarticulated, a widespread ritual in the French Neolithic (37). Consequently, to sample each individual only once, we used mandibular teeth without carious lesions and still fixed to the mandible. All mandibles still bearing teeth were collected. Molecular analyzes were thus performed on teeth from 53 individuals. Sampling was done by two laboratory members at the Natural History Museum of Toulouse (France), where the bone collection is preserved.
DNA Extraction.
The teeth were first decontaminated with bleach, rinsed with ultrapure water, exposed to UV light (254 nm) on each side during 30 min, and powdered in a grinder mill under liquid nitrogen. Two hundred millgrams of the tooth powder were suspended in an extraction buffer and incubated overnight at 50 °C. Purification and concentration steps were then performed as previously described (38). Between three and six extractions were carried out for each individual, depending on the powder quantity retrieved from each tooth.
Nuclear Quantification.
For one DNA extract per sample, a nuclear quantification was performed on an ABI Prism 7000 Sequence Detection System by using the Quantifiler Human DNA Quantification Kit (Applied Biosystems) according to the manufacturer's protocol.
Autosomal Analysis.
Sixteen autosomal STR loci were analyzed using the AmpFiSTR Identifiler Plus and the MiniFiler PCR Amplification Kits (Applied Biosystems). Capillary electrophoreses were performed on a 3500 Genetic Analyzer and the STRs profiles were analyzed with GeneMapper 4.1 software. Two amplifications were performed on three or four different DNA extracts for each sample.
mtDNA Analysis.
Mitochondrial haplogroups were determined for each ancient sample on the basis of the HVI haplotype and of SNPs chosen on the mtDNA coding region according to the latest mtDNA phylogeny (39). Three hundred eighty-one base pairs of the HVI region of the mtDNA were amplified and sequenced in two overlapping fragments (40). Twenty-one diagnostic SNPs of the mitochondrial coding region were typed to clarify the haplogroup status inferred from HVI sequences. Typing was performed using the iPLEX Gold technology (Sequenom) as described by Mendisco et al. (38). Two multiplexes containing a total of 28 SNPs located on mtDNA, the NRY, and the MCM6 gene were designed with MassArray Assay design software (version 4.0). The typing reactions were performed twice on two different DNA extracts.
Y-Chromosomal Analysis.
Y-chromosomal analyzes were made on the 22 ancient male samples. Haplotypes were obtained from the analysis of 17 Y-STRs loci using the AmpFiSTR Yfiler PCR Amplification Kit (Applied Biosystems). Haplogroups deduced with the haplogroup predictor software (41) were then tested by SNP typing by using iPLEX Gold technology (Sequenom). We chose the six Y-SNP markers characteristic of the haplogroups and subhaplogroups G (M201), G2 (M287) and G2a (P15) (42), and I (M170), I2 (M438), and I2a (P37.2) (43) to confirm the assignment to the haplogroups initially inferred.
Lactase Persistence Typing.
One SNP located in the MCM6 gene and found to be associate with hypolactasia, more commonly known as lactose intolerance in European Caucasian populations, was added into the multiplex 2 of the SNP typing (LP-C/T13910; Rs4988235).
Statistical Analysis.
All statistical analyses performed on the Treilles data are detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Natural History Museum of Toulouse (France), and more particularly Mr. Dalous, for access to ancient samples; Dr. Remi Hienne for his help with the DNA•VIEW Software; and Fanny Mendisco and Angela Gonzalez for technical help with the Sequenom technology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100723108/-/DCSupplemental.
References
- 1.Ammerman AJ, Cavalli-Sforza LL. The Neolithic Transition and the Genetics of Populations in Europe. Princeton, NJ: Princeton Univ Press; 1984. [Google Scholar]
- 2.Barbujani G, Chikhi L. Population genetics: DNAs from the European Neolithic. Heredity. 2006;97:84–85. doi: 10.1038/sj.hdy.6800852. [DOI] [PubMed] [Google Scholar]
- 3.Chikhi L, Destro-Bisol G, Bertorelle G, Pascali V, Barbujani G. Clines of nuclear DNA markers suggest a largely neolithic ancestry of the European gene pool. Proc Natl Acad Sci USA. 1998;95:9053–9058. doi: 10.1073/pnas.95.15.9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sampietro ML, et al. Palaeogenetic evidence supports a dual model of Neolithic spreading into Europe. Proc Biol Sci. 2007;274:2161–2167. doi: 10.1098/rspb.2007.0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bramanti B, et al. Genetic discontinuity between local hunter-gatherers and central Europe's first farmers. Science. 2009;326:137–140. doi: 10.1126/science.1176869. [DOI] [PubMed] [Google Scholar]
- 6.Deguilloux MF, et al. News from the West: Ancient DNA from a French megalithic burial chamber. Am J Phys Anthropol. 2010;144:108–118. doi: 10.1002/ajpa.21376. [DOI] [PubMed] [Google Scholar]
- 7.Ermini L, et al. Complete mitochondrial genome sequence of the Tyrolean Iceman. Curr Biol. 2008;18:1687–1693. doi: 10.1016/j.cub.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 8.Haak W, et al. Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science. 2005;310:1016–1018. doi: 10.1126/science.1118725. [DOI] [PubMed] [Google Scholar]
- 9.Malmström H, et al. Ancient DNA reveals lack of continuity between neolithic hunter-gatherers and contemporary Scandinavians. Curr Biol. 2009;19:1758–1762. doi: 10.1016/j.cub.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 10.Burger J, Kirchner M, Bramanti B, Haak W, Thomas MG. Absence of the lactase-persistence-associated allele in early Neolithic Europeans. Proc Natl Acad Sci USA. 2007;104:3736–3741. doi: 10.1073/pnas.0607187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haak W, et al. Ancient DNA, Strontium isotopes, and osteological analyses shed light on social and kinship organization of the Later Stone Age. Proc Natl Acad Sci USA. 2008;105:18226–18231. doi: 10.1073/pnas.0807592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malmström H, et al. High frequency of lactose intolerance in a prehistoric hunter-gatherer population in northern Europe. BMC Evol Biol. 2010;10:89. doi: 10.1186/1471-2148-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haak W, et al. Members of the Genographic Consortium Ancient DNA from European early neolithic farmers reveals their near eastern affinities. PLoS Biol. 8:e1000536. doi: 10.1371/journal.pbio.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duranthon F, et al. Etude Anthropologique de la Grotte des Treilles (Aveyron), Projet Collectif de Recherche. Toulouse, France: Muséum de Toulouse; 2008. [Google Scholar]
- 15.Itan Y, Powell A, Beaumont MA, Burger J, Thomas MG. The origins of lactase persistence in Europe. PLOS Comput Biol. 2009;5:e1000491. doi: 10.1371/journal.pcbi.1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Battaglia V, et al. Y-chromosomal evidence of the cultural diffusion of agriculture in Southeast Europe. Eur J Hum Genet. 2009;17:820–830. doi: 10.1038/ejhg.2008.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper A, Poinar HN. Ancient DNA: Do it right or not at all. Science. 2000;289:1139. doi: 10.1126/science.289.5482.1139b. [DOI] [PubMed] [Google Scholar]
- 18.Guilaine J, Zammit J. Le Sentier de la Guerre. Paris: Seuil; 2001. [Google Scholar]
- 19.Besaggio D, et al. Genetic variation in Northern Thailand Hill Tribes: Origins and relationships with social structure and linguistic differences. BMC Evol Biol. 2007;7(suppl 2):S12. doi: 10.1186/1471-2148-7-S2-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malyarchuk B, et al. The peopling of Europe from the mitochondrial haplogroup U5 perspective. PLoS ONE. 2010;5:e10285. doi: 10.1371/journal.pone.0010285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reidla M, et al. Origin and diffusion of mtDNA haplogroup X. Am J Hum Genet. 2003;73:1178–1190. doi: 10.1086/379380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richards M, et al. Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet. 2000;67:1251–1276. [PMC free article] [PubMed] [Google Scholar]
- 23.Achilli A, et al. The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am J Hum Genet. 2004;75:910–918. doi: 10.1086/425590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards M. The Neolithic invasion of Europe. Annu Rev Anthropol. 2003;32:135–162. [Google Scholar]
- 25.Soares P, et al. The archaeogenetics of Europe. Curr Biol. 2010;20:R174–R183. doi: 10.1016/j.cub.2009.11.054. [DOI] [PubMed] [Google Scholar]
- 26.Logan J. A comprehensive analysis on mtDNA haplogroup J. J Genet Geneol. 2008;4:104–124. [Google Scholar]
- 27.Rootsi S, et al. Phylogeography of Y-chromosome haplogroup I reveals distinct domains of prehistoric gene flow in europe. Am J Hum Genet. 2004;75:128–137. doi: 10.1086/422196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Semino O, et al. The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: A Y chromosome perspective. Science. 2000;290:1155–1159. doi: 10.1126/science.290.5494.1155. [DOI] [PubMed] [Google Scholar]
- 29.Cinnioğlu C, et al. Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet. 2004;114:127–148. doi: 10.1007/s00439-003-1031-4. [DOI] [PubMed] [Google Scholar]
- 30.Behar DM, et al. Contrasting patterns of Y chromosome variation in Ashkenazi Jewish and host non-Jewish European populations. Hum Genet. 2004;114:354–365. doi: 10.1007/s00439-003-1073-7. [DOI] [PubMed] [Google Scholar]
- 31.Vanek D, Saskova L, Koch H. Kinship and Y-chromosome analysis of 7th century human remains: Novel DNA extraction and typing procedure for ancient material. Croat Med J. 2009;50:286–295. doi: 10.3325/cmj.2009.50.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luca F, et al. Y-chromosomal variation in the Czech Republic. Am J Phys Anthropol. 2007;132:132–139. doi: 10.1002/ajpa.20500. [DOI] [PubMed] [Google Scholar]
- 33.Guilaine J, Manen C, Vigne JD. Pont de Roque-Haute. Nouveaux Regards sur la Néolithisation de la France Méditerranéenne. Toulouse, France: École des Hautes Études en Sciences Sociales; 2007. [Google Scholar]
- 34.Balaresque P, et al. A predominantly neolithic origin for European paternal lineages. PLoS Biol. 2010;8:e1000285. doi: 10.1371/journal.pbio.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myres NM, et al. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur J Hum Genet. 2011;19:95–101. doi: 10.1038/ejhg.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bersaglieri T, et al. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crubézy E, Mazière G. L‘hypogée II du Mont-Aimé à Val-Des-Marais (Marne). Premiers résultats. Bulletin de la Société d'Archéologie Champenoise. 1990;83:65–78. [Google Scholar]
- 38.Mendisco F, et al. Application of the iPLEX™ Gold SNP genotyping method for the analysis of Amerindian ancient DNA samples: Benefits for ancient population studies. Electrophoresis. 2011;32:386–393. doi: 10.1002/elps.201000483. [DOI] [PubMed] [Google Scholar]
- 39.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 40.Keyser-Tracqui C, Crubézy E, Ludes B. Nuclear and mitochondrial DNA analysis of a 2,000-year-old necropolis in the Egyin Gol Valley of Mongolia. Am J Hum Genet. 2003;73:247–260. doi: 10.1086/377005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Athey W. Haplogroup prediction from Y-STR values using an allele-frequency approach. J Genet Geneol. 2005;1:1–7. [Google Scholar]
- 42.Sims LM, Garvey D, Ballantyne J. Improved resolution haplogroup G phylogeny in the Y chromosome, revealed by a set of newly characterized SNPs. PLoS ONE. 2009;4:e5792. doi: 10.1371/journal.pone.0005792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karafet TM, et al. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.