Abstract
We report on the direct observation of a nanostructural transformation from a twisted ribbon to a helical ribbon in supramolecular assemblies of peptide amphiphiles. Using cryogenic electron microscopy, a peptide amphiphile molecule containing aromatic residues was found to first assemble into short twisted ribbons in the time range of seconds, which then elongate in the time scale of minutes, and finally transform into helical ribbons over the course of weeks. By synthesizing an analogous molecule without the aromatic side groups, it was found that a cylindrical nanostructure is formed that does not undergo any transitions during the same time period. The study of metastable states in peptide aggregation can contribute to our understanding of amyloid-related diseases such as Alzheimer’s.
Precise control of supramolecular architecture is an important challenge in the field of molecular self-assembly.1 Self-assembly of peptide-containing molecules has been an active area of research for more than a decade, and with amino acid building blocks it is possible to make nanotubes,2 ribbons,3–5 and helices.6–8 The varying shapes of these nanostructures arise from an interplay of several factors, including hydrophobic interactions, hydrogen bonding, crystallization, electrostatic interactions, steric effects and π-π stacking,9–11 along with the possible involvement of growth kinetics.12 This complex balance of forces makes a priori prediction of self-assembly behavior a difficult pursuit, and thus further understanding of how these forces interact to create different morphologies, including metastable structures, is an important focus of current research. In the context of peptide assembly, specific attention has been given to the amyloid β (Aβ) protein because of its role in Alzheimer’s disease and other neurodegenerative diseases. Efforts to find the minimum peptide segment necessary for Aβ protein fibrillization have yielded the KLVFF sequence.7,13,14 Phenylalanine residues are of particular importance in the aggregation of Aβ.15 In this context, Gazit et al demonstrated that only a dimer of phenylalanine is needed to assemble into nanotubes as a result of the π-π stacking.16–18 Here we studied the self-assembly behavior of a peptide amphiphile (PA) containing three phenylalanine residues (Scheme 1) and directly observed a morphological transition from short twisted ribbon segments to long twisted ribbons which then transform into helical ribbon. Studying the transitions and meta-stable states of peptide-based nanostructures can give insight into the nature of molecular self-assembly and contribute further to mechanistic research on amyloid fibrils.
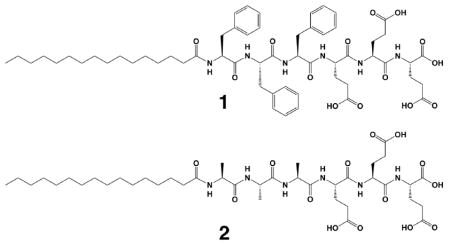
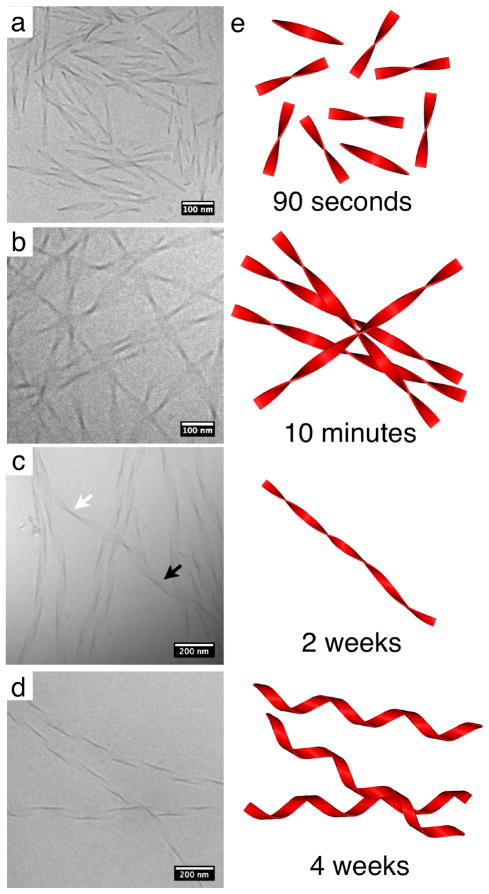
To monitor the assembly behavior at the earliest stages, PA 1 was first dissolved in hexafluoroisopropanol (HFIP) to disrupt any aggregation that occurs during peptide synthesis and purification. After removing the HFIP with rotary evaporation, the dried sample powder was resolubilized in ultrapure water and cryogenic transmission electron microscopy (cryoTEM) was done to observe the first steps of self-assembly. Thirty seconds after dissolution, very short twisted bilayer ribbons segments were observed to be the dominant morphology (Figure 1a). By three minutes, these structures rapidly combine to form long twisted ribbons (Figure S1). When PA 1 is completely dissolved in water at a 10 mM concentration a dense network of twisted ribbons are formed (Figure 1b). Interestingly these twisted ribbons undergo a morphological transformation to helical ribbons over four weeks when aged at 25°C, as seen in Figure 1c. This indicates that the twisted ribbon is a metastable state and not the thermodynamically stable endpoint. After two weeks of aging, roughly half of the twisted ribbons have undergone the transformation (Figure S2) and by four weeks they have all converted to helical ribbons (Figure 1d). These transitions are shown schematically in Figure 1e. Figure 1b reveals that these twisted ribbons are 5–6 nm thick, 20–40 nm wide, have a periodicity of 150–200 nm and are microns in length. The relative uniformity in nanostructure width is thought to be a balance of steric packing of chiral molecules and β-sheet hydrogen bonding. Both the ribbons and the helices have right handed twists and have similar periodicities for a complete ribbon rotation, 200–400 nm, which corresponds to two periods of the twisted ribbon or one turn of the helix. This twist and periodicity were also seen in AFM (Figure S4).
Figure 1.
CryoTEM of PA 1 a) forming short twists 30 seconds after the water was added to PA 1 after being molecularly dissolved b) long twisted ribbons when completely dissolved in water c) a twisted ribbon (white arrow) transforming into a helix (black arrow) and d) helical ribbons after being aged at 25°C for one month. Schematic illustrations are shown in part e).
In order to understand the role of the phenylalanine side chains in the self-assembly process we prepared PA 2, which lacks the aromatic groups found in PA 1. Under exactly the same solution conditions, PA 2 forms cylindrical nanofibers of 7–9 nm in diameter. Notably, these cylindrical nanofibers of PA 2 remain unchanged even after being aged for four weeks at 25°C (Figure S5). We performed X-ray diffraction (XRD) on the PA solutions to probe the arrangement of the molecules within the nanostructures. These experiments clearly demonstrate the presence of order in PA 1 both in freshly dissolved and aged solutions, with spacings of 4.8Å and 12.6 Å. The values of 4.8 Å and 12.6 Å are similar to those seen in fibrillar protein aggregates, such as Aβ plaques.13 Previous work has indicated that the 4.8 Å peak likely belongs to the hydrogen bonding distance in the β-sheet, whereas the 12.6 Å peak is attributed to the spacing between β-sheets.7 In contrast, PA 2 showed no significant diffraction in either the freshly dissolved solution or any of the aged solutions (Figure S6). This is not surprising, as it is difficult to obtain crystallinity in cylindrical PA nanofibers composed of β-sheets.19,20
To probe the secondary structure of these peptides, Fourier transform infrared spectroscopy (FTIR) and circular dichroism (CD) were carried out on the samples. In all cases, FTIR shows the major amide I peak was located near 1630 cm−1, which is indicative of a β-sheet secondary structure (Figure S7).19,21 The circular dichroism was more difficult to interpret, due to n-π* transitions from the aromatic stacking in the phenylalanine PAs (Figure S8a). It is, however, consistent with other reported aromatic peptides in the literature,22 and the increase in the intensity of the n-π* transition is indicative of increased interactions among aromatic sidegroups.23 A hypochromic shift of the absorbance maximum, which indicates that aromatic groups are in a more hydrophobic environment, is consistent with increased aromatic interactions (Figure S8b).25 Interestingly, fluorescence spectra of PA 1 (Figure S9) showed a greatly increased ability to bind thioflavin T (ThT) after four weeks, suggesting that the normal ThT binding pockets24 are not present initially, but open up after aging, likely due to aromatic sidegroup rearrangement. This is supported by UV fluorescence measurements, which show a blue shift of the fluorescence maximum (Figure S9a) Although both the XRD and FTIR indicate β-sheet formation, we were unable to experimentally determine if the β-sheets are perpendicular or parallel to the long axis of the nanostructure. However, previous work in our laboratory suggests that the β-sheets run parallel to the long axis of the ribbon, and this is likely true of the PAs studied here.3
We hypothesize that the observed structural transformation from twisted ribbons to helical ribbons is closely associated with changes towards a more stable molecular packing within the assemblies. One of the fingerprints of this process is the aromatic stacking rearrangement observed as the nanostructures transform from twisted to helical ribbons. Also, the alkyl tails are less likely to crystallize in the saddle-like, or Gaussian curvature of a twisted ribbon, but could do so upon transformation to a helix, which has cylindrical curvature.26 The seemingly unstable molecular packing within the twisted ribbon may be related to inhomogeneous twisting of β-sheets (see Figure S10). In the twisted ribbon, β-sheets at the periphery can be less twisted than those near the center, whereas in the helical ribbon they can have a similar twist throughout the width of the ribbon (due to the cylindrical curvature). A uniform and optimum amount of twist of β-sheets for molecular packing would favor the transformation of the kinetic twisted shape to the helical ribbon.
Supplementary Material
Acknowledgments
This work was funded by the National Institutes of Health (NIH)/NIBIB Award No. 5R01EB003806-04, and NIH/NIDCR award 5R01DE015920-05. The AFM and FTIR experiments were performed in the NIFTI and KECKII facilities, respectively, of the Northwestern University Atomic and Nanoscale Characterization Experimental Center (NUANCE) at Northwestern University. We are grateful for the use of instruments in the Keck Biophysics facility, the Biological Imaging Facility, the Institute for BioNanotechnology in Medicine, and the BioCARS X-ray diffraction facility at the Advanced Photon Source at Argonne National Lab. Finally, we are thankful to H. Cui, G. Darnell, J. Hulvat and L. Palmer, A. Cheetham and R. Bitton of the authors’ laboratory for useful discussions, D. Rozkiewicz for AFM assistance and M. Seniw for schematic illustrations.
Footnotes
Supporting Information Available:. Details of synthesis and purification, circular dichroism, UV-Vis, FTIR, cryoTEM, chemical structures of PA 1 and 2, and XRD data. This material is available free of charge via the Internet at http://pubs.ac.org.
References
- 1.Palmer LC, Velichko YS, de la Cruz MO, Stupp SI. Philos Trans R Soc A-Math Phys Eng Sci. 2007;365:1417–1433. doi: 10.1098/rsta.2007.2024. [DOI] [PubMed] [Google Scholar]
- 2.Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 3.Cui H, Muraoka T, Cheetham AG, Stupp SI. Nano Letters. 2009;9:945–951. doi: 10.1021/nl802813f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aggeli A, Nyrkova IA, Bell M, Harding R, Carrick L, McLeish TCB, Semenov AN, Boden N. Proceedings of the National Academy of Sciences of the United States of America; 2001. pp. 11857–11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aggeli A, Bell M, Boden N, Keen JN, Knowles PF, McLeish TCB, Pitkeathly M, Radford SE. Nature. 1997;386:259–262. doi: 10.1038/386259a0. [DOI] [PubMed] [Google Scholar]
- 6.Li LS, Jiang HZ, Messmore BW, Bull SR, Stupp SI. Angewandte Chemie-International Edition. 2007;46:5873–5876. doi: 10.1002/anie.200701328. [DOI] [PubMed] [Google Scholar]
- 7.Castelletto V, Hamley IW, Hule RA, Pochan D. Angewandte Chemie-International Edition. 2009;48:2317–2320. doi: 10.1002/anie.200805500. [DOI] [PubMed] [Google Scholar]
- 8.Muraoka T, Cui H, Stupp SI. Journal of the American Chemical Society. 2008;130:2946–2947. doi: 10.1021/ja711213s. [DOI] [PubMed] [Google Scholar]
- 9.Shimizu T, Masuda M, Minamikawa H. Chemical Reviews. 2005;105:1401–1443. doi: 10.1021/cr030072j. [DOI] [PubMed] [Google Scholar]
- 10.Tsai WW, Li LS, Cui HG, Jiang HZ, Stupp SI. Tetrahedron. 2008;64:8504–8514. [Google Scholar]
- 11.Yamamoto Y, Fukushima T, Suna Y, Ishii N, Saeki A, Seki S, Tagawa S, Taniguchi M, Kawai T, Aida T. Science. 2006;314:1761–1764. doi: 10.1126/science.1134441. [DOI] [PubMed] [Google Scholar]
- 12.Cui HG, Chen ZY, Zhong S, Wooley KL, Pochan DJ. Science. 2007;317:647–650. doi: 10.1126/science.1141768. [DOI] [PubMed] [Google Scholar]
- 13.Castelletto V, Hamley IW, Harris PJF. Biophys Chem. 2008;138:29–35. doi: 10.1016/j.bpc.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Balbach JJ, Ishii Y, Antzutkin ON, Leapman RD, Rizzo NW, Dyda F, Reed J, Tycko R. Biochemistry. 2000;39:13748–13759. doi: 10.1021/bi0011330. [DOI] [PubMed] [Google Scholar]
- 15.Gazit E. Faseb J. 2002;16:77–83. doi: 10.1096/fj.01-0442hyp. [DOI] [PubMed] [Google Scholar]
- 16.Reches M, Gazit E. Science. 2003;300:625–627. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]
- 17.Mahler A, Reches M, Rechter M, Cohen S, Gazit E. Advanced Materials. 2006;18:1365. [Google Scholar]
- 18.Smith AM, Williams RJ, Tang C, Coppo P, Collins RF, Turner ML, Saiani A, Ulijn RV. Advanced Materials. 2008;20:37–41. [Google Scholar]
- 19.Jiang HZ, Guler MO, Stupp SI. Soft Matter. 2007;3:454–462. doi: 10.1039/b614426h. [DOI] [PubMed] [Google Scholar]
- 20.Paramonov SE, Jun HW, Hartgerink JD. Journal of the American Chemical Society. 2006;128:7291–7298. doi: 10.1021/ja060573x. [DOI] [PubMed] [Google Scholar]
- 21.Byler DM, Susi H. Biopolymers. 1986;25:469–487. doi: 10.1002/bip.360250307. [DOI] [PubMed] [Google Scholar]
- 22.Krysmann MJ, Castelletto V, Hamley IW. Soft Matter. 2007;3:1401–1406. doi: 10.1039/b709889h. [DOI] [PubMed] [Google Scholar]
- 23.Mahalakshmi R, Shanmugam G, Polavarapu PL, Balaram P. Chembiochem. 2005;6:2152–2158. doi: 10.1002/cbic.200500152. [DOI] [PubMed] [Google Scholar]
- 24.Wu C, Wang ZX, Lei HX, Duan Y, Bowers MT, Shea JE. J Mol Biol. 2008;384:718–729. doi: 10.1016/j.jmb.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar CV, Buranaprapuk A. Journal of the American Chemical Society. 1999;121:4262–4270. [Google Scholar]
- 26.Oda R, Huc I, Schmutz M, Candau SJ, MacKintosh FC. Nature. 1999;399:566–569. doi: 10.1038/21154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.