Abstract
DNA double-strand breaks (DSBs) are extremely cytotoxic lesions with a single unrepaired DSB being sufficient to induce cell death. A complex signaling cascade, termed the DNA damage response (DDR), is in place to deal with such DNA lesions and maintain genome stability. Recent work by us and others has found that the signaling cascade activated by DSBs in mitosis is truncated, displaying apical, but not downstream, components of the DDR. The E3 Ubiquitin ligases RNF8, RNF168 and BRCA1, along with the DDR mediator 53BP1, are not recruited to DSB sites in mitosis, and activation of downstream checkpoint kinases is also impaired. Here, we show that RNF8 and RNF168 are recruited to DNA damage foci in late mitosis, presumably to prime sites for 53BP1 recruitment in early G1. Interestingly, we show that, although RNF8, RNF168 and 53BP1 are excluded from DSB sites during most of mitosis, they associate with mitotic structures such as the kinetochore, suggesting roles for these DDR factors during mitotic cell division. We discuss these and other recent findings and suggest how these novel data collectively contribute to our understanding of mitosis and how cells deal with DNA damage during this crucial cell cycle stage.
Key words: mitosis, DNA damage response, DNA double-strand breaks, signaling cascade, chromatin
Signaling Cascades in Response to DNA DSBs During Interphase
The DNA damage response (DDR) comprises an intricate network of pathways that coordinates cellular reactions to DNA insults. Cellular responses to DNA damage span from arrest of the cell cycle, repair of the DNA lesions, to the re-establishment of cellular homeostasis.1,2 At the apex of the DDR, ‘sensor’ proteins, such as the Mre11-Rad50-Nbs1 (MRN) complex,3 detect damaged DNA and recruit PIKKs (phosphatidylinositol-3-kinase-like kinases) such as ATM (ataxia telangiectasia mutated) and DNA-dependent protein kinase (DNA-PK) to DSBs. A prime PIKK target is the histone H2A variant H2AX. Phosphorylation of H2AX on Ser-139, referred to as γH2AX, is a hallmark of DSB responses and can be cytologically visualized by fluorescent microscopy as discrete nuclear ‘dots,’ called ionizing radiation-induced foci (IRIF).4 γH2AX acts as a docking site for the recruitment of MDC1, a DDR-mediator protein5 that in turn promotes phospho-dependent recruitment of MRN-ATM, inducing a positive feedback loop to amplify and sustain DNA damage signaling.6 ATM-mediated phosphorylations of MDC1 on ‘TQXF’ motifs then allow the recruitment of RNF8, a RING-finger E3 Ubiquitin ligase,7–9 whose activity brings about the focal accumulation of a second E3 Ubiquitin ligase, RNF168, to DSB sites. RNF8/RNF168-mediated ubiquitylation is then believed to be required for remodelling of DSB-flanking chromatin compartment and retention of 53BP1 (p53-binding protein 1) and BRCA1 (breast cancer protein 1),10,11 to DSBs regions.
The DNA damage-signaling cascade can thus be broadly divided into two phases: an early, phosphorylation-driven cascade comprising PIKK activation, γH2AX generation and MDC1 plus MRN recruitment, followed by a later, ubiquitylation-dependent cascade, induced by RNF8 that mediates the retention of RNF168, BRCA1 and 53BP1 to DSB regions. By concentrating DSB repair and signaling components in these regions, the ensuing IRIF may promote both DNA repair and amplification of DDR signaling that, amongst other things, activates the checkpoint kinases Chk1 and Chk2, which phosphorylate downstream targets to induce transient cell cycle arrest while the DNA damage is repaired (reviewed in ref. 12). Alternatively, under certain circumstances, sustained DNA damage signaling from unrepairable lesions can activate apoptosis or can trigger permanent cell cycle withdrawal, termed senescence.13
Impact of DNA Damage During Mitosis
Although DNA damage induces defined points of arrest in progression through the cell cycle, termed checkpoints, a striking observation is that irradiation of mitotic cells generally fails to halt cell cycle progression, with such cells proceeding to yield two daughter cells.14,15 The observation that mitotic cells seemed unresponsive to ionizing radiation (IR) prompted speculations that no checkpoint, and therefore no DNA damage responses, existed in mitosis. Nevertheless, recent studies have shown that vertebrate cells can delay mitosis, or even reverse mitotic progression if exposed to IR during a newly defined ‘antephase’ stage, a point between late G2 and mid prophase when chromatin condensation is actively taking place in preparation for entry into mitosis and cell division.16–18 The antephase checkpoint acts to complement the prior G2/M checkpoint—which monitors absence of DNA damage before proceeding into mitosis—and the spindle assembly checkpoint (SAC) that ensures accurate microtubule attachment on mitotic chromosomes before allowing transition into anaphase and mitotic exit. Following antephase, mammalian cells become committed to completing mitosis even in the presence of DNA damage,19 indicating that DNA breaks per se do not hinder mitotic progression and do not trigger cell cycle checkpoint activation.20 Nonetheless, the rate of mitotic progression can be affected by the amount of DNA damage: while relatively low levels of damage do not delay M-phase exit, more substantial levels of DNA damage can interfere with the structure and functions of kinetochores, resulting in a significantly prolonged mitosis due to a need to satisfy the SAC.21 Importantly, recent evidence in cultured mammalian cells suggests that a threshold of 10–20 DSBs is the minimum required to elicit activation of the G2/M checkpoint and prevent entry into mitosis.22 However, it is currently unclear whether the presence of a few DSBs is compatible with error-free segregation, or if it leads to chromosomal damage and imbalances in the following cell cycles.
Regulation of the Secondary DDR During Mitosis
Although mitotic cells do not delay cell cycle progression in the presence of DSBs, our recent findings23 challenged the pre-existing view that the absence of a mitotic checkpoint correlates with the absence of a DDR in mitosis. This work showed that mitotic cells do in fact respond to the presence of DSBs by initiating a primary DDR, comprising activation of the apical PIKK kinases ATM and DNA-PK, phosphorylation of γH2AX and recruitment of MDC1 and MRN to sites of DNA damage.23 The mitotic DDR, however, exibits detectable recruitment of neither the E3 Ubiquitin ligases RNF8, RNF168 and BRCA1, nor of the DDR mediator protein 53BP1. This correlates with lack of detectable DDR-induced Ubiquitin conjugates at the DSB sites in mitotic cells.23 Notably, the observed active exclusion of mitotic 53BP1 from DSB regions generated by treatment with exogenous genotoxic agents23,24 is consistent with a recent report showing that 53BP1 dissociates from endogenously-arising DSBs once cells have passed the G2/M boundary.25
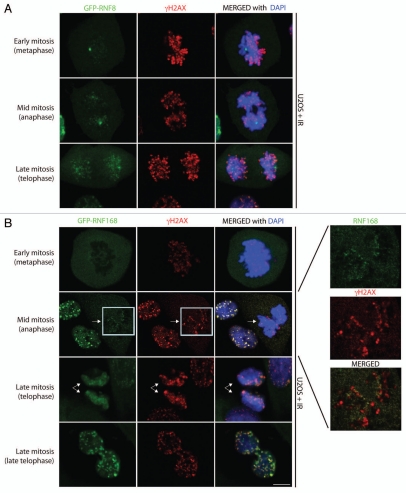
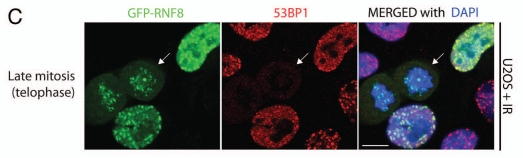
Time-course analyses of irradiated mitotic cells revealed that RNF8 and RNF168 follow similar localization kinetics in their resumed recruitment to IRIF in late mitosis. Thus, while these factors are excluded from DSB sites during mitosis until the cell passes the metaphase-to-anaphase transition, they become recruited to DNA damage foci and co-localize with γH2AX once mitotic cells enter telophase (Fig. 1A and B). It is tempting to speculate that the pre-emptive recruitment of RNF8 and RNF168 to DSB sites during mitosis serves to prepare chromatin for conformational re-arrangements anticipatory to 53BP1 binding once the nuclear envelope reforms and the cell enters G1. Indeed, 53BP1 is not recruited to RNF8-marked IRIF in late mitosis (Fig. 1C), yet localizes to IRIF in early G1.23 These observations infer the existence of multi-layered control mechanisms to prevent activation of secondary DDR responses by IRIF during mitosis.
Figure 1.
Analysis of U2OS cells stably expressing GFP-RNF89 and GFP-RNF168 show that these proteins are excluded from IRIF in early and mid-mitosis and re-associate with IRIF in late mitosis, prior to 53BP1 recruitment. (A) GFP-RNF8 forms bright dots, localizing to centrosomes in mitosis. In telophase, GFP-RNF8 starts to form IRIF and co-localizes with γH2AX. (B) GFP-RNF168 is largely excluded from chromosomes and IRIF until late mitosis, when it co-localizes with γH2AX foci. A fine GFP-RNF168 staining localized in punctate dots can be observed in mitosis, reminiscent of kinetochores. These dots do not co-localize with γH2AX. At the right are zoomed-in views of white boxes shown on the main parts. (C) Association of RNF8 with IRIF in late mitosis does not trigger recruitment of 53BP1 until cells re-enter G1. White arrows point to mitotic cells. Scale bars 10 µm.
RNF8-mediated responses at DSB sites, which likely include alterations in chromatin status, may be infeasible in the context of highly compacted mitotic chromosomes in early and mid mitosis, and may be blocked at several levels until cells have completed karyokinesis. Additionally, the delaying of 53BP1 recruitment until G1, while RNF8 and RNF168 localize to DSB foci in late mitosis, points to the existence of distinct regulatory mechanisms to prevent localization of such factors to DSBs during mitosis. Mitosis-specific post-translational modifications, including hyper-phosphorylations typical of many DDR proteins,26 and additional control processes, may also play important roles in modulating access of 53BP1 to mitotic IRIF. Moreover, the ability of RNF8 and RNF168 to bind to DSB regions within the compacted mitotic chromatin in late mitosis infers that chromatin compaction is unlikely to play a major role in the exclusion of these factors in earlier mitotic stages and that other, perhaps more direct, mechanisms influence the timing of these factors' recruitment during and following mitosis.
Localization and Function of DDR Factors During Unperturbed Mitosis
Regulation of DDR factors during mitosis may reflect mitosis-specific changes in the sub-cellular localization of certain DDR proteins. Although difficult to analyze experimentally, it is possible that sequestration of such proteins into mitotic structures prevents their recruitment and/or accumulation at sites of damaged chromatin. For instance, 53BP1 is loaded onto the kinetochore during prophase,27 while RNF8 not only localizes to the midbody, where it regulates mitotic exit,28,29 but is also recruited to other two mitotic structures: centrosomes and kinetochores.23 Further analysis of RNF168 localization during mitosis revealed that, similarly to RNF8, it displayed punctate staining at kinetochores but was excluded from γH2AX foci (Fig. 1B; insert). The observation that RNF8,23 RNF168 and 53BP1,27 localize to kinetochores during mitosis raises the possibility that their recruitment to such structures takes place through mechanisms related to those controlling their localization at DNA damage sites during interphase.
Notably, the observation that spindle-generated DSBs are preferentially found in centromere-containing micronuclei30 suggests that DSBs may be generated at centromeres, because they represent particularly vulnerable sites subjected to spindle-induced tensional forces during mitosis. Because centromere-kinetochore regions are subjected to such forces, one could envisage a scenario whereby genome surveillance mechanisms would be preferentially active in such regions to monitor these sites of chromosomal fragility and thus promote rapid recognition and mending of spontaneously arising DNA breaks during mitosis. On the other hand, it may be that mitotic structures that selectively harbour RNF8, such as centrosomes and the midbody, have different binding requirements from those that operate during DNA damage responses and could serve different, mitotic-specific functions and/or act to sequester certain DDR proteins away from sites of DNA damage during mitosis.
The above issues add growing evidence to the idea that, in addition to having DDR functions, certain DDR components may have roles under unchallenged conditions in the monitoring and enabling processes such as mitosis. Indeed, various DDR components are now known to be localized to mitotic structures in absence of apparent DNA damage. For instance, ATM, ATR, DNA-PKcs, p53, TopBP1, BRCA1, Chk1 and Chk2 have been found to associate with centrosomes in mitosis.31–35 Furthermore, 53BP1 is loaded to the kinetochore during prophase, where it colocalizes with CENP-E; and in metaphase, it is released only if all chromosomes are aligned correctly on the equatorial plate, suggesting that 53BP1 may play a physiological role in controlling mitotic progression into anaphase.27 Perhaps reflecting such a function, Xenopus 53BP1 has been identified as a suppressor of mitotic catastrophe.36 In addition, Chk1 is directly regulated by CDK1-dependent phosphorylation37 and is required for several aspects of cell division, ranging from regulation of the SAC, transcriptional inhibition of mitotic cyclin/CDK genes, to controlling chromosome segregation and cytokinesis.38–40 MDC1 was also recently shown to directly bind the APC/C (anaphase-promoting complex/cyclosome),41 and to regulate mitotic progression by enabling Cdc20-mediated activation of the APC during the metaphase-to-anaphase transition.42 Thus, localization of DDR factors to mitotic structures often correlates with functional activity of these proteins in the process of cell division. In this regard, it is notable that siRNA-mediated depletion of RNF8 caused failure of nocodazole-induced mitotic arrest and led to cell death (Giunta S, unpublished observations), suggesting that RNF8 plays additional roles to that of enabling mitotic exit, possibly functioning in cellular entry into and/or progression through mitosis. In light of these findings, it will be interesting to investigate the precise mitotic roles that RNF8 and other DDR proteins play at mitotic structures and to untangle the complex interplay between the regulation of cell cycle progression and DNA damage signaling.
Lack of DNA Damage Checkpoint in Mitosis: The Art of Prioritizing
In addition to lacking the “secondary” DDR, mitotic cells also do not display DNA damage checkpoint activation. For instance, following treatment with DSB-inducing agents such as phleomycin and IR, mitotic cells do not delay progression through mitosis and, instead, they enter G1 in a synchronous manner.19,21,23 This correlates with lack of DNA damage induced phosphorylation of the checkpoint kinases Chk1 and Chk2 on their prime ATM target sites, Ser-345 and Thr-68, respectively,23 in spite of DNA damage-dependent activation of ATM taking place during mitosis.23,43 Lack of Chk2 phosphorylation on Thr-68 in mitosis was recently reported to be due to inhibitory phosphorylation of Chk2 by Polo-like kinase (Plk1) that prevents activation of the DNA damage checkpoint in mitosis.43 Notably, Cdk1 phosphorylation of 53BP1 on Ser-380 during mitosis is required for its binding to Plk1, with 53BP1 possibly serving to bring Plk1 into juxtaposition with Chk2, mediating its inactivation.43 The inability of 53BP1 to be recruited to DNA damage foci in mitosis, and the associated lack of ATM-mediated phosphorylation of 53BP1 on Ser-25 in damaged mitotic cells, correlates not only with lack of Chk2 activation but also with the absence of phosphorylation of other ATM targets, including Chk1 and Smc1.23
It is therefore possible that absence of 53BP1 at DSBs during mitosis compromises the positive feedback loop that, in interphase cells, acts to enhance DDR signaling and convey signals to Chk1 and Chk2 for DNA damage checkpoint activation. The parallel between lack of recruitment of 53BP1 to IRIF and lack of Chk2 Thr-68 phosphorylation following DSB induction in mitosis might reflect the previously described inter-dependencies between 53BP1 and Chk2 in checkpoint activation.44 While it is possible that the absence of the secondary DDR during mitosis could reflect a widespread disruption of signal transduction by ATM, the presence of DNA damage induced phosphorylation of ATM substrates such as KAP1 (Ser-824) and MDC1 (Thr-719) in mitosis23 suggests that some ATM targets have different requirements and/or thresholds to be maintained in their phosphorylation status. Such considerations raise the prospect that understanding the DDR during mitosis might provide key insights into DDR signaling and regulation in other cell cycle phases.
It appears that several mechanisms have evolved to restrict secondary DDR activation in mitosis, by inhibiting ubiquitylation-dependent chromatin remodelling, downstream mediator recruitment and Chk1 and Chk2-mediated activation of downstream DDR events. It is thus tempting to speculate that, if it were activated during mitosis, the secondary DDR would have deleterious consequences. Indeed, in the complex scenario of mitosis, where the nuclear envelope has broken down and chromatin is compacted, if a secondary DDR did take place, associated chromatin-remodelling events might result in local dismantling of mitotic chromosomes that could interfere with karyokinesis. Furthermore, if DSBs led to activation of cell cycle checkpoint processes during mitosis, these could delay mitotic progression, leading to mis-coordination of accurately timed mitotic events, potentially inducing aneuploidy in daughter cells and/or mitotic catastrophe. It has been suggested that activation of cell cycle checkpoint events in mitosis could also lead to premature Cdk1/Cyclin B inactivation, resulting in aberrant mitotic exit and tetraploidy.43 The prevention of such an outcome would thus be particularly important for long-lived multicellular organisms, because unstable tetraploid cells can be precursors of aneuploid cancer cells.
Biological Significance of the Mitotic DDR in the Context of Chromatin
Mitotic cells mark DSB sites by employing γH2AX, MDC1 and MRN, suggesting that this has a functional importance. Indeed, we have obtained evidence that activation of the primary DDR in mitosis promotes cell survival.23 We hypothesise that the effect of the primary DDR in mitosis is two-fold. Firstly, γH2AX mediates marking of DSB sites in mitosis, which may serve to facilitate the identification of damaged sites in the subsequent G1 by accelerating the recruitment of DDR factors and prompting more rapid DDR activation and repair. In addition, the recruitment of MRN to mitotic DSBs could be instrumental in helping to hold the broken DNA ends together until G1, through the established DNA end-tethering activity of Mre11.45 It is tempting to speculate that, while simpler lesions could be held together by compacted mitotic chromatin structures, tethering of more complex lesions would benefit from MRN-dependent DNA-end bridging activity, facilitating repair processes in the ensuing G1. In line with this hypothesis, a subset of DSB foci remain unrepaired 24 hours after DNA damage induction when primary DDR activation is inhibited in mitotic cells,23 suggesting that these persistent DSBs are likely to represent lesions that would have benefited from marking and end tethering during mitosis. Such persistent DSBs would subsequently lead to increased mortality as the cell progressed through the cell cycle. Thus, we suggest that, while secondary DDR responses are inhibited during mitosis, mitotic primary responses act to favour DNA damage recognition and repair in the following cell cycle, thereby helping to maintain genome integrity and promote cell survival.
It is noteworthy that mitotic cells are considerably more radio-sensitive to IR than interphase cells.23,46 This intrinsic radio-sensitivity might not only be because of mitotic cells having a truncated DDR, but also because the physical chromatin conformation of mitotic chromosomes directly or indirectly impedes DNA repair events. Consistent with this idea, chromatin plasticity seems to facilitate genomic surveillance, while chromatin relaxation, caused by experimentally reducing the expression of the linker histone H1, yields hyper-responsiveness to DSBs and resistance to DNA damaging agents.47 Consistent with these findings, work in budding yeast has shown that deletion of the linker histone Hho1 leads to an enhanced capacity for homologous recombination.48 It is also noteworthy that transcriptional upregulation of chromatin remodelling components in pluripotent stem cells49 increases chromatin plasticity, which may be linked to the hyper-responsiveness of stem cells to DNA lesions.50 In addition, there is increasing evidence that the DDR is adapted to deal with various chromatin environments, with somewhat different responses being used to repair DSBs within euchromatin and heterochromatin.51
Conclusions and Future Directions
The studies discussed here highlight the uniqueness of the mitotic phase and indicate how DNA damage responses are affected by the specific physiological requirements of this cell cycle phase. In this regard, it seems that activation of the secondary DDR and associated chromatin changes during mitosis are not compatible with active mitotic progression. Both the brevity of mitosis and the need to prioritize mitotic progression over activation of a full DDR cascade may represent evolutionarily selected traits, helping to ensure that the intrinsic DNA damage hyper-sensitivity of mitotic cells is exhibited for a short window of time. The ability of mitotic cells to activate the primary DDR may nevertheless be biologically important to promote cell viability, radio-resistance and ultimately genome stability.
Although a picture is starting to emerge as to how signaling responses elicited by DSBs unfold during mitosis, several important questions remain unanswered. For instance, a key challenge for the future will be to understand how RNF8 is regulated during mitosis and what is the mechanism(s) to prevent its recruitment to IRIF in mitotic cells. In addition to explaining events during mitosis, this understanding would also likely help us gain more insights into RNF8 functions and the regulation of DDR events during interphase. Additionally, work into how the DNA damage checkpoint is modulated during mitosis would be of major interest. In this regard, defining the mechanisms and functions by which DDR mediator proteins and checkpoint kinases communicate will be particularly relevant. More studies are also required to understand the physiological roles of DDR factors within mitotic structures, with the recruitment of RNF8, RNF168 and 53BP1 to kinetochores being particularly worthy of investigation. Answering these and other questions regarding the DDR and mitosis would represent substantial progress toward gaining a better understanding of the signaling cascade initiated by DSBs and the processes and regulation of mitosis. Lastly, such work might allow us to gain a better understanding of the actions of anti-mitotic drugs on cancer cells and such an understanding could be exploited to improve the use of DNA-damaging chemotherapeutics in the treatment of cancer.
Acknowledgements
We thank Dr. Kate Dry for critical reading of this manuscript and Yaron Galantry for the GFP-RNF168 cell line. S. Giunta is supported by a UK Biotechnology and Biological Sciences Research Council (BBSRC) Cooperative Award in Science and Engineering (CASE) studentship with KuDOS Pharmaceuticals/AstraZeneca (BB/D526 129). S.P. Jackson is salaried by the University of Cambridge, supplemented by Cancer Research UK. Research in the Jackson laboratory is supported by core infrastructure funding from Cancer Research UK and the Wellcome Trust, and by funding from the European Community (EU Projects GENICA and DDResponse).
References
- 1.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 2.Rouse J, Jackson SP. Interfaces between the detection, signaling and repair of DNA damage. Science. 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 3.Mirzoeva OK, Petrini JH. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol Cell Biol. 2001;21:281–288. doi: 10.1128/MCB.21.1.281-288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 5.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 6.Stucki M, Jackson SP. MDC1/NFBD1: A key regulator of the DNA damage response in higher eukaryotes. DNA Repair. 2004;3:953–957. doi: 10.1016/j.dnarep.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, et al. RNF8 transduces the DNA damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, et al. Orchestration of the DNA damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 10.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 11.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 12.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 13.Jackson SP, Bartek J. The DNA damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazia D. How cells divide. Scientific American. 1961;205:100–120. doi: 10.1038/scientificamerican0961-100. [DOI] [PubMed] [Google Scholar]
- 15.Swann MM. The control of cell division; a review. I. General mechanisms. Cancer Res. 1957;17:727–757. [PubMed] [Google Scholar]
- 16.Chin CF, Yeong FM. Safeguarding Entry into Mitosis: the Antephase Checkpoint. Molecular and cellular biology. 2009 doi: 10.1128/MCB.00687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pines J, Rieder CL. Re-staging mitosis: A contemporary view of mitotic progression. Nat Cell Biol. 2001;3:3–6. doi: 10.1038/35050676. [DOI] [PubMed] [Google Scholar]
- 18.Matsusaka T, Pines J. Chfr acts with the p38 stress kinases to block entry to mitosis in mammalian cells. J Cell Biol. 2004;166:507–516. doi: 10.1083/jcb.200401139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rieder CL, Cole RW. Entry into mitosis in vertebrate somatic cells is guarded by a chromosome damage checkpoint that reverses the cell cycle when triggered during early but not late prophase. J Cell Biol. 1998;142:1013–1022. doi: 10.1083/jcb.142.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rieder CL, Salmon ED. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–318. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikhailov A, Cole RW, Rieder CL. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr Biol. 2002;12:1797–1806. doi: 10.1016/s0960-9822(02)01226-5. [DOI] [PubMed] [Google Scholar]
- 22.Deckbar D, Birraux J, Krempler A, Tchouandong L, Beucher A, Walker S, et al. Chromosome breakage after G2 checkpoint release. J Cell Biol. 2007;176:749–755. doi: 10.1083/jcb.200612047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giunta S, Belotserkovskaya R, Jackson SP. DNA damage signaling in response to double-strand breaks during mitosis. J Cell Biol. 2010;190:197–207. doi: 10.1083/jcb.200911156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura AJ, Rao VA, Pommier Y, Bonner WM. The complexity of phosphorylated H2AX foci formation and DNA repair assembly at DNA double-strand breaks. Cell Cycle. 2010;9:389–397. doi: 10.4161/cc.9.2.10475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson G, Buhmann M, von Zglinicki T. DNA damage foci in mitosis are devoid of 53BP1. Cell Cycle. 2009;8:3379–3383. doi: 10.4161/cc.8.20.9857. [DOI] [PubMed] [Google Scholar]
- 26.Poon RY. Mitotic phosphorylation: breaking the balance of power by a tactical retreat. Biochem J. 2007;403:5–7. doi: 10.1042/BJ20070290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jullien D, Vagnarelli P, Earnshaw WC, Adachi Y. Kinetochore localisation of the DNA damage response component 53BP1 during mitosis. J Cell Sci. 2002;115:71–79. doi: 10.1242/jcs.115.1.71. [DOI] [PubMed] [Google Scholar]
- 28.Plans V, Guerra-Rebollo M, Thomson TM. Regulation of mitotic exit by the RNF8 ubiquitin ligase. Oncogene. 2008;27:1355–1365. doi: 10.1038/sj.onc.1210782. [DOI] [PubMed] [Google Scholar]
- 29.Tuttle RL, Bothos J, Summers MK, Luca FC, Halazonetis TD. Defective in mitotic arrest 1/ring finger 8 is a checkpoint protein that antagonizes the human mitotic exit network. Mol Cancer Res. 2007;5:1304–1311. doi: 10.1158/1541-7786.MCR-07-0388. [DOI] [PubMed] [Google Scholar]
- 30.Guerrero AA, Gamero MC, Trachana V, Futterer A, Pacios-Bras C, Diaz-Concha NP, et al. Centromere-localized breaks indicate the generation of DNA damage by the mitotic spindle. Proc Natl Acad Sci USA. 2009;107:4159–4164. doi: 10.1073/pnas.0912143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reini K, Uitto L, Perera D, Moens PB, Freire R, Syvaoja JE. TopBP1 localises to centrosomes in mitosis and to chromosome cores in meiosis. Chromosoma. 2004;112:323–330. doi: 10.1007/s00412-004-0277-5. [DOI] [PubMed] [Google Scholar]
- 32.Zhang S, Hemmerich P, Grosse F. Centrosomal localization of DNA damage checkpoint proteins. Journal of cellular biochemistry. 2007;101:451–465. doi: 10.1002/jcb.21195. [DOI] [PubMed] [Google Scholar]
- 33.Tsvetkov L, Xu X, Li J, Stern DF. Polo-like kinase 1 and Chk2 interact and co-localize to centrosomes and the midbody. J Cell Biol. 2003;278:8468–8475. doi: 10.1074/jbc.M211202200. [DOI] [PubMed] [Google Scholar]
- 34.Hsu LC, White RL. BRCA1 is associated with the centrosome during mitosis. Proc Natl Acad Sci USA. 1998;95:12983–12988. doi: 10.1073/pnas.95.22.12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tritarelli A, Oricchio E, Ciciarello M, Mangiacasale R, Palena A, Lavia P, et al. p53 localization at centrosomes during mitosis and postmitotic checkpoint are ATM-dependent and require serine 15 phosphorylation. Mol Biol Cell. 2004;15:3751–3757. doi: 10.1091/mbc.E03-12-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia Z, Morales JC, Dunphy WG, Carpenter PB. Negative cell cycle regulation and DNA damage-inducible phosphorylation of the BRCT protein 53BP1. J Biol Chem. 2001;276:2708–2718. doi: 10.1074/jbc.M007665200. [DOI] [PubMed] [Google Scholar]
- 37.Shiromizu T, Goto H, Tomono Y, Bartek J, Totsukawa G, Inoko A, et al. Regulation of mitotic function of Chk1 through phosphorylation at novel sites by cyclin-dependent kinase 1 (Cdk1) Genes Cells. 2006;11:477–485. doi: 10.1111/j.1365-2443.2006.00955.x. [DOI] [PubMed] [Google Scholar]
- 38.Kramer A, Mailand N, Lukas C, Syljuasen RG, Wilkinson CJ, Nigg EA, et al. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol. 2004;6:884–891. doi: 10.1038/ncb1165. [DOI] [PubMed] [Google Scholar]
- 39.Peddibhotla S, Lam MH, Gonzalez-Rimbau M, Rosen JM. The DNA damage effector checkpoint kinase 1 is essential for chromosome segregation and cytokinesis. Proc Natl Acad Sci USA. 2009;106:5159–5164. doi: 10.1073/pnas.0806671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zachos G, Black EJ, Walker M, Scott MT, Vagnarelli P, Earnshaw WC, et al. Chk1 is required for spindle checkpoint function. Dev Cell. 2007;12:247–260. doi: 10.1016/j.devcel.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coster G, Hayouka Z, Argaman L, Strauss C, Friedler A, Brandeis M, et al. The DNA damage response mediator MDC1 directly interacts with the anaphase-promoting complex/cyclosome. J Biol Chem. 2007;282:32053–32064. doi: 10.1074/jbc.M705890200. [DOI] [PubMed] [Google Scholar]
- 42.Townsend K, Mason H, Blackford AN, Miller ES, Chapman JR, Sedgwick GG, et al. Mediator of DNA damage checkpoint 1 (MDC1) regulates mitotic progression. J Biol Chem. 2009;284:33939–33948. doi: 10.1074/jbc.M109.009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Vugt MA, Gardino AK, Linding R, Ostheimer GJ, Reinhardt HC, Ong SE, et al. A mitotic phosphorylation feedback network connects Cdk1, Plk1, 53BP1 and Chk2 to inactivate the G(2)/M DNA damage checkpoint. PLoS Biol. 2010;8:1000287. doi: 10.1371/journal.pbio.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298:1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- 45.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stobbe CC, Park SJ, Chapman JD. The radiation hypersensitivity of cells at mitosis. Int J Radiat Biol. 2002;78:1149–1157. doi: 10.1080/09553000210166570. [DOI] [PubMed] [Google Scholar]
- 47.Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, et al. Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol. 2007;178:1101–1108. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Downs JA, Kosmidou E, Morgan A, Jackson SP. Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol Cell. 2003;11:1685–1692. doi: 10.1016/s1097-2765(03)00197-7. [DOI] [PubMed] [Google Scholar]
- 49.Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maynard S, Swistowska AM, Lee JW, Liu Y, Liu ST, Da Cruz AB, et al. Human embryonic stem cells have enhanced repair of multiple forms of DNA damage. Stem Cells. 2008;26:2266–2274. doi: 10.1634/stemcells.2007-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goodarzi AA, Noon AT, Jeggo PA. The impact of heterochromatin on DSB repair. Biochem Soc Trans. 2009;37:569–576. doi: 10.1042/BST0370569. [DOI] [PubMed] [Google Scholar]