Abstract
The spatial and temporal organization of molecules within a cell is critical for coordinating the many distinct activities carried out by the cell. In an increasing number of biological signaling processes, scaffold proteins have been found to play a central role in physically assembling the relevant molecular components. Although most scaffolds use a simple tethering mechanism to increase the efficiency of interaction between individual partner molecules, these proteins can also exert complex allosteric control over their partners and are themselves the target of regulation. Scaffold proteins offer a simple, flexible strategy for regulating selectivity in pathways, shaping output behaviors, and achieving new responses from preexisting signaling components. As a result, scaffold proteins have been exploited by evolution, pathogens, and cellular engineers to reshape cellular behavior.
Introduction
Mammalian cells contain an estimated one billion individual protein molecules, with as many as 10 percent of these involved in signal transduction (1). Given this enormous number of molecules, it seems remarkable that cells can accurately process the vast array of signaling information they constantly receive. How can signaling proteins find their correct partners, and avoid their incorrect partners, among so many other proteins?
A principle that has emerged over the last two decades is that cells achieve specificity in their molecular signaling networks by organizing discrete subsets of proteins in space and time (Fig. 1A) (2–4). For example, functionally interacting signaling components can be sequestered into specific subcellular compartments (e.g. organelles) or at the plasma membrane. Another solution is to assemble functionally interacting proteins into specific complexes. Over 15 years ago, the first scaffold proteins were discovered – proteins that coordinate the physical assembly of components of a signaling pathway or network (5–10). These proteins have captured the attention of the signaling field because they appear to provide a simple and elegant solution for determining the specificity of information flow in intracellular networks (2, 11, 12). We review our current understanding of these organizational proteins: the types of pathways that they coordinate, the ways that they shape signaling responses, their biochemical and structural mechanisms, and how they are used in evolution or engineering to change signaling behaviors.
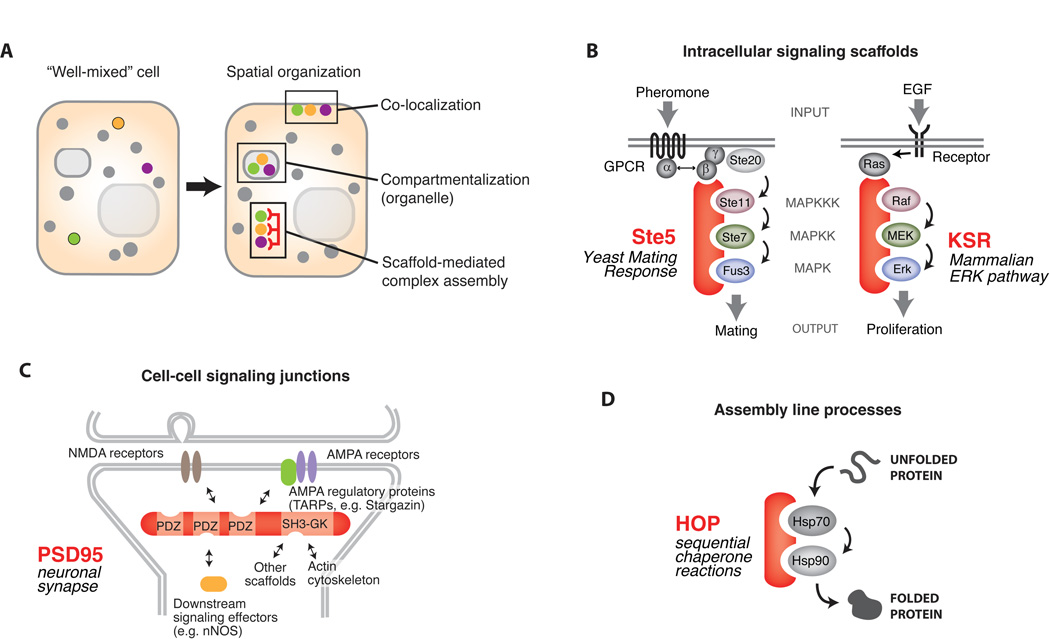
Fig. 1. Scaffold proteins organize cellular information flow.
A) Spatial organization is necessary to achieve high fidelity intracellular information transfer. Proteins can be assembled into specific complexes by compartmentalization (organelle targeting), membrane localization, and by scaffold proteins. B) Intracellular signaling pathways often use scaffold proteins. Canonical examples include Ste5, essential to the yeast mating MAPK pathway, and KSR, which directs signaling in the mammalian Ras-Raf-MEK-MAPK pathway. C) Scaffold proteins also play an important role in organizing cell-cell communication junctions, such as neuronal synapses. The PDZ scaffold, PSD-95, controls NMDA and AMPA glutamate receptor targeting to the synapse. D) Assembly line processes such as protein folding use scaffold proteins. The HOP protein promotes transfer of unfolded proteins between Hsp70 and Hsp90 chaperones.
Scaffolds Proteins: Versatile Tools to Assemble Diverse Pathways
Scaffolds are extremely diverse proteins, many of which are likely to have evolved independently. Nevertheless, they are conceptually related in that they are usually composed of multiple modular interaction domains or motifs (for example, see Box 1A). Their exact domain composition and order, however, can vary widely depending on the pathway that they organize.
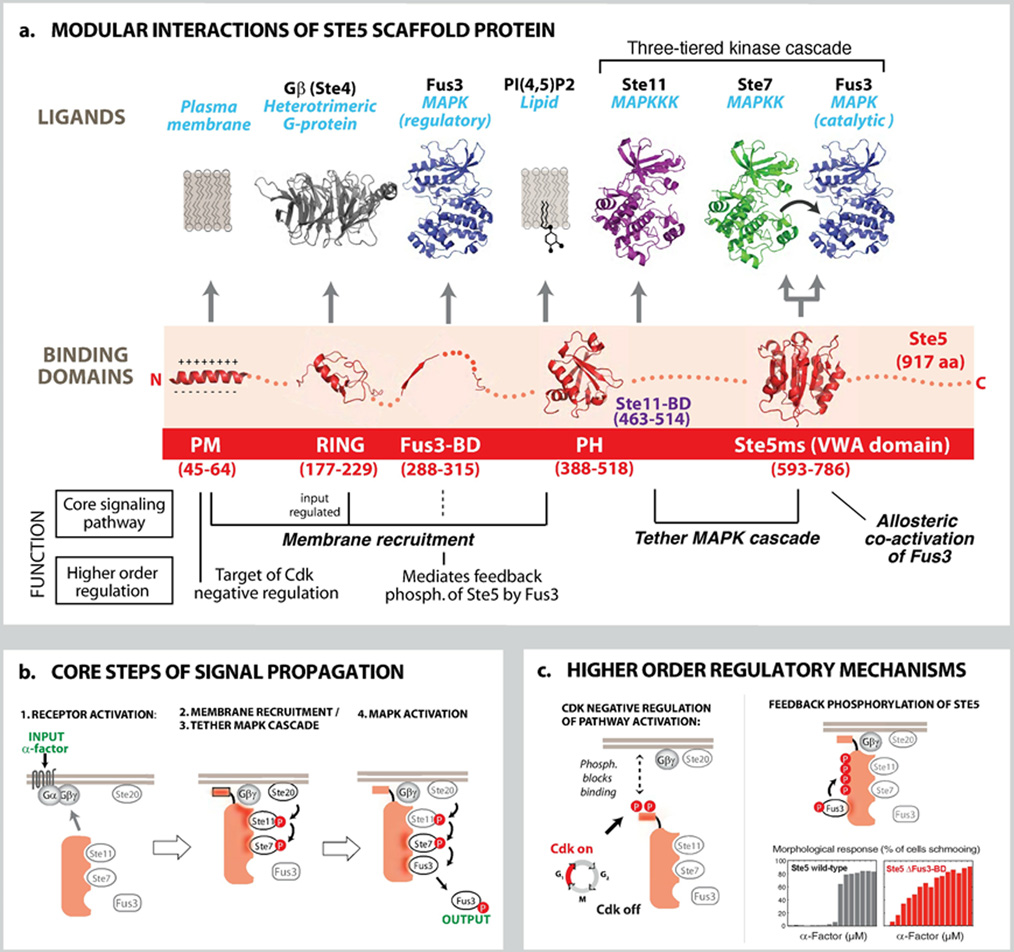
Box 1. Structure, Interactions and Mechanisms of Ste5: MAPK Pathway Scaffold Protein.
In some cases, homologous individual interaction motifs can be found in scaffolds associated with particular signaling proteins. For example, the AKAPs (A-kinase anchoring proteins), which link protein kinase A (PKA) to diverse signaling processes, all share a common short peptide motif that binds to the regulatory subunit of PKA (4). However, the other domains in individual AKAPs are highly variable, depending on what inputs and outputs the scaffold protein coordinates with PKA. Thus, scaffold proteins are flexible platforms assembled through mixing and matching of interaction domains.
There are a number of examples of convergent scaffold evolution. For example, the Ste5 protein in yeast and the kinase suppressor of Ras (KSR) protein in mammals (Fig. 1B) both coordinate mitogen-activated protein kinase (MAPK) pathways, but do not appear to be related in sequence. The Ste5 and KSR scaffold proteins both carry out a similar set of functions: they physically assemble the individual kinases that make up their cognate MAPK cascades (as well as upstream regulators) (Fig. 1B) (5, 7, 8), they control MAPK pathway localization (e.g. membrane anchoring) (8, 13, 14), they can insulate MAPK signaling proteins from competing inputs, such as components from functionally distinct MAPK pathways (15, 16), and they are required for efficient signaling (17, 18). Box 1 summarizes the mechanisms used by the Ste5 scaffold protein to control yeast MAPK signaling. Thus different molecular implementations of scaffolds can be used to play a similar role in signaling.
Box 1. Structure and Mechanisms of a Canonical Scaffold: the MAPK Scaffold Protein Ste5.
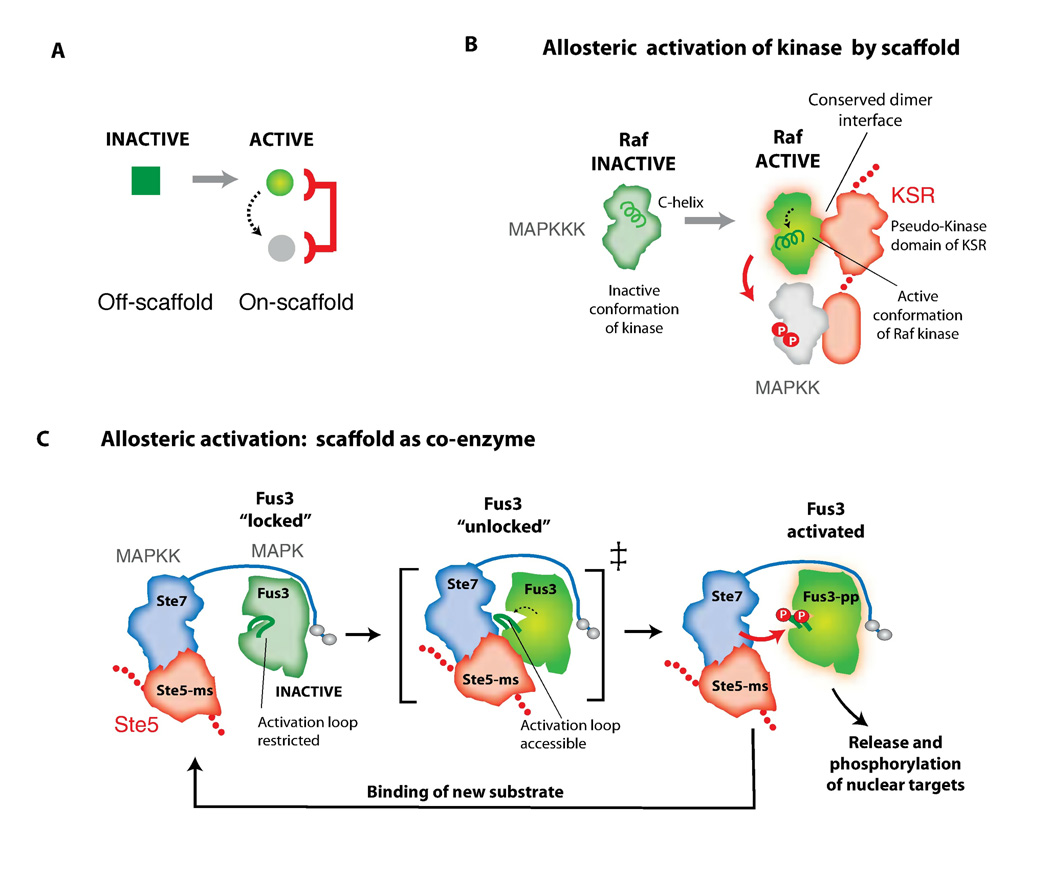
A) The Ste5 scaffold protein is composed of modular interaction domains, some of which mediate essential steps in the three-tiered mating MAPK signaling cascade, and some of which function in higher order regulatory behaviors. B) Core steps of mating pathway (animated in online interactive figure): Binding of α-factor peptide to its receptor (Ste2) leads to activation of the guanine nucleotide binding protein (G-protein), and dissociation of Gβγ (Ste4 and Ste18) from the Gα subunit (Gpa1). The Ste5 RING domain binds to the free Gβγ complex (52, 53), triggering rapid recruitment of the scaffold to the membrane. Stabilization and discrete localization of Ste5 at the plasma membrane also requires the interaction of its PM domain (an amphipathic helix) (14, 25) and a cryptic plekstrin homology (PH) domain with the lipid bilayer or anchored phosphoinositides (13). A region on Ste5 that overlaps with the PH domain binds to the MAPKKK Ste11 (5, 54), and upon pathway activation, co-localizes the MAPKKK Ste11 with its activator, Ste20 (a MAPKKKK, similar to the p21-activated kinase (PAK)), which is localized to the membrane in a pre- activated state. Phosphorylation of the MAPKKK Ste11 by Ste20 initiates the MAPK cascade. The MAPKK Ste7 is assembled into the mating signaling complex through the VWA domain of Ste5 (3FZE.pdb), and can be efficiently phosphorylated by the co-localized and activated MAPKKK Ste11. The minimal VWA also functions as a co-activator that permits Ste7 activation of the MAPK Fus3, which is tethered to Ste7 via docking motifs (18, 55) (see Fig. 4c). C) The Ste5 scaffold also takes part in higher order regulatory processes. Phosphorylation of the PM helix by Cdk blocks Ste5 membrane-binding (25), thus preventing activation of the mating response at specific stages of the cell cycle. The Fus3-binding domain (Fus3-BD) (2F49.pdb) appears to be important for regulatory feedback phosphorylation of Ste5 by Fus3, rather than for core signal transmission through the MAPK cascade (27). Phosphorylation of at least 4 sites on the Ste5 scaffold is dependent on recruitment of Fus3 though the Fus3-BD. These regulatory phosphorylation sites on the scaffold inhibit pathway activity and are thought to help shape the ultrasensitive cell morphology response (shmooing) that occurs during mating. Mutation of the Fus3-BD does not prevent mating but rather leads to a much more graded shmooing response when stimulated by α-factor (27, 28, 39). Thus this regulatory interaction may shape this switch-like cell-fate decision. Structures not described by Protein Data Bank (PDB) numbers in (B & C) were created using homology models. (56)
Scaffold proteins that direct intracellular signaling are not limited to coordinating kinase cascades - they can organize other classes of molecules, such as those involved in guanosine triphosphate hydrolyase (GTPase) signaling. For example, the yeast protein Bem1 directs the interaction between the guanine nucleotide exchange factor (GEF) Cdc24 and its downstream GTPase substrate, Cdc42 (19). Such coordinated GTPase regulation controls precise morphological behaviors such as polarized budding of yeast cells.
Scaffold proteins can also coordinate communication at cell-cell signaling junctions, such as neuronal synapses. Synaptic scaffolds, such as the PDZ domain containing protein, PSD-95, contain a set of domains that bind to neuronal receptors (such as N-methyl-D aspartate [NMDA] receptors), to other scaffold proteins, and to the actin cytoskeleton. They build preformed assemblies that are precisely anchored at sites of cell-cell contact (e.g. the post-synaptic density), thus allowing cells to respond efficiently to stimuli (e.g. neurotransmitter release from the partner cell) (20). PSD-95 also helps determine the output of synaptic activation by co-localizing key downstream effectors, such as neuronal nitric oxide synthase (nNOS) (which is activated by local calcium influx upon NMDA receptor activation) (Fig. 1C). These scaffolds also mediate key functional changes at the synapse – they coordinate the stimulus-induced recruitment of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors to synapses, a process thought to be required for long-term potentiation and memory. These critical regulatory changes at properly stimulated synapses are mediated by direct interaction of PSD-95 and other scaffolds with a class of proteins known as transmembrane AMPA receptor regulatory proteins (TARPs) (21). The scaffold proteins that function in cell-cell communication not only coordinate the signaling molecules that they interact with, but also target or anchor the complexes at the appropriate cellular location for receiving specific inputs. For example, the related PDZ domain scaffold, InaD, organizes the visual response cascade in the fruit fly (9, 22).
Scaffold proteins can also coordinate assembly-line processes – situations where a molecule must be passed from one partner to another in a sequential manner. Proper folding of some client proteins, such as steroid hormone receptors, appears to require a sequential handoff from Hsp70 to Hsp90 chaperones, a process that is coordinated by the scaffold, Hsp70 and Hsp90 Organizing Protein (HOP) (Fig. 1D) (23). Thus, scaffold proteins can have functions in controlling the flow of different classes of biological information that extend beyond what is traditionally considered signal transduction.
Circuit Boards for Wiring Pathway Connectivities and Shaping Pathway Responses
Scaffold proteins can be thought of as molecular circuit boards that can organize a wide variety of circuit relationships between signaling proteins (Fig. 2A). Despite their diversity in detailed molecular implementation, there appear to be common themes to the types of functional circuit topologies organized by scaffold proteins.
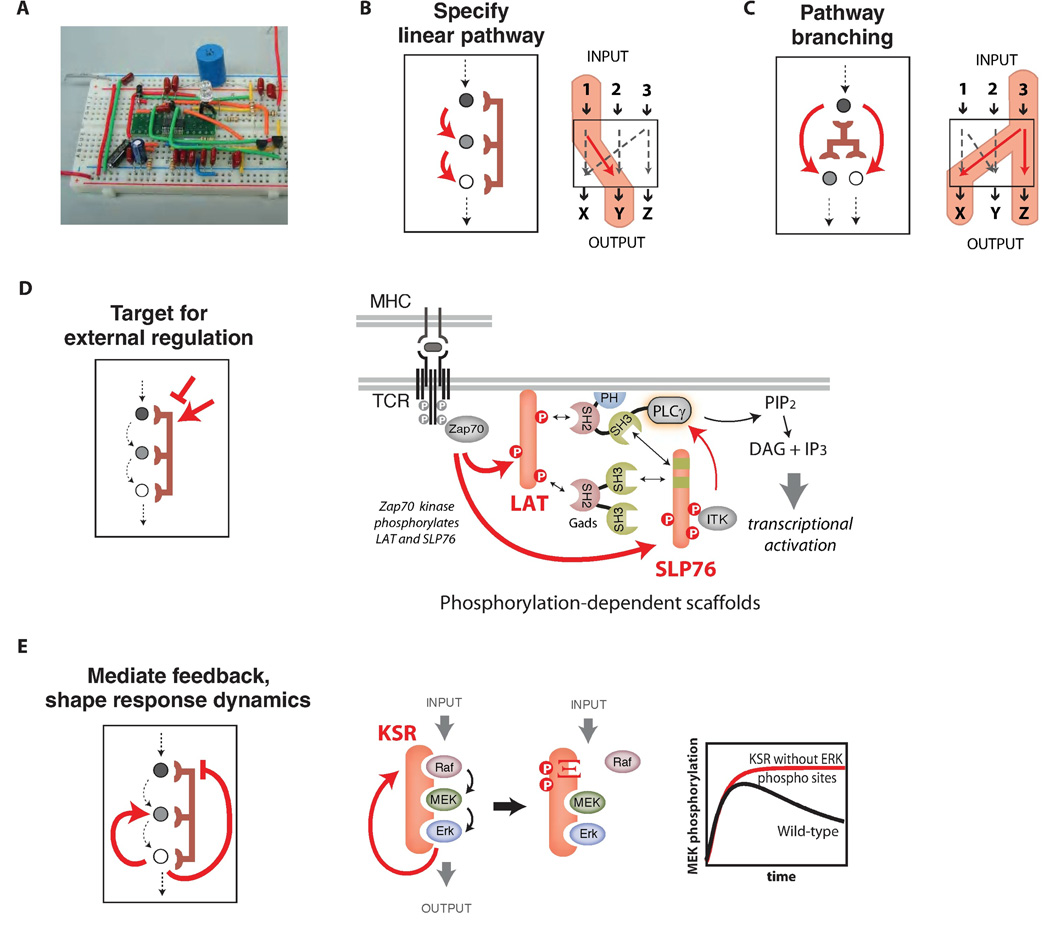
Fig. 2. Scaffold proteins can mediate pathway regulation and feedback to shape complex signaling responses.
A) Scaffold proteins are analogous to circuit boards - modular platforms that wire together components and direct the flow of information - and can program complex signaling behaviors. B) Scaffold proteins function to wire pathway input and output through alternative possible routes. C) Scaffold proteins can mediate branching of pathways to multiple outputs. D) Scaffold proteins are themselves the targets of regulation. In T-cell signaling, activation of the T-cell receptor causes phosphorylation of the LAT and Slp76 scaffolds by Zap70, and phosphorylation-dependent recruitment of substrates leads to PLCγ activation and PIP2 hydrolysis. E) Scaffold proteins can be the target of feedback phosphorylation that tunes pathway responses. Feedback phosphorylation of the KSR scaffold by activated ERK blocks Raf (MAPKKK) binding and attenuates MEK activation, thereby decreasing pathway output. Plot adapted from (24).
The conceptually simplest scaffold proteins determine a specific linear input-output pathway among a set of potential partner proteins (Fig. 2B). Scaffold proteins can also mediate pathway branching - fanning out of signaling information to multiple outputs that are part of the assembled complex (Fig. 2C).
In an increasing number of cases, scaffold proteins are direct targets for regulation – pathways can be turned on or off by inputs that modify the scaffold protein rather than the actual signaling enzymes (Fig. 2D) (24, 25). One of the clear benefits of using scaffold proteins to organize signaling complexes is that protein recruitment, and thus pathway function, can be easily regulated by external signals that modify association of other proteins with the scaffold. The scaffold proteins LAT and SLP-76, which help organize T cell signaling (Fig. 2D), provide an elegant example of this positive regulation. Under basal conditions, LAT and SLP-76 do not assemble active signaling complexes. However, upon T cell activation, the tyrosine kinase Zap70 is activated and recruited to the T cell receptor and phosphorylates a number of tyrosine motifs within the scaffold proteins LAT and SLP-76. These phosphorylated sites act as docking motifs for several Src Homology 2 (SH2) domain-containing proteins (10, 26). The phosphorylation-induced assembly of this complex triggers the major downstream pathways of T cell activation.
Scaffold phosphorylation can also be inhibitory and block protein-protein and protein-lipid interactions. Phosphorylation of the yeast Ste5 scaffold protein by the cell-cycle regulated kinase, cyclin-dependent kinase 1 (Cdk1), blocks association of the scaffold with the plasma membrane, thereby specifically attenuating mating signaling after cells have committed to the ‘Start’ (G1-S transition) of the cell cycle (Box 1C) (25).
Finally, perhaps the most sophisticated role of scaffold proteins is to coordinate complex feedback loops in signaling pathways by, for example, coordinating mechanisms that can turn off the pathway (Fig. 2E) (27, 28). In these cases, the scaffold appears to play a central role in precisely shaping signaling response properties, such as dynamics or dose-response.
For example, the KSR scaffold protein assembles a three-tiered MAP kinase pathway in mammalian cells, and activation of the terminal MAPK creates a feedback loop that phosphorylates the KSR scaffold and the MAPKKK Raf (Fig. 2E). These modifications disrupt binding of the MAPKKK to the KSR scaffold and shut down pathway activation (Fig. 1D) (24). Mutation of the KSR phosphorylation sites results in dissociation of the scaffold from the plasma membrane and abnormal pathway dynamics, including sustained pathway activation.
A recurring theme is that scaffold proteins increase the flexibility of a cell’s signaling responses. Scaffold proteins can serve as targets for many forms of regulatory modulation, which allows the cell to achieve a wide range of behaviors from a limited set of components.
Molecular Mechanisms of Scaffold Proteins: Tethering, Orientation, and Allosteric Regulation
How do scaffolds physically direct communication between the appropriate signaling partners? The most primitive scaffold proteins likely exert their effects through simple tethering of partner molecules. Tethering increases the effective concentration of enzymes and their substrates (Fig. 3A). For an enzyme that brings together two small-molecule substrates, the effective concentration may be as large as 108 M (29). This large effect is a consequence of avoiding the entropic penalty (the loss of translational and rotational degrees of freedom) for the molecules finding one another in solution, made possible by an enzyme that binds and prepositions its substrates. Similarly, a scaffold protein that binds and orients two weakly-interacting protein substrates is expected to provide a large entropic advantage. Theoretical and experimental estimates for the entropic penalty for bringing two protein molecules together vary widely, however (30, 31). The length and flexibility of the scaffold tether will also affect reaction rates, and these factors are only beginning to be systematically explored.
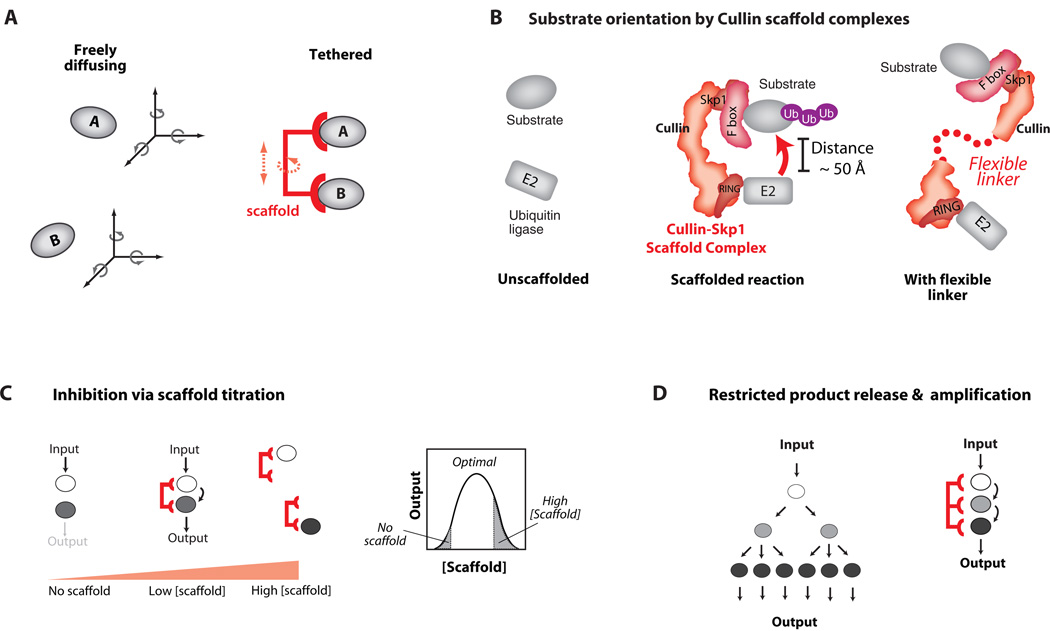
Fig. 3. Benefits and Costs of Scaffold Tethering Mechanisms.
A) By co-localizing enzyme and substrate, scaffold proteins can lower the entropic cost of signaling interactions – the loss of independent translational and rotational degrees of freedom is paid through binding interactions with the scaffold. The size of the advantage gained depends on the flexibility of the scaffold structure. B) By restricting the conformational freedom of interacting proteins, scaffolds can orient these molecules to enhance the rate of signal transfer. The rigid Cullin scaffold proteins tether E2 ubiquitin conjugating enzymes and their substrates. If the Cullin backbone is made flexible by mutation, the rate of substrate ubiquitination is greatly decreased. C) Tethering has potential drawbacks: at high concentrations scaffolds may titrate enzyme and substrate away from one another. D) Increased affinities can restrict substrate release and diffusion throughout the cell, potentially limiting signal amplification and spatial redistribution (e.g. nuclear localization).
Scaffold proteins that direct protein ubiquitination appear to function in part through properly orienting target proteins with upstream enzymes. Efficient ubiquitination of target substrates for proteosomal degradation requires the Cullin-RING-F Box complex, which acts as a scaffold to tether the target substrate and the E2 ubiquitin-conjugating enzyme together. However, simple tethering alone is not sufficient to stimulate substrate ubiquitination - the Cullin scaffold backbone must be rigid to function properly (Fig. 3B) (32). Mutations that introduce flexibility into the scaffold greatly limit substrate ubiquitination (33), presumably because of loss of orientational specificity.
Although nearly all evidence suggests that simple enforced proximity is an important mechanism for scaffold proteins, there are functional tradeoffs that emerge when only this type of mechanism is used for wiring the interactions of signaling components. For example, a simple tethering scaffold can exhibit concentration-dependent titration effects. Mathematical models predict that increasing the concentration of scaffold protein will first favor increased interaction between partner proteins, but then, at higher concentrations (in excess of the component partner proteins), will titrate partner proteins into separate complexes, thus inhibiting their interaction (Fig. 3C) (34). Some hint of this biphasic effect has been demonstrated experimentally for the scaffold protein Ste5 in the yeast mating MAPK pathway response (35), although the degree of inhibition at high scaffold protein expression is small. Inhibition of JNK MAPK signaling due to over-expression of the JNK interacting protein 1 (JIP1) scaffold also demonstrates the biphasic effect (36).
A second potential tradeoff in using a simple tethering scaffold is the potential reduction in signal amplification. Many pathways are thought to amplify input signals by having multiple stages of signal transfer, as in a kinase cascade. Since each enzyme can modify many substrates, a single input signal (such as a peptide or hormone binding to a receptor) can be converted through a three-tiered kinase cascade into a greatly magnified output response. But if a tethering scaffold protein is required for the cascade, substrate turnover and signal amplification could be dramatically reduced (Fig. 3D) (37), assuming binding to individual components is tight and dissociation is slow. A related problem is that high-affinity tethering could prevent the diffusion of substrates away from their site of activation.
In principle, scaffold proteins could use more sophisticated mechanisms to overcome the tradeoffs of tethering. Cooperative or allosteric assembly of pathway components to the scaffold, for example, could mitigate the biphasic effect. Biochemical and structural studies of scaffold proteins are beginning to reveal that these proteins often utilize allosteric mechanisms that can minimize the tradeoffs of tethering (Fig. 4A). The MAPK scaffold protein KSR promotes signaling through the three-tiered ERK MAP kinase cascade through both tethering and allosteric mechanisms. In addition to co-localizing the MAPKKK Raf and the MAPKK MEK, the KSR scaffold protein also allosterically activates the kinase domain of Raf (Fig. 4B) (38). This allosteric effect of KSR is mediated by a kinase-like domain in KSR which binds to and activates Raf. This scaffold function appears to be distinct from canonical tethering because there is no indication that Raf activity biphasically decreases with increasing concentrations of KSR (38). Instead, binding of increased amounts of KSR to Raf monotonically increases the amount of active Raf enzyme. This allosteric mechanism may increase the efficiency of signal transmission relative to that which occurs simply through tethering the MAPKKK and MAPKK proteins together.As this review was in press, data was published showing that the KSR scaffold contains a functional kinase domain that is allosterically regulated by the MAPKKK Raf and whose kinase activity may play a role in signaling from Raf to the MAPKK MEK (Brennan et al 2011, Hu et al 2011).
Fig. 4. Allosteric regulation by scaffold proteins.
A) Scaffolds can allosterically modulate the conformation of enzymes and substrates to gate information flow. B) In MAPK ERK signaling, KSR can directly bind to the MAPKKK Raf and influence its activity toward the MAPKK MEK. The pseudokinase domain of KSR dimerizes with Raf, altering the conformation of the C-helix on Raf so that its kinase domain becomes catalytically active (thereby allowing Raf to phosphorylate MEK). C) The VWA domain of Ste5 promotes phosphorylation of the MAPK Fus3 by the MAPKK Ste7. The scaffold may unlock the activation loop of the MAPK Fus3 to make it a better substrate for MAPKK Ste7.
Allosteric regulation has also been observed for the yeast mating response MAPK scaffold protein, Ste5. A von Willebrand factor type A (VWA) domain in this scaffold protein is required to allosterically unlock the MAPK Fus3 so that it becomes a good substrate for the MAPKK Ste7 - in the absence of scaffold, activated Ste7 cannot phosphorylate the MAPK Fus3, although it is catalytically competent to phosphorylate other potential substrates (Fig. 4C and Box 1) (18). This scaffold mechanism may be important for signaling specificity in S. cerevisiae since the MAPKK Ste7 can be activated by inputs other than mating pheromone stimulation. The Ste5 scaffold VWA domain functions as a co-activator – it does not appear to modulate the association of the MAPKK Ste7 with the MAPK Fus3; instead it specifically enhances the phosphorylation of Fus3 by Ste7 (Fig. 4C). This type of allosteric control may be physiologically important for the pathway: because this mechanism does not appear to require tight binding between the scaffold and the MAPK Fus3, it may avoid problems associated with substrate release, thus allowing both signal amplification and MAPK translocation to the nucleus (18, 39).
Scaffolds can be used as platforms for redirecting information flow in evolution and engineering
Perhaps the most powerful feature of scaffold proteins is their potential to facilitate the evolution of new pathways: by having a separate, genetically-encoded entity controlling the interaction of signaling components, the creation of new or recombined scaffolds could provide a simple mechanism for linking pre-existing components in novel ways (2). The ability to recombine pathways and regulate signaling behaviors with scaffolds can be likened to how the modular architecture of promoters gives rise to the diverse transcriptional responses that differentiate cell and tissue types.
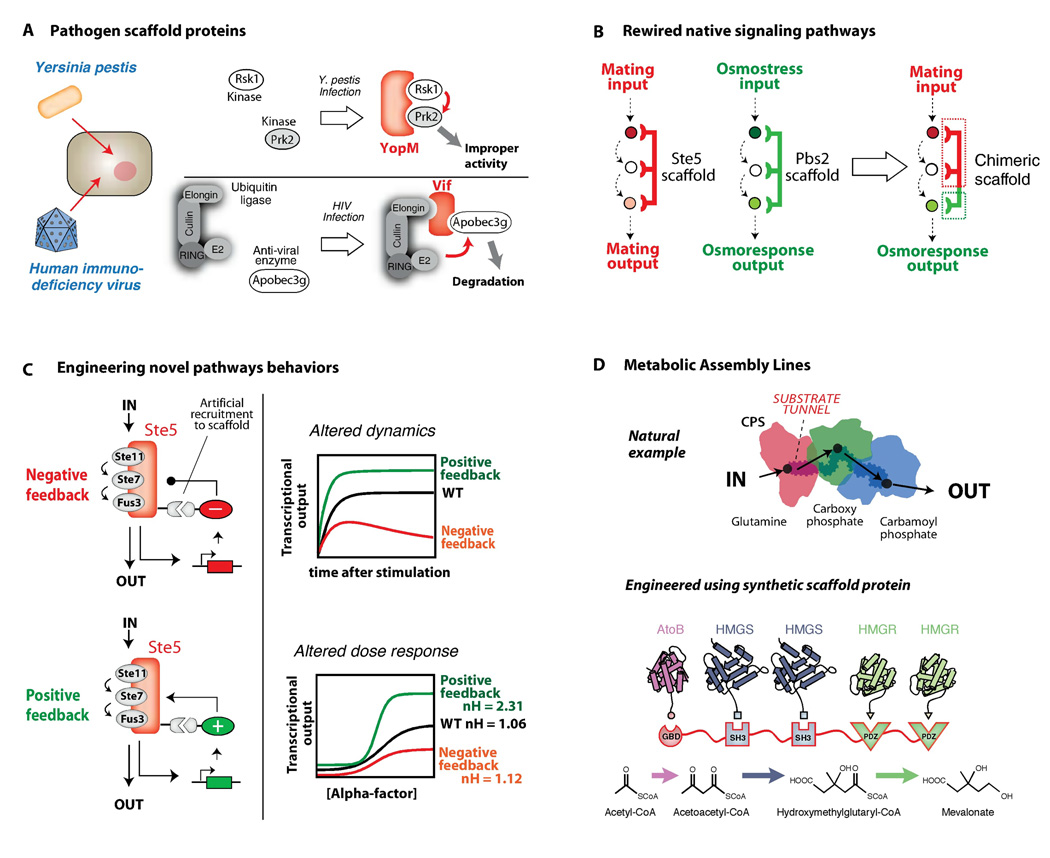
The modular structure of most scaffold proteins implies an evolutionary history involving recombination of interaction domains. Although we are just beginning to trace the evolution of scaffold proteins through genomic comparisons, one example of rapid evolution of new pathway linkages through scaffolds is in pathogen-host interactions. Pathogens often hijack and rewire host signaling pathways, and pathogen scaffold proteins have been described (Fig. 5A) (40). The pathogenic bacteria, Yersinia pestis, produces a scaffold-like protein (YopM) that artificially links together two kinases (Rsk1 and Prk2) that do not normally interact (Fig. 5A). YopM is secreted into host immune cells, and although the exact consequence of linking these two kinases is unclear, the binding and activation of the Rsk1 and Prk2 kinases by YopM is necessary for Y. pestis virulence (41, 42). Similarly, a number of viruses encode scaffold proteins that act to target specific host anti-viral proteins for ubiquitination and degradation. The human immunodeficiency virus (HIV) destroys the host APOBEC3G protein (a cytidine deaminase that interferes with viral replication) by producing a scaffold protein, Vif, which binds both APOBEC3G and a Cullin-E2 complex (43). A similar mechanism is used by the respiratory syncitial virus (RSV) to downregulate the host STAT2 protein (44).
Fig. 5. Scaffold proteins are modular and can be used as platforms for redirecting information in evolution and engineering.
A) Pathogens can use scaffold-like proteins to rewire host signaling responses. The YopM scaffold from Yersinia pestis forces the interaction of the host Rsk1 and Prk2 kinases. The inappropriate activation is necessary for virulence. Viral scaffold proteins, such as HIV Vif, can target antiviral host proteins, such as the cytidine deaminase APOBEC3G, for degradation by targeting them to Cullin-E2 ubiquitin ligases. B) Engineered scaffolds can direct new cell signaling behaviors. A chimera of the Ste5 and Pbs2 yeast MAPK scaffold proteins can redirect mating pathway input to osmolarity pathway output. C) Synthetic feedback loops can be engineered by controlling recruitment of positive and negative effectors to the Ste5 MAPK scaffold protein. Such loops can be used to precisely shape the dynamics and dose-response of the yeast mating MAPK pathway to produce a wide range of signaling behaviors. D) Natural metabolic pathways are often organized into multi-enzyme complexes that function like an assembly line to enhance the rate and yield of metabolite production. Engineered scaffold proteins can link together novel combinations of metabolic enzymes to more efficiently synthesize desired chemical products. Adapted from (51).
In principle, cellular engineers could also mimic evolution and wire new signaling pathways and cellular behaviors by building synthetic scaffolds. One simple approach to rewiring native signaling pathways is to fuse the functional elements of two different scaffold proteins into a chimeric scaffold protein. This strategy was successfully used in S. cerevisiae to link the mating and high osmolarity stress MAPK pathway scaffold proteins so that a mating input resulted in an osmolarity response (Fig. 5B) (45).
More complex signaling behaviors can theoretically be achieved by engineering recruitment of pathway regulators, thus generating feedback. A design strategy using modular recruitment of positive and negative effectors to the Ste5 scaffold generated feedback loops that result in a diverse array of MAPK pathway responses (46). By tethering components such as phosphatases to a scaffold-kinase signaling complex, the dynamics and dose-response behaviors of the yeast mating MAPK pathway were altered dramatically (Fig. 5C).
Like cellular signaling, metabolic pathways are composed of a series of enzymes that are often assembled into complexes. Although these assemblies might not be considered true scaffold complexes, they use the same principle of enforced proximity. Substrate channeling in carbamoyl phosphate synthase, polyketide synthase, and tryptophan synthase is used to prevent loss of low-abundance intermediates, to protect unstable intermediates from interacting with solvent, and to increase the effective concentration of reactants (Fig. 5D) (47, 48). Moreover, pathway flux can be controlled by regulating assembly of these complexes, just as signaling can be regulated by assembly of a scaffolded complex (49). Given these parallels, an important question to industry and medicine is whether metabolic enzymes can be wired together into functional assemblies by engineered scaffolds, to create designer metabolic pathways and small molecule products. Remarkably, an artificial scaffold protein that tethered together three enzymes in a synthetic metabolic pathway in E. coli was found to enhance mevalonate production by ~100-fold (Fig. 5D) (50). Thus, the principles of modular complex assembly can be used to flexibly control the flow of information in both signaling and metabolic pathways.
Conclusions
Scaffold proteins function in a diverse array of biological processes. Simple mechanisms such as tethering are layered with more sophisticated mechanisms like allosteric control so that scaffolds can precisely control the specificity and dynamics of information transfer. Scaffold proteins can also control the wiring of more complex network configurations – they can integrate feedback loops and regulatory controls to generate precisely controlled signaling behaviors.
The versatility of scaffold proteins comes from their modularity, which allows recombination of protein interaction domains to generate new signaling pathways. Cells use scaffolds to diversify signaling behaviors and to evolve new responses. Pathogens can create scaffold proteins that are to their advantage: their virulence depends on rewiring host signaling pathways to turn off or avoid host defenses. In the lab, scaffolds are being used to build new, predictable signaling or metabolic networks to program useful cellular behaviors.
REFERENCES
- 1.Milo R, Jorgensen P, Moran U, Weber G, Springer M. Nucleic Acids Res. Jan;38:D750. doi: 10.1093/nar/gkp889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharyya RP, Remenyi A, Yeh BJ, Lim WA. Annu Rev Biochem. 2006;75:655. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- 3.Kholodenko BN. Nat Rev Mol Cell Biol. 2006 Mar;7:165. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott JD, Pawson T. Science. 2009 Nov 27;326:1220. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi KY, Satterberg B, Lyons DM, Elion EA. Cell. 1994 Aug 12;78:499. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 6.Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Science. 1995 Sep 22;269:1737. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 7.Printen JA, Sprague GF., Jr Genetics. 1994 Nov;138:609. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Therrien M, Michaud NR, Rubin GM, Morrison DK. Genes Dev. 1996 Nov 1;10:2684. doi: 10.1101/gad.10.21.2684. [DOI] [PubMed] [Google Scholar]
- 9.Tsunoda S, et al. Nature. 1997 Jul 17;388:243. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- 10.Zhang W, Sloan-Lancaster J, Kitchen J, Trible RP, Samelson LE. Cell. 1998 Jan 9;92:83. doi: 10.1016/s0092-8674(00)80901-0. [DOI] [PubMed] [Google Scholar]
- 11.Morrison DK, Davis RJ. Annu Rev Cell Dev Biol. 2003;19:91. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 12.Shaw AS, Filbert EL. Nat Rev Immunol. 2009 Jan;9:47. doi: 10.1038/nri2473. [DOI] [PubMed] [Google Scholar]
- 13.Garrenton LS, Young SL, Thorner J. Genes Dev. 2006 Jul 15;20:1946. doi: 10.1101/gad.1413706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winters MJ, Lamson RE, Nakanishi H, Neiman AM, Pryciak PM. Mol Cell. 2005 Oct 7;20:21. doi: 10.1016/j.molcel.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Patterson JC, Klimenko ES, Thorner J. Sci Signal. 3:ra75. doi: 10.1126/scisignal.2001275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz MA, Madhani HD. Curr Genet. 2006 Jun;49:351. doi: 10.1007/s00294-006-0061-6. [DOI] [PubMed] [Google Scholar]
- 17.Flatauer LJ, Zadeh SF, Bardwell L. Mol Cell Biol. 2005 Mar;25:1793. doi: 10.1128/MCB.25.5.1793-1803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Good M, Tang G, Singleton J, Remenyi A, Lim WA. Cell. 2009 Mar 20;136:1085. doi: 10.1016/j.cell.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozubowski L, et al. Curr Biol. 2008 Nov 25;18:1719. doi: 10.1016/j.cub.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Funke L, Dakoji S, Bredt DS. Annu Rev Biochem. 2005;74:219. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- 21.Schnell E, et al. Proc Natl Acad Sci U S A. 2002 Oct 15;99:13902. doi: 10.1073/pnas.172511199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ranganathan R, Ross EM. Curr Biol. 1997 Dec 1;7:R770. doi: 10.1016/s0960-9822(06)00401-5. [DOI] [PubMed] [Google Scholar]
- 23.Taipale M, Jarosz DF, Lindquist S. Nat Rev Mol Cell Biol. Jul;11:515. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 24.McKay MM, Ritt DA, Morrison DK. Proc Natl Acad Sci U S A. 2009 Jul 7;106:11022. doi: 10.1073/pnas.0901590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strickfaden SC, et al. Cell. 2007 Feb 9;128:519. doi: 10.1016/j.cell.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin J, Weiss A. J Biol Chem. 2001 Aug 3;276:29588. doi: 10.1074/jbc.M102221200. [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharyya RP, et al. Science. 2006 Feb 10;311:822. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- 28.Malleshaiah MK, Shahrezaei V, Swain PS, Michnick SW. Nature. May 6;465:101. doi: 10.1038/nature08946. [DOI] [PubMed] [Google Scholar]
- 29.Page MI, Jencks WP. Proc Natl Acad Sci U S A. 1971 Aug;68:1678. doi: 10.1073/pnas.68.8.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finkelstein AV, Janin J. Protein Eng. 1989 Oct;3:1. doi: 10.1093/protein/3.1.1. [DOI] [PubMed] [Google Scholar]
- 31.Yu YB, Privalov PL, Hodges RS. Biophys J. 2001 Sep;81:1632. doi: 10.1016/S0006-3495(01)75817-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saha A, Deshaies RJ. Mol Cell. 2008 Oct 10;32:21. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng N, et al. Nature. 2002 Apr 18;416:703. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 34.Levchenko A, Bruck J, Sternberg PW. Proc Natl Acad Sci U S A. 2000 May 23;97:5818. doi: 10.1073/pnas.97.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chapman SA, Asthagiri AR. Mol Syst Biol. 2009;5:313. doi: 10.1038/msb.2009.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dickens M, et al. Science. 1997 Aug 1;277:693. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- 37.Locasale JW, Shaw AS, Chakraborty AK. Proc Natl Acad Sci U S A. 2007 Aug 14;104:13307. doi: 10.1073/pnas.0706311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, Therrien M. Nature. 2009 Sep 24;461:542. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 39.Maeder CI, et al. Nat Cell Biol. 2007 Nov;9:1319. doi: 10.1038/ncb1652. [DOI] [PubMed] [Google Scholar]
- 40.Selyunin AS, et al. Nature. Jan 6;469:107. [Google Scholar]
- 41.McCoy MW, Marre ML, Lesser CF, Mecsas J. Infect Immun. Jun;78:2584. doi: 10.1128/IAI.00141-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDonald C, Vacratsis PO, Bliska JB, Dixon JE. J Biol Chem. 2003 May 16;278:18514. doi: 10.1074/jbc.M301226200. [DOI] [PubMed] [Google Scholar]
- 43.Yu X, et al. Science. 2003 Nov 7;302:1056. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 44.Elliott J, et al. J Virol. 2007 Apr;81:3428. doi: 10.1128/JVI.02303-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park SH, Zarrinpar A, Lim WA. Science. 2003 Feb 14;299:1061. doi: 10.1126/science.1076979. [DOI] [PubMed] [Google Scholar]
- 46.Bashor CJ, Helman NC, Yan S, Lim WA. Science. 2008 Mar 14;319:1539. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- 47.Miles EW, Rhee S, Davies DR. J Biol Chem. 1999 Apr 30;274:12193. doi: 10.1074/jbc.274.18.12193. [DOI] [PubMed] [Google Scholar]
- 48.Thoden JB, Holden HM, Wesenberg G, Raushel FM, Rayment I. Biochemistry. 1997 May 27;36:6305. doi: 10.1021/bi970503q. [DOI] [PubMed] [Google Scholar]
- 49.An S, Kumar R, Sheets ED, Benkovic SJ. Science. 2008 Apr 4;320:103. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- 50.Dueber JE, et al. Nat Biotechnol. 2009 Aug;27:753. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 51.DeLisa MP, Conrado RJ. Nat Biotechnol. 2009 Aug;27:728. doi: 10.1038/nbt0809-728. [DOI] [PubMed] [Google Scholar]
- 52.Inouye C, Dhillon N, Thorner J. Science. 1997 Oct 3;278:103. doi: 10.1126/science.278.5335.103. [DOI] [PubMed] [Google Scholar]
- 53.Pryciak PM, Huntress FA. Genes Dev. 1998 Sep 1;12:2684. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inouye C, Dhillon N, Durfee T, Zambryski PC, Thorner J. Genetics. 1997 Oct;147:479. doi: 10.1093/genetics/147.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Remenyi A, Good MC, Bhattacharyya RP, Lim WA. Mol Cell. 2005 Dec 22;20:951. doi: 10.1016/j.molcel.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 56.We thank the following people for helpful discussions and comments: P. Pryciak, A. Weiss, R. Nicoll, R. Phillips, N. Pierce, J. Iwasa, J. Dueber, A. Remenyi, M. Borovinskaya, S. Peisajovich, J. Garbarino, A. Won, C. Bashor, N. Helman, S.H. Park, S. Coyle, P. Wei, J. Toettcher, and other members of the Lim Lab. This work was supported by a Miller Fellowship (M.C.G.), a Damon Runyan Fellowship (J.G.Z.), NIH grants RO1GM055040, RO1GM062583, PN2EY016546, P50GM081879 (W.A.L.) NSF Synthetic Biology and Engineering Research Center (W.A.L.), the Packard Foundation (W.A.L.), and the Howard Hughes Medical Institute (W.A.L.)