Abstract
Motivation: Accurate prediction of genes encoding small proteins (on the order of 50 amino acids or less) remains an elusive open problem in bioinformatics. Some of the best methods for gene prediction use either sequence composition analysis or sequence similarity to a known protein coding sequence. These methods often fail for small proteins, however, either due to a lack of experimentally verified small protein coding genes or due to the limited statistical significance of statistics on small sequences.
Our approach is based upon the hypothesis that true small proteins will be under selective pressure for encoding the particular amino acid sequence, for ease of translation by the ribosome and for structural stability. This stability can be achieved either independently or as part of a larger protein complex. Given this assumption, it follows that small proteins should display conserved local protein structure properties much like larger proteins. Our method incorporates neural-net predictions for three local structure alphabets within a comparative genomic approach using a genomic alignment of 22 closely related bacteria genomes to generate predictions for whether or not a given open reading frame (ORF) encodes for a small protein.
Results: We have applied this method to the complete genome for Escherichia coli strain K12 and looked at how well our method performed on a set of 60 experimentally verified small proteins from this organism. Out of a total of 11 407 possible ORFs, we found that 6 of the top 10 and 27 of the top 100 predictions belonged to the set of 60 experimentally verified small proteins. We found 35 of all the true small proteins within the top 200 predictions. We compared our method to Glimmer, using a default Glimmer protocol and a modified small ORF Glimmer protocol with a lower minimum size cutoff. The default Glimmer protocol identified 16 of the true small proteins (all in the top 200 predictions), but failed to predict on 34 due to size cutoffs. The small ORF Glimmer protocol made predictions for all the experimentally verified small proteins but only contained 9 of the 60 true small proteins within the top 200 predictions.
Contact: jsamayoa@jhu.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Increasingly, small proteins have been found to be important functional elements in cellular biology. Members of this class of molecules have been associated with a diverse set of functions including the regulation of amino acid metabolism (Yanofsky, 2000), iron-homeostasis (Sela, 2008), spore development (Cutting et al., 1997; Ruvolo et al., 2006) and antimicrobial activity (Gallo and Nizet, 2003). Despite the recent discoveries of functionally relevant small proteins, there is still relatively little known about how widespread and critical small proteins are. Furthermore, the advent of next-generation sequencing technologies has enabled transcriptional profiling of complete genome sequences (Cloonan and Grimmond, 2008). In the yeast genome, these efforts have led to the expansion of the transcriptome (Nagalakshmi et al., 2008). The problem of discriminating small protein-encoding sequences from other small RNAs will only increase as transcriptional profiling efforts expand.
Due to the lack of introns and alternative splicing mechanisms, prokaryotic organisms represent a unique setting for the elucidation of novel small proteins. Within this context, any open reading frame (ORF) is potentially a protein-encoding gene. For prokaryotic genomes, the most accurate way to predict a gene is via similarity to a protein in another genome. This technique is problematic, however, due to limited numbers of experimentally verified small proteins in sequence databases. Further complicating the problem is a contamination of sequence databases caused by the propagation of dubious ORF predictions via homology-based annotation efforts (Hemm et al., 2008).
In situations where there is no matching protein, sequence composition-based methods are traditionally used. There are several automated gene finders that fall into this category such as GLIMMER (Delcher et al., 1999; Salzberg et al., 1998), ORPHEUS (Frishman et al., 1998), GeneMark (Besemer and Borodovsky, 1999; Besemer et al., 2001; Lukashin and Borodovsky, 1998) and EasyGene (Larsen and Krogh, 2003; Nielsen and Krogh, 2005). GLIMMER uses interpolated Markov models to distinguish coding from non-coding DNA. The program combines first through eighth order Markov models and weights them by their predictive power. ORPHEUS begins by a database similarity search. The genes with matches in the database are then used to create a statistical profile of protein coding regions and ribosome binding sites. This profile is then used to make genome-wide predictions for protein-coding genes. The original GeneMark program used non-homogeneous Markov models to distinguish coding from non-coding sequences. The newer GeneMark-hmm program embeds the original GeneMark models into a hidden Markov model (HMM) framework. EasyGene uses sequences that match a protein in Swiss-Prot to estimate an HMM for a given genome. The HMM is then used to score putative genes.
The multivariate entropy distance (MED) algorithm combines a comprehensive statistical model of protein coding ORFs with a model of prokaryotic translation initiation sites (TISs) (Zhu et al., 2007). A novel feature of this algorithm is that the statistical model is based on a linguistic Entropy Distance Profile (EDP), which is inferred from observed amino acid probabilities. This profile is then used to map a given sequence within a 20-dimensional EDP phase space. Coding and non-coding sequences are then discriminated in part by how they cluster within this 20-dimensional space.
All the gene finders described above work on single genomes, taking little advantage of conservation signals available with comparative genomics. Howard Ochman investigated the ability to identify small bacterial proteins via a method that measured the ratio of nucleotide substitution rates between non-synonymous and synonymous mutation sites (Ochman, 2002). This method is based on a prior observation that among a set of closely related protein-coding sequences, divergence at synonymous sites is greater than at non-synonymous sites (Kumar and Gadagkar, 2000; Shimmin et al., 1997). Ochman's study only looked at previously annotated ORFs in bacterial genomes.
Small proteins represent a particularly difficult problem for all methods using sequence composition. Given that the ORFs are small, sequence composition analysis yields weak statistics, making it hard to discriminate a protein-encoding ORF from an ORF occurring due to chance. Genome annotators are often left with a difficult decision: to predict or not to predict. Using a large minimum size for predictions reduces the false positives but yields severe under-annotation for small proteins. Conversely, lower minimum sizes lead to over-annotation of prokaryotic genome sequences for small putative ORFs. An analysis in 2001 by Skovgaard et al. estimated that as much as 10% of the original annotation for the Escherichia coli genome published in 1997 was a result of over-annotation, particularly for small ORFs (Skovgaard et al., 2001). If using a minimum ORF size, the cutoff should probably be a function of GC content, as the expected length of ORFs in random DNA increases as GC content increases.
We have developed a method to discriminate true small protein-coding ORFs from an ensemble of all possible unannotated ORFs in a given genome, without using ORF length cutoffs. Our method combines traditional gene annotation techniques with a novel application of local protein structure prediction tools within a comparative genomic framework. The novel assumptions in our method are that ORFs encoding small proteins will be selected for structural stability of the protein and for high frequency codons in a given genome. Just as for larger proteins, we hypothesize that structural stability will be critical to protein function. Therefore, we should be able to recognize conservation for properties related to structural stability in multi-genome alignments of closely related organisms. We validated our method on the complete genome sequence of E.coli strain K12 MG1655. Approximately 60 small proteins have been annotated and experimentally validated for this organism, representing the largest repertoire of validated small proteins described thus far (Hemm et al., 2008). Somewhat surprisingly, a large proportion of these proteins have been found to be associated with membranes.
2 APPROACH
In order to determine whether a given sequence codes for a small protein, we begin with a multiple genome alignment for our target organism. We then use this alignment to generate scores for each sequence based on three categories of analysis. The first analysis we perform is to analyze the observed codon composition for a given sequence according to a log-odds score. We score each sequence for agreement with its genome's codon biases on long protein genes. Second, we analyze each sequence for protein-like conservation patterns in the multiple sequence alignment. We score an alignment of a homologous sequence to the target sequence according to a BLOSUM90 substitution matrix. We then compare this to the score of the target sequence aligned to itself. We expect homologous sequences that code for proteins to have a score similar to the target self-alignment score. Finally, we look for prediction strength and consistency among a set of local structure alphabets. For each sequence, we generate three independent predictions for a given local structure alphabet and measure their overall agreement. We hypothesize that sequences coding for a protein will generate more consistent predictions than sequences not coding for a protein. We combine these scores, for a set of positive and negative training examples, to generate a model which we use to predict on new sequences.
3 METHODS
3.1 Data compilation
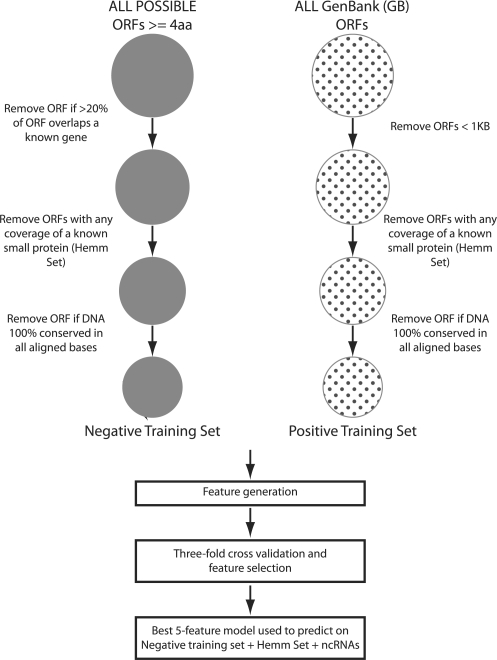
To take advantage of an existing wealth of prokaryotic comparative genomic data and analysis tools at UC Santa Cruz, we obtained all sequence data and gene annotation directly from the UCSC Microbial Genome Browser (http://microbes.ucsc.edu/) (Schneider et al., 2006). For this analysis (Fig. 1), we generated a set of all possible ORFs, four amino acids or longer, in the entire genome for E.coli K12. This set was then filtered to remove ORFs with >20% of their sequence overlapping an annotated gene in GenBank. We then removed any ORFs that shared any sequence with the experimentally verified set of 60 small proteins (Hemm et al., 2008) so that we could use them for validating the method. The final set of ORFs contained 12 514 sequences. Our assumption is that this set consists primarily of spurious ORFs and not true protein-coding genes and therefore can be used as a source of negative training examples (negative set). For a representative set of true protein encoding genes, we chose all genes annotated in GenBank as protein coding that were 1000 bases or longer. This list consisted of 1625 sequences (positive set). We also looked at all ncRNAs annotated in GenBank. This set consisted of 168 sequences. Because our positive set consisted entirely of long ORFs, and our negative set entirely of small ORFs, we needed to select features that are not length dependent. Otherwise, we could separate our training set trivially, without having any predictive value for finding small protein-coding genes.
Fig. 1.
Flow diagram starting from the generation of the negative and positive training sets through the feature generation, 3-fold cross-validation, and validation experiment.
3.2 Multiple alignment generation
To make predictions for E.coli K12, we started with a multiple-genome alignment including 15 unique E.coli strains and 7 other Enterobacteriaceae: Blochmannia floridanus, Buchnera aphidicola, Enterobacter 638, Salmonella enterica ATCC 9150, Salmonella enterica CT18, Shigella flexneri and Yersinia pestis. These genomes were selected on the basis of their relationship to E.coli K12. We included both closely and more distantly related species in our analysis, including the most distant genomes that could be easily aligned. The multiple genome alignment file (MA) used in this study was created using the program Threaded Blockset Aligner (TBA) (Blanchette et al., 2004), which used a phylogenetic tree derived from an analysis of 23S rRNA. One of the advantages of using TBA is the ability to make any organism in a multiple alignment the reference genome, ensuring a 1:1 alignment mapping for all regions in the reference genome. For our study, E.coli K12 was the reference genome.
All sequences that were perfectly conserved in the DNA multiple alignment were omitted from further analysis. The lack of any mutations inhibited measurement of protein-like conservation, thus removing a critical component of our study. This filter reduced the number of ORFs analyzed to 11 185 non-annotated ORFs, 1585 GenBank annotated ORFs greater than 1000 bases, 163 annotated ncRNAs and 59 experimentally validated small proteins.
3.3 Codon bias calculations
For each genome, we made two models of codon probabilities: one based on observed counts in all GenBank-annotated protein genes for that genome, and the other on the GC-richness of the genome (provided by the genome browser). We then made a log-odds scoring system for each codon c,
and averaged it over all codons in the ORF and over all aligned genomes. This codon-bias term measures selection for high expression and common amino acids in each genome, and turned out to be our strongest single predictor of protein-encoding ORFs. In order to investigate the impact of MA data on the method's performance, we also calculated the codon bias in the absence of any alignment data, using only the data for E.coli K12.
3.4 Amino acid conservation and BLOSUM-loss
To look for protein-like conservation, we converted the nucleotide alignments to amino acid alignments. For each column in the multiple alignment, we scored each pairwise target-sequence-to-homolog-sequence alignment according to a BLOSUM90 substitution matrix. We then computed a weighted average for this target-to-homolog score across all homologs in the alignment. To measure protein conservation in a given alignment column, we compared the weighted average for the target-to-homolog score to the BLOSUM90 score for the target sequence aligned to itself:
| (1) |
where h ranges over all homologs in the alignment, Wh is a weight for each homolog, i is the amino acid in the target sequence for the column, j is the amino acid in sequence h, Sij is the target-to-homolog BLOSUM90 score and Sii is the target-to-self BLOSUM90 score. Because we were specifically interested in mutations that are consistent with protein coding, we averaged these column scores over all codon positions that had at least one base substitution in at least one genome, ignoring codon positions which were invariant over all genomes or had only indel changes. We are more interested in whether the conservation is protein-like than how much conservation there is, so including invariant positions would just dilute our signal. This choice turned out to be important, as performance was substantially worse when other positions were included in the average (data not shown). Furthermore, ORFs which only contained silent mutations, i.e. synonymous substitutions, were excluded from the training data. Silent mutations are the most extreme type of protein-like conservation and usually are a good signature of protein-coding genes. However, given the lengths of the ORFs being evaluated in this study, we thought these instances would more likely be artifacts than actual examples of high protein conservation. In fact, many of the excluded ORFs were instances of a perfectly conserved sequence except for a single synonymous base substitution. To ensure the robustness of the method however, predictions for ORFs with only a silent mutation were included in both the validation experiment and the side-by-side Glimmer comparison.
The homolog-specific weight Wh was set to 1 minus the computed DNA sequence identity between h and the target sequence. Therefore, sequences that are very closely related had their scores down-weighted while scores from more distantly related sequences were given a higher weight (See Supplementary Materials for a complete list of weights).
According to this scoring scheme, positions that are perfectly conserved at the amino acid level yield a ratio of 1, conservative mutations will decrease the ratio somewhat and non-conservative mutations will decrease it substantially. To simplify plotting with log scales, we subtracted this average ratio from 1.01 to generate a BLOSUM-loss measure. A loss of 0.01 indicates perfect conservation at the protein level and large losses indicate non-protein-like mutations.
3.5 Local structure predictions
All local structure predictions were generated using Predict-2nd (Katzman et al., 2008), using the amino acid multiple alignments derived from the original DNA multiple alignment as inputs. We generated predictions for a set of 15 local structure alphabets, though only 3 of the 15 ended up being used in our final predictor of protein-coding ORFs. To test the value of the comparative genomics input, we also made predictions using the same neural nets (NNs), but with only the E.coli K12 translated ORFs as inputs, without the other genomes.
For each alphabet, we generated predictions from three independently trained NNs. Each was trained on the same set of proteins, but using different multiple sequence alignments and different starting conditions for the optimization. The NNs were not specially built for finding small proteins—they were available from the structure prediction work done for CASP8.
We then compared the output probability vectors for each target position to look for agreement among the three predictions (a, b and c). To measure agreement, we calculated the ‘dot product’ of the resulting probability vectors for each position, ∑iaibici, and took the average across the entire target sequence. This value is maximized when all three NNs generate strong, consistent predictions for the local structure—a signal we expected to be indicative of protein-like peptides.
3.6 Three-fold cross-training
We performed 3-fold cross-validation experiments using a logistic regression model implemented in R (R Development Core Team, 2011) and the negative and positive training data described in Section 3.1 and illustrated in Figure 1. The training data was split into three parts and two parts were used to train a logistic regression model, which was then tested on the remaining part. The training and test was repeated three times, once for each held-out third of the data.
To determine what combination of features to use in the logistic regression model, we used a simple greedy algorithm. We started by looking at the performance on the test set of all 17 features (15 local structure alphabet agreement scores, 1 codon bias score and 1 BLOSUM-loss score) independently of one another. Then we selected the best performing single feature where performance was determined by how many false positives were produced at a threshold that accepted half the real protein ORFs as true positives. We then repeated the analysis with all possible pairs of features containing the best individual feature, then took the best performing pair of features and looked at all possible three-feature combinations containing this pair. We repeated this process, selecting the best 2-, 3-, 4-, 5-, 6- and 7-feature combinations. We did not go beyond seven features because performance saturated at this point—in fact, we had to look at other thresholds besides TP = half the proteins ORFs in order to continue the greedy algorithm past five features, as no further changes at that threshold were visible when adding a sixth feature.
3.7 Validation run
Because our model selection method used all the training data to help select the models, it does not directly tell us what to expect when applied to new data. Also, we defined all small ORFs as negatives for training purposes, but we were trying to find small ORFs that do code for proteins. We used the five features selected for the best five-feature model in cross-validation to make a predictor for validation with the experimentally validated set of small proteins.
The actual predictions were made by taking the average of 20 logistic regression models. Each model was trained on a different dataset containing 1000 positive and 1000 negative examples randomly selected from the training data. After the training, each model was used to predict all training data not used in building the model as well as the 60 experimentally validated small protein sequences (which had been excluded from both the negative and the positive training data). As a negative control, we used the same 20 regression models to make predictions for all 163 GenBank-annotated ncRNAs. All the predictions were ranked according to the average probability from the 20 independent regression models used for prediction. The total number of ORFs scored in this set was 11 407.
3.8 Glimmer predictions
In order to benchmark our performance versus another common microbial gene prediction tool, we generated a set of predictions for the entire E.coli K12 genome using the Glimmer program (Delcher et al., 1999; Salzberg et al., 1998). Glimmer was developed at The Institute for Genomic Research (TIGR) and is their primary gene finder. Glimmer version 3.02 was obtained directly from the program web site (http://www.cbcb.umd.edu/software/glimmer/). The set of all GenBank-annotated genes of length >1 kb were used to train a genome-specific probability model of coding sequences. The same genes were also used to create a position weight matrix representing the ribosome binding site. None of the experimentally validated small proteins were included in the training set. Glimmer was run with two minimum gene sizes, 110 bases (default Glimmer protocol) and 30 bases (small ORF Glimmer protocol). Standard settings were used for maximum overlap (50) and maximum entropy distance scores (30) in both runs. The default and small ORF Glimmer protocols generated 5296 and 8544 predictions, respectively.
In order to make a direct comparison of performance between Glimmer and our method, we filtered out any Glimmer predictions that were either a complete match to a GenBank-annotated gene or if 20% or greater of the ORF sequence overlapped a known gene. The filter was applied to both sets of Glimmer predictions (default and small ORF Glimmer protocols) as well as the original validation set described above. Predictions for any of the experimentally validated small proteins remained in the sets. In addition, the initial validation set only scored the correct reading frame for all the experimentally validated small proteins and ignored any other possible ORFs that overlapped the known small proteins. These possible ORFs were scored and included in the side-by-side comparison with Glimmer. The predictions made according to both Glimmer protocols, default and small ORF, were ranked according to the ‘raw score’ which is a per-base log-odds ratio of the in-frame coding score versus a null-model score (i.e. non-coding). The total number of predictions after this filtering step were 216 and 1606 for the default and small ORF Glimmer protocols, respectively. The comparable validation set of predictions with our method now consisted of 11 676 ORFs.
4 RESULTS
4.1 Distributions of individual scoring features
We plotted the distribution of scores for positive and negative examples for all of our features (See Supplementary Materials), and looked at how well each feature discriminated the two training populations. It was clear that by all measures the set of true protein coding sequences had a very distinguishable distribution. This information was very useful during the development and optimization of our scoring features.
4.2 Cross-training results
The results from the systematic analysis of all 17 features was that the codon bias calculation was the best single feature. We generated true positive versus false positive curves (ROC plots) for each feature (see the Supplementary Materials). At 800 TPs, roughly half of positive training examples, the codon bias calculation has only 85 FPs (Table 1). The next best single feature was the BLOSUM-loss score with 394 FPs after the first 800 TPs, a significant drop-off in performance. The ROC plots show improved performance as we move from one to two, three, four and even five features. After this point, however, we seem to have saturated our performance. The best five-feature combination included codon bias, CB-burial-14-7, BLOSUM-loss, CB8-sep9 and n-notor2, added in that order. For a more detailed explanation of the features included in the optimal five-feature logistic regression model, please see the Supplementary Materials.
Table 1.
Table of the number of false positives at 800 true positives for each single-feature logistic regression model
| Alphabet | NUM FP |
|---|---|
| Codon bias | 85 |
| BLOSUM-loss | 394 |
| str4 | 673 |
| Strand-sep | 677 |
| Alpha | 714 |
| Bystroff | 881 |
| CB-burial-14-7 | 1004 |
| str2 | 1050 |
| Protein blocks | 1287 |
| n-notor2 | 2118 |
| n-notor | 2206 |
| Near-backbone-11 | 2683 |
| n-sep | 2794 |
| CB8-sep9 | 3097 |
| o-notor | 3197 |
| o-notor2 | 3632 |
| o-sep | 4162 |
All 17 features are listed in order of performance on the cross-validation test. See Supplementary Materials for explanation of features.
4.3 Validation
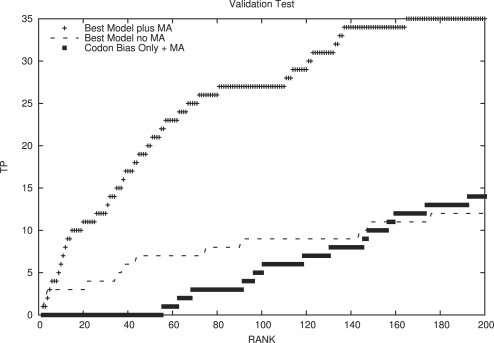
The validation experiment showed a surprisingly large number of the experimentally validated set of small proteins among the top prediction ranks. Using the best five-feature combination from the cross-training experiment, 6 of the top 10, 20 of the top 50, 27 of the top 100 and 35 of the top 200 predictions were from the set of experimentally validated small proteins (Fig. 2). Finding more than half the known small proteins within the top 200 out of 11 407 small ORFs was much better enrichment than we expected.
Fig. 2.
Validation results on experimentally verified small proteins. The use of multiple alignment (MA) data improved the performance substantially, but even without MA data there are 12 experimentally validated small ORFs in the top 200 hits (out of 11 407 ORFs scored). Using the best single feature, codon-bias, we found 14 out of the top 200 predictions were experimentally verified. However, when we calculated the codon bias score without MA data, we did not find any of the experimentally validated small proteins in the top 200 predictions (data not shown).
The GenBank-annotated ncRNAs produced both expected and not-so-expected results. Overall, the set of annotated ncRNAs were correctly identified as not protein coding by the five-feature predictor using multiple alignment data—the median rank for all ncRNAs was 3047. However, 7 annotated ncRNAs ranked among the top 200 predictions (Table 2). Five of the seven (b4603, b4451, b4434, b1574 and b4439) were annotated as sRNAs which modulate expression of a target gene by complementary base pairing to regions of the target RNA. The b4434 gene encompasses one of the experimentally verified small proteins, azuC, providing a possible explanation for why it scored so well. Although both sequences were ranked high by our method, the small protein (azuC) encoding sequence ranked higher (13) than the ncRNA (b4434) sequence (53). The two tRNAs occurring within the top 200 predictions, b1032 and b0883, coincided with a complete ORF devoid of any premature stop codons and, most likely as a result of that, had reasonable scores for all five features used in the predictor.
Table 2.
GenBank-annotated ncRNAs appearing within top 200 predictions for the five-feature predictor using multiple alignment data
| ID | Annotated function | Prediction rank |
|---|---|---|
| b4603 | sRNA | 17 |
| b4451 | sRNA | 36 |
| b4434 | sRNA | 53 |
| b1574 | sRNA | 104 |
| b1032 | Ser tRNA | 122 |
| b0883 | Ser tRNA | 143 |
| b4439 | sRNA | 161 |
The median rank for all GenBank-annotated ncRNAs was 3047.
We were curious how well we could do if we did not have any comparative genomic data, as would be the case for a newly sequenced genome that is not closely related to other bacterial genomes. We know that our NN predictors are less accurate when given only single sequences as inputs, rather than alignments, so we expected a considerable drop in performance. Using only the single-genome-based codon bias data as a single-feature model did not get us any of the true small proteins within the top 200 predictions, much worse than 14 true proteins found in the top 200 with the multiple alignment-based codon bias single-feature model. The best combination of features determined by cross-validation, generated in the absence of any multiple alignment data, was able to find 12 out of 60 true small proteins in the top 200. Together, these results suggest that the use of multiple features increases selectivity (fewer false positives in the first few ranks), while the use of multiple alignment information increases sensitivity (more true positives in the top 200). Using both is essential to good performance.
4.4 Length bias
We investigated how prediction accuracy correlated with sequence length. ORF length is typically a good predictor of whether a given gene is real or an artifact of random chance. The longer the ORF, the less likely it is to have occurred randomly, and the more likely it is to be under selection pressure to maintain a protein-coding signal. Using length as a signal, however, makes it difficult to find genuine small proteins, and so we attempted to avoid length bias in our predictor.
When we plotted the rank of all test predictions (i.e. all un-annotated ORFs plus the validated small proteins set) against the length of each sequence, we observed a strong correlation between the length and prediction ranks, Spearman's ρ=−0.71 and Kendall's τ=−0.52 (see the Supplementary Materials). Specifically, very small ORFs tend to rank poorly. However, if we focus on the top prediction ranks, the area we are most interested in, the correlation goes away. Within the top 1000 predictions, the Spearman's ρ=0.02 and Kendall's τ=0.02 as well. Looking at the top 200 predictions, the Spearman's ρ=−0.05 and Kendall's τ=−0.03.
4.5 Comparison to Glimmer
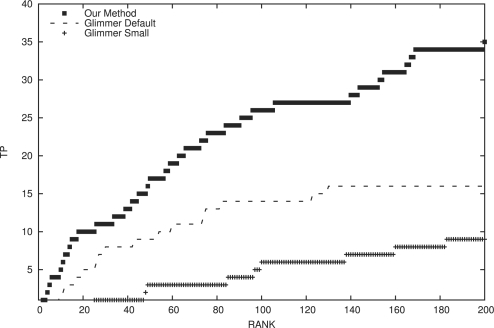
The default Glimmer protocol correctly identified only 16 of the experimentally validated small proteins within the top 200 predictions and was unable to perform better than our method at any cutoff on predictions (Fig. 3). See Supplementary Table S3 for a side-by-side comparison of the Glimmer protocol and our predictions for all 60 known small proteins.
Fig. 3.
Our approach identified 35 of the experimentally validated small proteins within the top 200 predictions in our ensemble. Conversely, the default Glimmer protocol found 16 from this dataset within the top 200 predictions. Using a lower minimum size cutoff for Glimmer introduces more false positive into the ensemble, resulting in reduced performance compared with the standard Glimmer protocol. Note: results for our method in this figure are slightly poorer than the results shown in Figure 2, because this test included ORFs that overlapped the true small proteins and some overlapping ORFs scored quite well.
Running Glimmer to look for smaller ORFs did not yield good results, because the ensemble of predictions had grown considerably with the lower minimum gene size. The modified Glimmer protocol identified only nine of the experimentally validated small proteins within the top 200. This example illustrates the classic problem with using existing tools to find small proteins. Using a higher size cutoff yielded better results but failed to predict over half (34 out of 60) of the experimentally validated small proteins. When we expanded the search to include really small ORFs, a lot of junk fell into the prediction pool. This made the task of ‘fishing’ out the right ones increasingly difficult.
4.6 Identification of novel small proteins
Several small ORFs not previously annotated as protein encoding were among the highest scoring in the validation experiment (see Supplementary Materials for a comprehensive table of all novel predictions including prediction rank). Based on the observed enrichment of validated small proteins among the top prediction ranks, we expect that these novel predictions should be highly enriched for true protein encoding sequences.
A hallmark of protein encoding sequences in prokaryotes is the presence of the Shine-Dalgarno (SD) motif which is involved in the recruitment and correct orientation of the ribosome to a gene's start codon (Laursen et al., 2005). The presence of an SD motif was one of the highest contributing factors used to identify the set of annotated small proteins in E.coli K12 (Hemm et al., 2008). To ensure a clean validation experiment, we intentionally omitted the absence or presence of an SD motif from our set of predictive features. However, this signal can now be used to filter the most promising candidate small proteins in our prediction set.
For each putative small protein in our validation set, we searched within the immediate 15 bp upstream of the start codon for the best scoring SD motif match. The scores were obtained directly from the UCSC Microbial Genome Browser (http://microbes.ucsc.edu/) (Schneider et al., 2006) and were generated by creating a position-specific scoring matrix (PSSM) of the SD motifs found in known genes in E.coli K12. The PSSM was then used for genome-wide scanning starting at every base.
The score of a 10 nt motif at each position was converted to a 0-to-1 scale with scores closer to 1 representing better matches (P.P.Chan and T.M.Lowe, manuscript in preparation). The Supplementary Table lists the score of the best SD motif match for each putative small protein. While not all of the top predictions have a high-scoring match to an SD motif, this does not completely rule out the possibility of protein expression. Examples of leaderless or non-SD mediated translation continue to be identified in a growing number of organisms (Brock et al., 2008; Chang et al., 2006) including one example, Carsonella ruddii, devoid of any sequence in its 16S rRNA complementary to the SD motif (Clark et al., 2001).
5 DISCUSSION
We have developed a method that uses traditional measures, protein conservation and codon bias, as well as local protein structure properties to generate predictions that a small ORF encodes for a protein. The framework for all our calculations is comparative genomics using whole genome alignments between closely related species. We have shown that within this context, multiple alignment information can be very valuable.
As we expected, sequence composition and protein-like conservation alone do not perform as well as our combined approach. Specifically, we were unable to identify any of the validated small proteins within the top 200 predictions when we used a single-genome codon bias metric as our lone data source. This is especially interesting because this approach is one of the standard ‘off-the-shelf’ tools used to distinguish protein-coding sequences. While this approach may work for longer sequences, our results show that small proteins will be missed by such efforts.
For our analysis of the E.coli K12 genome, we intentionally omitted all ribosome binding site data as a possible scoring feature. This was done for two reasons. In several bacterial genomes, there are a large number of leaderless protein genes, so we created a method that did not rely on strong ribosome binding sites. Furthermore, a majority of the experimentally validated small proteins in this genome were identified in large part due to a strong ribosome binding site prediction near the ORF start site. Therefore, inclusion of this information would have given us a less stringent test of our method. On the other hand, programs such as Glimmer incorporate this signal into their prediction algorithm. This makes the observed improvement in performance over Glimmer even more dramatic. We plan to incorporate the absence or presence of a ribosome binding site into future predictions, which should reduce the observed enrichment for ncRNAs in our top prediction ranks, as none of the 7 ncRNAs found within the top 200 prediction contained a strong ribosome binding site prediction.
Our method has two main preconditions required in order to generate a set of predictions. First is a multiple genome alignment of closely related species containing the organism of interest. Note however, that this is a soft requirement as single-genome predictions are possible, though of substantially lower quality. The second requirement is a set of positive and negative sequence examples in order to train a logistic regression model. Minimally, we can use the 1600 longest ORFs as a positive training set. These sequences should be almost entirely true protein-coding sequences. Conversely, a negative training set could be built from all possible ORFs below an arbitrary size cutoff, say 30 amino acids, the large majority of which should be non-protein coding.
As presently constructed, our method is highly adaptable to new genome sequences. We are in the process of adding our method to the UCSC Microbial Browser creation pipeline. This should enable us to produce predictions for any completed genome in the public databases.
We are extremely encouraged by the performance of our method in silico. However, we know that the final validation of our predictions must come from experimental techniques. Therefore, we are currently designing experiments to validate high confidence predictions in other bacterial genomes, specifically the human pathogenic species Vibrio cholerae.
Supplementary Material
ACKNOWLEDGEMENTS
Thanks to Brian Raney for assistance with the generation of multiple genome alignments and Grant Thiltgen, who designed and maintained many of the local structure alphabets used in this study (Thlitgen, 2010).
Funding: (Conducted from 2005 to 2009); NIH IMSD (grant 2 R25 GM58903); NIH-NHGRI grant (5P41HG002371-09).
Conflict of Interest: none declared.
REFERENCES
- Besemer J., Borodovsky M. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 1999;27:3911–3920. doi: 10.1093/nar/27.19.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J., et al. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock J.E., et al. Ribosomes bind leaderless mRNA in Escherichia coli through recognition of their 5′-terminal AUG. RNA. 2008;14:2159–2169. doi: 10.1261/rna.1089208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B., et al. Analysis of SD sequences in completed microbial genomes: non-SD-led genes are as common as SD-led genes. Gene. 2006;373:90–99. doi: 10.1016/j.gene.2006.01.033. [DOI] [PubMed] [Google Scholar]
- Clark M.A., et al. Degenerative minimalism in the genome of a psyllid endosymbiont. J. Bacteriol. 2001;183:1853–1861. doi: 10.1128/JB.183.6.1853-1861.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N., Grimmond S.M. Transcriptome content and dynamics at single-nucleotide resolution. Genome Biol. 2008;9:234. doi: 10.1186/gb-2008-9-9-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutting S., et al. SpoVM, a small protein essential to development in Bacillus subtilis, interacts with the ATP-dependent protease FtsH. J. Bacteriol. 1997;179:5534–5542. doi: 10.1128/jb.179.17.5534-5542.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher A.L., et al. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frishman D., et al. Combining diverse evidence for gene recognition in completely sequenced bacterial genomes. Nucleic Acids Res. 1998;26:2941–2947. doi: 10.1093/nar/26.12.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo R.L., Nizet V. Endogenous production of antimicrobial peptides in innate immunity and human disease. Curr. Allergy Asthma Rep. 2003;3:402–409. doi: 10.1007/s11882-003-0074-x. [DOI] [PubMed] [Google Scholar]
- Hemm M.R., et al. Small membrane proteins found by comparative genomics and ribosome binding site models. Mol. Microbiol. 2008;70:1487–1501. doi: 10.1111/j.1365-2958.2008.06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman S., et al. PREDICT-2ND: a tool for generalized protein local structure prediction. Bioinformatics. 2008;24:2453–2459. doi: 10.1093/bioinformatics/btn438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Gadagkar S.R. Efficiency of the neighbor-joining method in reconstructing deep and shallow evolutionary relationships in large phylogenies. J. Mol. Evol. 2000;51:544–553. doi: 10.1007/s002390010118. [DOI] [PubMed] [Google Scholar]
- Larsen T.S., Krogh A. EasyGene–a prokaryotic gene finder that ranks ORFs by statistical significance. BMC Bioinformatics. 2003;2003:21. doi: 10.1186/1471-2105-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen B.S., et al. Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin A.V., Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U., et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P., Krogh A. Large-scale prokaryotic gene prediction and comparison to genome annotation. Bioinformatics. 2005;21:4322–4329. doi: 10.1093/bioinformatics/bti701. [DOI] [PubMed] [Google Scholar]
- Ochman H. Distinguishing the ORFs from the ELFs: short bacterial genes and the annotation of genomes. Trends Genet. 2002;18:335–337. doi: 10.1016/s0168-9525(02)02668-9. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- Ruvolo M.V., et al. Proteolysis of the replication checkpoint protein SDA is necessary for the efficient initiation of sporulation after transient replication stress in Bacillus subtilis. Mol. Microbiol. 2006;60:1490–1508. doi: 10.1111/j.1365-2958.2006.05167.x. [DOI] [PubMed] [Google Scholar]
- Salzberg S.L., et al. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K.L., et al. The UCSC Archaeal Genome Browser. Nucleic Acids Res. 2006;34:D407–D410. doi: 10.1093/nar/gkj134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela B.A. Hepcidin–the discovery of a small protein with a pivotal role in iron homeostasis. Harefuah. 2008;147:261–266. [PubMed] [Google Scholar]
- Shimmin L.C., et al. Sequences and evolution of human and squirrel monkey blue opsin genes. J. Mol. Evol. 1997;44:378–382. doi: 10.1007/pl00006157. [DOI] [PubMed] [Google Scholar]
- Skovgaard M., et al. On the total number of genes and their length distribution in complete microbial genomes. Trends Genet. 2001;17:425–428. doi: 10.1016/s0168-9525(01)02372-1. [DOI] [PubMed] [Google Scholar]
- Thiltgen G. PhD dissertation. Santa Cruz: University of California Santa Cruz; 2010. Creating New Local Structure Alphabets for Protein Structure Prediction. [Google Scholar]
- Yanofsky C. Transcription attenuation: once viewed as a novel regulatory strategy. J. Bacteriol. 2000;182:1–8. doi: 10.1128/jb.182.1.1-8.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., et al. MED: a new non-supervised gene prediction algorithm for bacterial and archaeal genomes. BMC Bioinformatics. 2007;8:97. doi: 10.1186/1471-2105-8-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.