Abstract
Autophagy is a conserved degradative process that is crucial for cellular homeostasis and cellular quality control via the selective removal of subcellular structures such as mitochondria. We demonstrate that a regulatory link exists between mitochondrial function and autophagy in Saccharomyces cerevisiae. During amino-acid starvation, the autophagic response consists of two independent regulatory arms—autophagy gene induction and autophagic flux—and our analysis indicates that mitochondrial respiratory deficiency severely compromises both. We show that the evolutionarily conserved protein kinases Atg1, target of rapamycin kinase complex I, and protein kinase A (PKA) regulate autophagic flux, whereas autophagy gene induction depends solely on PKA. Within this regulatory network, mitochondrial respiratory deficiency suppresses autophagic flux, autophagy gene induction, and recruitment of the Atg1–Atg13 kinase complex to the pre-autophagosomal structure by stimulating PKA activity. Our findings indicate an interrelation of two common risk factors—mitochondrial dysfunction and autophagy inhibition—for ageing, cancerogenesis, and neurodegeneration.
Keywords: autophagy regulation, mitochondria, protein kinase A, TOR
Introduction
Autophagy is a highly conserved and regulated process essential for the degradation of long-lived proteins and organelles in eukaryotic cells. During autophagy, cytosolic content is sequestered via de novo formation of double-membrane-bounded structures termed autophagosomes. The autophagosomal outer membrane docks and fuses with the vacuole and the inner membrane vesicle is subsequently released and degraded by resident hydrolases to generate metabolic building blocks for biosynthesis. In yeast, the autophagy machinery comprised ∼30 autophagy-related (ATG) genes, including highly conserved core components. Most Atg proteins dynamically assemble in a hierarchical manner at pre-autophagosomal structures (PAS), which are putative sites for autophagosome formation (Suzuki et al, 2007; Xie and Klionsky, 2007; Kawamata et al, 2008; Cebollero and Reggiori, 2009; He and Klionsky, 2009; Nakatogawa et al, 2009).
The extent and rate of autophagy are controlled by the availability of nutrients. Starvation strongly induces autophagy and this regulation is essential for long-term cellular survival (Takeshige et al, 1992; Tsukada and Ohsumi, 1993; Komatsu et al, 2005). Multiple signalling pathways regulate autophagy including the conserved serine/threonine protein kinase target of rapamycin kinase complex I (TORC1) and cAMP-dependent protein kinase A (PKA) pathways, which also sense nutrient availability and control cellular growth (Noda and Ohsumi, 1998; Budovskaya et al, 2004; Yorimitsu et al, 2007, 2009; Stephan et al, 2009; Yang et al, 2010). In yeast, TORC1 and PKA negatively regulate autophagy at least in part by directly and independently phosphorylating the conserved kinase Atg1 (ULK1/2 in mammals) and Atg13 (mammalian Atg13), and by blocking the assembly and localization of an active Atg1–Atg13 complex to the PAS. Localization of an active Atg1–Atg13 complex to the PAS is required to initiate autophagosome formation and subsequent vacuolar turnover or autophagic flux (Budovskaya et al, 2005; Kabeya et al, 2005; Chan et al, 2007; Cheong et al, 2008; Hara et al, 2008; Stephan et al, 2009; Kamada et al, 2010).
Autophagy is also regulated via the transcriptional up-regulation or induction of autophagy genes by ill-defined mechanisms (Abeliovich et al, 2000). In yeast, starvation induces the transcription of ATG1 and ATG13 as well as ATG14, which encodes a component of the phosphatidylinositol-3 kinase complex I (Hardwick et al, 1999; Chan et al, 2001). ATG8, which encodes an ubiquitin-like PAS-localized protein required for autophagosome formation, and the genes encoding the Atg8 ubiquitin-like modification system (ATG3, ATG4, ATG5, ATG7, and ATG12) are also transcriptionally regulated during starvation (Hardwick et al, 1999; Kirisako et al, 1999; Huang et al, 2000). Induction of this ATG8 module is functionally relevant, as the level of Atg8 has been shown to determine the size of forming autophagosomes and, thus, to positively influence the magnitude of the autophagic response (Abeliovich et al, 2000; Xie et al, 2008).
Although autophagy is considered non-specific in yeast, selective forms have been described including receptor-mediated targeting of the autophagy machinery to mitochondria (mitophagy), but the functional role of mitophagy in yeast is not known (Kissova et al, 2004, 2007; Tal et al, 2007; Kanki et al, 2009; Okamoto et al, 2009). In mammalian cells, the selective autophagic removal of depolarized mitochondria is consistent with a quality control mechanism (Priault et al, 2005; Nowikovsky et al, 2007; Narendra et al, 2008, 2010; Twig et al, 2008; Geisler et al, 2010; Suen et al, 2010; Vives-Bauza et al, 2010). A connection between autophagy and maintenance of mitochondrial function is also implied by the fact that defects in either are associated with a set of common, but diverse diseases, such as cancer, ageing, and neurodegeneration (Hara et al, 2006; Komatsu et al, 2006; Huang and Klionsky, 2007; Banerjee et al, 2009; Meijer and Codogno, 2009; Morselli et al, 2009; Cardoso et al, 2010; Galluzzi et al, 2010; Lee et al, 2010; Wong and Cuervo, 2010). Alternatively, the common denominator of autophagy and mitochondria in disease might be that mitochondrial dysfunction directly affects and regulates the autophagy capacity of eukaryotic cells. We addressed this latter possibility in Saccharomyces cerevisiae and demonstrate that an intimate link exists between mitochondrial function and autophagy regulation during amino-acid starvation.
Results
Mitochondrial respiratory deficiency impairs autophagy gene induction and autophagic flux
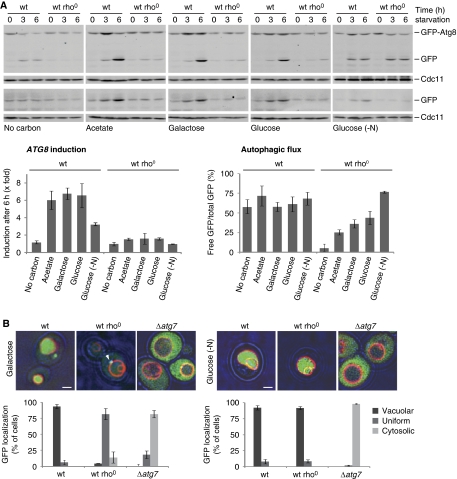
We investigated the role of mitochondria in the regulation of autophagy by monitoring the behaviour of the GFP-Atg8 reporter under the control of the endogenous ATG8 promoter in yeast (pRS416-prATG8-GFP-ATG8) (Abeliovich et al, 2003). Using this reporter, we assayed two critical components of the autophagic response: autophagic flux and ATG8 induction (Kirisako et al, 1999; Abeliovich et al, 2000; Shintani and Klionsky, 2004; Xie et al, 2008). Autophagic flux, which is the vacuolar transfer and degradation of autophagosomes over time, was quantified by measuring the vacuolar degradation of the Atg8 domain of the reporter (ratio of free GFP to total GFP signal) over time by western blot analysis (Shintani and Klionsky, 2004). Similarly, we measured ATG8 induction by quantifying the fold increase of total GFP signal (GFP-Atg8 and free GFP signal) normalized to a non-induced protein (Cdc11). In addition, ATG8 induction was monitored independently from autophagic flux using a GFP-only reporter under the control of the endogenous ATG8 promoter (pRS426-prATG8-GFP).
To examine the role of mitochondria, we compared the autophagic response under starvation conditions of wild-type cells to wild-type cells lacking mitochondrial genomes (rho0 cells). Cells were grown in galactose as opposed to glucose as a carbon source to allow fermentative growth of respiratory-deficient rho0 cells without glucose repression of mitochondrial biogenesis. Subsequently, autophagy was induced by exposing cells to amino-acid starvation, which was faithfully produced using a relatively low concentration of yeast extract (0.1% (w/v)) (Supplementary Figure S1) or standard nitrogen starvation conditions. We also varied the carbon source during amino-acid starvation to determine the degree to which the autophagic response depends on mitochondrial function. Specifically, we examined the effects of no carbon source (carbon starvation); acetate as a non-fermentable carbon source; and galactose or glucose as fermentable carbon sources that are non-repressive or repressive for mitochondrial biogenesis, respectively.
Analysis of wild-type and rho0 cells indicates that under amino-acid starvation, ATG8 induction was strictly dependent on both the presence of a carbon source and mitochondrial function (Figure 1A, ATG8 induction). Specifically, we found that in the absence of a carbon source, neither wild-type nor rho0 cells displayed significant ATG8 induction (Figure 1A, no carbon). In contrast, in the presence of the non-fermentable carbon source acetate or the fermentable carbon sources galactose and glucose, ATG8 expression in wild-type cells was significantly stimulated (Figure 1A, acetate, galactose, or glucose). Remarkably, we did not observe any ATG8 induction in rho0 cells in the presence of either non-fermentable or fermentable carbon sources, demonstrating that mitochondrial respiratory deficiency blocks ATG8 induction in response to amino-acid starvation (Figure 1A).
Figure 1.
Mitochondrial respiratory deficiency impairs ATG8 induction and autophagic flux. (A) Wild-type and rho0 cells harbouring prATG8-GFP-ATG8 (upper panels) or prATG8-GFP (lower panels) were exposed to amino-acid or nitrogen starvation (-N) medium supplemented with indicated carbon sources. Cells were analysed at indicated time points by whole cell extraction and western blot analysis using α-GFP and α-Cdc11 antibodies. Quantification of ATG8 induction is shown in the lower left panel. Total GFP signals (GFP-Atg8 and free GFP) were quantified and normalized to Cdc11 signals. Normalized values at 0 h were set as one and relative changes are shown after 6 h starvation. Quantification of autophagic flux is shown in the lower right panel as ratio of free GFP to total GFP signals (GFP-Atg8 and free GFP) after 6 h starvation. The means and s.d. of four (n=4) independent experiments are indicated. (B) Fluorescence microscopical analysis. Wild-type, rho0, and Δatg7 cells expressing prATG8-GFP-ATG8 were grown as described in (A) and exposed to amino-acid (left panel, galactose) or nitrogen starvation (right panel, glucose (-N)) for 6 h. Vacuoles were visualized by over night FM4-64 (1 μM) staining (red). Arrowhead indicates a punctate GFP-Atg8 structure. Transmission and fluorescence light microscopy images were superimposed to visualize cellular boundaries. Cellular localization of GFP signal was analysed in at least 150 cells (n⩾150) for each strain and condition. Scale bars represent 1.5 μm.
To test whether mitochondrial function also regulates the expression of other key autophagy factors under amino-acid starvation, we analysed ATG14, which encodes another highly induced essential component of the autophagy machinery (Chan et al, 2001). We analysed the expression of a plasmid-encoded GFP reporter under the control of the endogenous ATG14 promoter in wild-type, rho0, and Δatg7 cells. In wild-type and Δatg7 cells, we observed a strong induction of the ATG14 promoter-driven GFP reporter upon amino-acid starvation (Supplementary Figure S2). Importantly, ATG14 induction was completely absent in rho0 cells (Supplementary Figure S2), indicating that mitochondrial function is a more general regulator of expression-regulated autophagy components. Interestingly, we observed a slightly elevated basal level of ATG14 promoter-controlled GFP expression in rho0 cells (0 h starvation) as compared with wild-type cells that might reflect compensatory adaptations to mitochondrial dysfunction (Supplementary Figure S2).
In contrast to ATG8 induction, autophagic flux was not dependent on carbon source in wild-type cells during amino-acid starvation (Figure 1A, autophagic flux). In rho0 cells, however, autophagic flux was significantly decreased in comparison with wild-type cells, even in the presence of galactose or glucose (Figure 1A, galactose or glucose), but the extent of inhibition varied with the type of carbon source (Figure 1A). These observations indicate that the strong defect in the overall autophagic response observed in respiratory-deficient cells is due to a combined inhibitory effect on both ATG8 induction and autophagic flux. Interestingly, wild-type and rho0 cells starved for all nitrogen sources instead of only amino acids displayed an indistinguishable autophagic flux (Figure 1A, glucose (-N)). This demonstrates that the defects in autophagic flux observed in rho0 cells under amino-acid starvation are not due to intrinsic defects in the autophagy machinery or in vacuolar biogenesis. This conclusion is consistent with published data, indicating that vacuolar morphology and acidification is normal in rho0 cells (Chen et al, 2008) and is further supported by our phenotypical analysis showing that rho0 cells are resistant to stress conditions that inhibit growth of a vacuolar acidification mutant (Δvma2) (Supplementary Figure S3). However, as with amino-acid starvation, a severe defect in ATG8 induction was observed in rho0 cells under nitrogen starvation in comparison with the three-fold induction of ATG8 in wild-type cells, which is consistent with published results (Figure 1A) (Kirisako et al, 1999; Huang et al, 2000). These findings indicate that there are differences in the regulation of autophagy under amino-acid versus nitrogen starvation conditions, consistent with recently published work, indicating a differential role for Gcn2 and Gcn4 in autophagy regulation in response to amino-acid and nitrogen starvation (Ecker et al, 2010). Importantly, our data point to an essential role for mitochondrial function in ATG8 induction and the autophagic response.
To further substantiate our conclusions, we analysed GFP-ATG8-expressing cells by fluorescence microscopy. Consistently, under amino-acid starvation, wild-type cells exhibited a strong vacuolar GFP signal, as assessed by the vital vacuolar dye FM4-64, that was dependent on the essential autophagy component, Atg7, indicating autophagic turnover of GFP-Atg8 (Figure 1B) (Tanida et al, 1999; Huang et al, 2000). In contrast, in rho0 cells under identical conditions, a uniformly distributed weak GFP signal was observed, which supports our biochemical data showing impaired ATG8 induction and autophagic flux in rho0 cells (Figure 1B, galactose). GFP-Atg8 was observed in punctate structures in amino-acid starved rho0 cells (Figure 1B, galactose), raising the possibility that Atg8 recruitment to the PAS is not affected (Kirisako et al, 1999). In contrast, under nitrogen starvation, rho0 cells displayed a vacuolar GFP signal, which indicates autophagic turnover under these conditions—a conclusion also supported by our biochemical analysis (Figure 1A and B). Together, these data indicate that ATG8 induction and autophagic flux are differentially regulated and demonstrate that mitochondria have important roles in both ATG8 induction and in the modulation of autophagic flux in the autophagic response to amino-acid starvation.
We also tested whether mitochondrial function regulates autophagy induced during stationary phase. Wild-type cells displayed a significant autophagic response after 2–4 days in stationary phase (Supplementary Figure S4). Autophagic flux was blocked in Δatg7 cells as expected; however, ATG8 induction was observed (Supplementary Figure S4). In contrast, we did not detect either autophagic flux or ATG8 induction in rho0 cells under these conditions (Supplementary Figure S4). Thus, mitochondrial function is a general regulator of the autophagic response.
Characterization of the role of mitochondria in the autophagic response
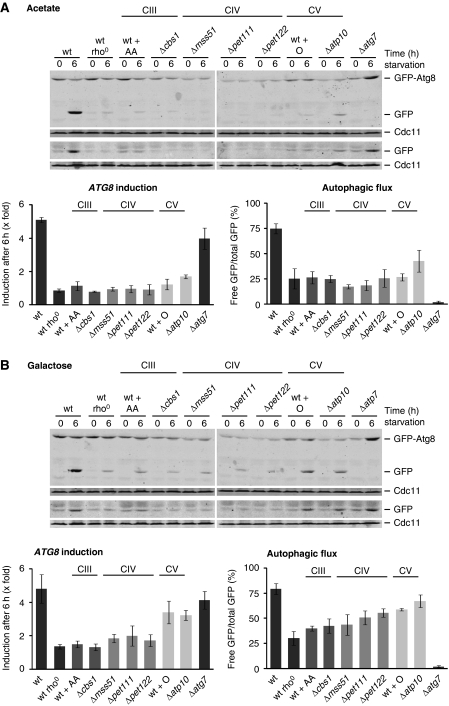
The rho0 cells lack the mitochondrial-encoded core subunits of the respiratory chain and mitochondrial F1Fo-ATP synthase and are, therefore, unable to utilize non-fermentable carbon sources to generate a proton gradient across the inner mitochondrial membrane or produce ATP by respiration. To determine whether specific defects in the respiratory chain complex III (CIII), complex IV (CIV), or in the F1Fo-ATP synthase (CV) are linked to the regulation of autophagy, we compared autophagy induction in wild-type, rho0, and Δatg7 cells with cells that maintain mtDNA, but are selectively impaired in CIII, CIV, or CV activity. Specifically, CIII was blocked in wild-type cells by the addition of the selective inhibitor antimycin A or in a mutant lacking Cbs1 (Δcbs1), a nuclear-encoded factor specifically required for the translation of Cob1, the mitochondrial-encoded core subunit of CIII (Rodel, 1997). Similarly, CIV function was impaired in mutants lacking the nuclear-encoded factors Mss51 (Δmss51), Pet111 (Δpet111), or Pet122 (Δpet122) required for the translation of the mitochondrial-encoded CIV core subunits Cox1, Cox2, or Cox3, respectively (Poutre and Fox, 1987; Costanzo and Fox, 1988; Decoster et al, 1990; Herrmann and Funes, 2005). CV activity was inhibited in wild-type cells by the addition of the selective inhibitor oligomycin or in a mutant lacking Atp10 (Δatp10), a nuclear-encoded factor required for the assembly of the Fo-sector of F1Fo-ATP synthase (Ackerman and Tzagoloff, 1990; Tzagoloff et al, 2004). We analysed autophagy in the presence of the non-fermentable carbon source acetate or the non-repressive fermentable carbon source galactose during amino-acid starvation to vary dependence on mitochondrial function.
In the presence of the non-fermentable carbon source acetate, inhibition of CIII, CIV, or CV blocked the induction of ATG8 expression and autophagic flux in response to amino-acid starvation to a similar degree as observed in rho0 cells (Figure 2A). Similarly, induction of ATG14 was strongly inhibited in the presence of antimycin A or oligomycin under these conditions (Supplementary Figure S2). Thus, respiratory activity that results in the production of mitochondrial ATP is required for the autophagic response under amino-acid starvation in the presence of a non-fermentable carbon source. Significantly, in the presence of the fermentable carbon source galactose, inhibition of CIII or CIV had different effects on the autophagic response as compared with those observed upon inhibition of the F1Fo-ATP synthase (CV). Specifically, a block of CIII or CIV activity strongly impaired ATG8 induction and partially affected autophagic flux in response to starvation (Figure 2B). In contrast, cells inhibited for CV activity displayed over three-fold induction of ATG8, which was similar to that observed for untreated wild-type or Δatg7 cells, and a level of autophagic flux similar to that observed for wild-type cells (Figure 2B). These data indicate that mitochondrial ATP production per se is not required for the autophagic response during amino-acid starvation.
Figure 2.
Autophagic response in cells with compromised respiratory chain complex III, IV, or V activity during amino-acid starvation. Wild-type, rho0, Δatg7 cells and mutants that are selectively inhibited in the biogenesis of respiratory chain complex III (CIII: Δcbs1), complex IV (CIV: Δmss51, Δpet111, Δpet122), or the F1Fo-ATP synthase (complex V; CV: Δatp10) were exposed to amino-acid starvation medium supplemented with acetate (A) or galactose (B). When indicated, wild-type cells were exposed to antimycin A (AA) or oligomycin (O) during the amino-acid starvation period. All strains expressed prATG8-GFP-ATG8 (upper panels) or prATG8-GFP (lower panels). Samples were analysed as described in Figure 1A. The means and s.d. of five (n=5) independent experiments are indicated.
Given our data demonstrating that mitochondrial respiratory activity is a critical regulator of autophagy induction during amino-acid starvation, we investigated the nature of the regulatory signal(s). Inhibition of respiratory complexes has been associated with an increased generation of reactive oxygen species, previously implicated in mammalian autophagy regulation (Scherz-Shouval et al, 2007; Lambert and Brand, 2009). However, overexpression of cytosolic (Sod1) or mitochondrial (Sod2) superoxide dismutase did not affect the autophagy response in wild-type or rho0 cells or in wild-type cells treated with antimycin A or oligomycin during amino-acid starvation in the presence of galactose (Supplementary Figure S5A). Thus, ROS do not have a significant role under these conditions.
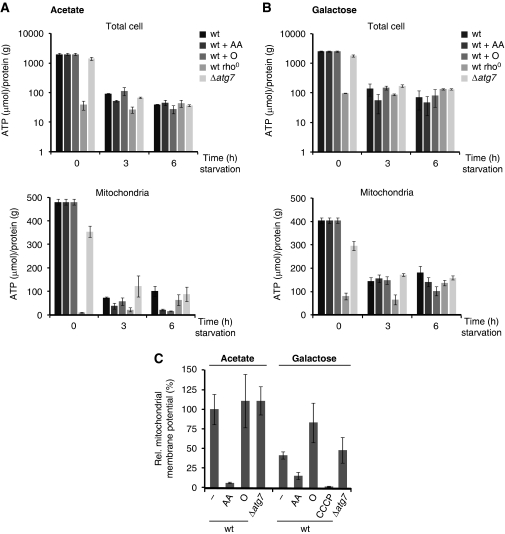
Next, we tested whether ATP levels and/or mitochondrial membrane potential could function as mitochondrial signals for autophagy regulation during amino-acid starvation. Therefore, we examined the effects of mitochondrial respiratory deficiency on cellular and mitochondrial ATP levels during amino-acid starvation in the presence of acetate or galactose (Figure 3A and B). In rho0 cells, the cellular and mitochondrial ATP levels were significantly lower under growing conditions than those observed in wild-type cells (Figure 3A and B, time 0 h), and did not change during amino-acid starvation in the presence of acetate or galactose (Figure 3A and B, time 3 and 6 h). In contrast, the levels of cellular and mitochondrial ATP significantly decreased in wild-type cells, antimycin A (block of CIII)- or oligomycin (block of CV)-treated wild-type cells, and Δatg7 cells during amino-acid starvation (Figure 3A and B, time 0, 3, and 6 h). Interestingly, under all conditions, the reduction of ATP levels occurred independently of mitochondrial function and levels were similar to those observed in rho0 cells under both growing and amino-acid starvation conditions. This raises the possibility that regulatory mechanisms exist that maintain ATP levels in a relatively narrow range. Although our data indicate that ATP production is required for the autophagy response during amino-acid starvation (Figure 2A and B), modulation of ATP level was independent of mitochondrial function and, therefore, ATP levels are unlikely to represent a direct mitochondrial signal for autophagy regulation.
Figure 3.
ATP level and mitochondrial membrane potential during amino-acid starvation. (A, B) Wild-type, rho0, and Δatg7 cells were exposed to amino-acid starvation medium supplemented with acetate (A) or galactose (B). When indicated, wild-type cells were exposed to antimycin A (AA) or oligomycin (O) during the amino-acid starvation period. ATP and protein from total cells (upper panels) or isolated mitochondria (lower panels) were determined at indicated time points. (C) Wild-type and Δatg7 cells were exposed to amino-acid starvation medium supplemented with acetate or galactose for 3 h. Wild-type cells were exposed to antimycin A (AA), oligomycin (O), or CCCP (50 μM) during the amino-acid starvation period when indicated. Cells were treated with the mitochondrial membrane potential-dependent dye DiOC6(3) and examined by fluorescence microscopy. Average pixel intensities for at least 10 mitochondrial tubules (n=10) in 5 representative cells were determined for each condition. Values for wild-type mitochondria in the presence of acetate were defined as 100% and relative values were calculated accordingly. The means and s.d. are shown.
During amino-acid starvation in the presence of a fermentable carbon source, ATG8 induction and, to a lesser extent, autophagic flux require functional CIII and CIV respiratory chain complexes. Blocking the function of these complexes has been correlated with a decrease in mitochondrial membrane potential (Johnson et al, 1981; Chen, 1988; Petit et al, 1996; Ludovico et al, 2001), raising the possibility that maintenance of a mitochondrial membrane potential by respiration under fermentative amino-acid starvation conditions is required for autophagy induction. To test this possibility, we monitored mitochondrial membrane potential during amino-acid starvation by fluorescence microscopy of living cells using the mitochondrial membrane potential-dependent dye DiOC3(6). After 3 h of amino-acid starvation, wild-type cells treated with antimycin A (block of CIII) showed a strong reduction in DiOC3(6) fluorescence compared with wild-type or Δatg7 cells in the presence of acetate or galactose as carbon sources, indicating that mitochondrial membrane potential is significantly reduced (Figure 3C). In contrast, oligomycin treatment of wild-type cells (block of CV) resulted in a similar or increased DiOC3(6) fluorescence compared with untreated wild-type cells in the presence of acetate or galactose, respectively, indicating maintenance of mitochondrial membrane potential (Figure 3C). Hence, mitochondrial defects that result in a reduced mitochondrial membrane potential are correlated with a strong inhibition of the autophagy response, even under fermentative conditions. To substantiate a role for mitochondrial membrane potential changes in the regulation of authophagy, we monitored autophagy induction in wild-type cells in the presence of increasing concentrations of the membrane uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP) during amino-acid starvation (Supplementary Figure S5B). As expected, CCCP strongly reduced mitochondrial membrane potential as assessed by DiOC3(6) fluorescence and, consistently, impaired the induction of ATG8 in a dose-dependent manner to a similar extent as that observed in rho0 cells (Figure 3C; Supplementary Figure S5B). Similarly, autophagic flux was strongly impaired in the presence of CCCP, but we cannot exclude the possibility that CCCP treatment impaired acidification of the vacuole, essential for autophagic turnover (Supplementary Figure S5B) (Nakamura et al, 1997). Together, these observations support a model where mitochondrial membrane potential signals the functional state of mitochondria and predominantly modulates the ATG8 induction component of the autophagic response in cells.
Mitochondria regulate autophagy primarily via PKA
To understand how mitochondria regulate the autophagic response, we assessed the role of two major signalling pathways involved in autophagy regulation. The TORC1 and the cAMP-dependent PKA independently control autophagy by regulating the assembly, activity, and PAS localization of the Atg1–Atg13 complex by direct phosphorylation (Kamada et al, 2000, 2010; Budovskaya et al, 2005; Stephan et al, 2009). TORC1 and PKA activity suppresses both ATG8 induction and autophagic flux, indicating that both are negative regulators of the autophagic response (Noda and Ohsumi, 1998; Budovskaya et al, 2004).
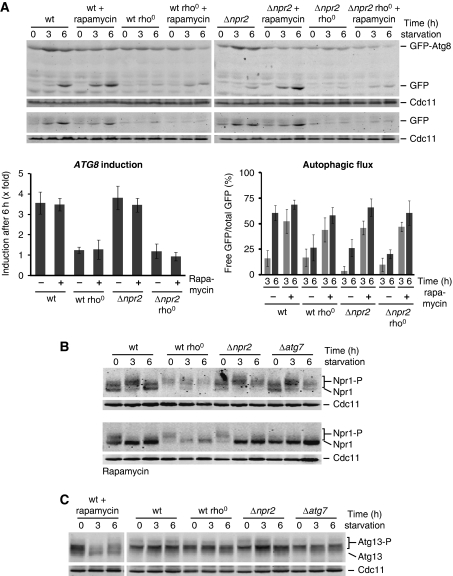
We examined the role of TORC1 in mitochondria-regulated autophagy during amino-acid starvation by monitoring the autophagic response in wild-type, wild-type rho0, Δnpr2, and Δnpr2 rho0 cells in the absence or presence of the TORC1-specific inhibitor rapamycin. NPR2 encodes a component of a conserved Npr2/Npr3 complex recently identified to have a critical role in TORC1 inhibition and cellular adaptation upon amino-acid starvation (Neklesa and Davis, 2009). Autophagic flux was strongly impaired in the Δnpr2 mutant compared with wild-type cells under amino-acid starvation (Figure 4A), indicating that inhibition of TORC1 via Npr2/3 is critical for the positive regulation of autophagic flux. Consistently, TORC1 inhibition by rapamycin treatment resulted in an indistinguishable autophagic flux in wild-type and Δnpr2 cells (Figure 4A). Interestingly, autophagic flux was significantly accelerated in rapamycin-treated wild-type cells compared with untreated wild-type cells, suggesting that TORC1 is only partially inactivated upon amino-acid starvation (Figure 4A, 3 versus 6 h). Significantly, we observed a similar rapamycin-mediated stimulation of autophagic flux in rho0 and Δnpr2 rho0 cells (Figure 4A), indicating that inhibition of TORC1 can suppress decreased autophagic flux caused by mitochondrial respiratory deficiency. It is interesting to note that wild-type and rho0 cells displayed remarkably similar autophagic flux during nitrogen starvation in the presence and absence of rapamycin compared with rapamycin-treated cells during amino-acid starvation (Figure 4A; Supplementary Figure S6A). Further, the autophagic response to nitrogen starvation was completely absent in the Δnpr2 mutant (Supplementary Figure S6A).
Figure 4.
Role of TORC1 in autophagy regulation under amino-acid starvation. (A) Wild-type, rho0, Δnpr2, and Δnpr2 rho0 cells expressing prATG8-GFP-ATG8 (upper panel) or prATG8-GFP (lower panel) were exposed to amino-acid starvation medium supplemented with galactose in the absence or presence of rapamycin. Samples were analysed as described in Figure 1A. The means and s.d. of four (n=4) independent experiments are indicated. (B) Wild-type, rho0, Δnpr2, and Δatg7 cells expressing prNPR1-NPR1-HA were exposed to amino-acid starvation medium supplemented with galactose in the absence (upper panels) or presence (lower panels) of rapamycin. The hyperphosphorylated (Npr1-P) and dephosphorylated (Npr1) forms of Npr1 are indicated. Cells were analysed at indicated time points by whole cell extraction and western blot analysis using α-HA and α-Cdc11 antibodies. (C) Wild-type, rho0, Δnpr2, and Δatg7 cells were exposed to amino-acid starvation medium supplemented with galactose. Additionally, wild-type cells were treated with rapamycin during starvation (left panel). Phosphorylated (Atg13-P) and dephosphorylated (Atg13) Atg13 was monitored at indicated time points by whole cell extraction and western blot analysis using α-Atg13 and α-Cdc11 antibodies.
To assess TORC1 activity in cells, we monitored phosphorylation of Npr1, a TORC1 effector that is hyperphosphorylated under growing conditions and dephosphorylated under nitrogen starvation or rapamycin treatment in a TORC1-dependent manner (Schmidt et al, 1998; Gander et al, 2008). Although the significance is not clear, the steady-state level of Npr1 was decreased in rho0 cells (Figure 4B). More importantly, however, we observed no significant differences in the phosphorylation pattern of Npr1 in wild-type, rho0, Δnpr2, or Δatg7 cells during amino-acid starvation (Figure 4B). In contrast, we observed an accumulation of the dephosphorylated form of Npr1 during nitrogen starvation in wild-type, rho0, and Δatg7, indicating a strong inhibition of TORC1 activity (Supplementary Figure S6B). Similarly, upon amino-acid starvation in the presence of rapamycin, we observed a shift towards the dephosphorylated form of Npr1 in all strains (Figure 4B). Consistent with published results, the phosphorylation pattern of Npr1 was unchanged in Δnpr2 cells during nitrogen starvation (Neklesa and Davis, 2009), but chemical inhibition of TORC1 with rapamycin caused the accumulation of the dephosphorylated Npr1 form. In contrast, rapamycin treatment did not further alter the phosphorylation status of Npr1 in wild-type, rho0, or Δatg7 cells (Supplementary Figure S6B). Similarly, the phosphorylation pattern of Atg13 did not change in wild-type, rho0, Δnpr2, or Δatg7 cells during amino-acid starvation (Figure 4C). In contrast, a strong decrease in the phosphorylated forms and appearance of the dephosphorylated form of Atg13 were observed in rapamycin-treated wild-type cells, consistent with inhibition of TORC1 (Figure 4C). These data demonstrate a dominant regulatory role for TORC1 activity during nitrogen starvation in contrast to amino-acid starvation and indicate that there are significant differences in autophagy regulation under these two conditions.
In contrast to autophagic flux, ATG8 induction was not affected in Δnpr2 cells compared with wild-type cells during amino-acid starvation (Figure 4A). Furthermore, rapamycin-mediated TORC1 inhibition did not alter ATG8 induction in wild-type or Δnpr2 cells or the defect in ATG8 induction caused by mitochondrial dysfunction in rho0 and Δnpr2 rho0 cells (Figure 4A). These results indicate that, under amino-acid starvation, the induction of ATG8 expression is regulated in a TORC1-independent manner. Thus, under conditions of amino-acid starvation, our results support the existence of two regulatory arms of the autophagic response that differentially control autophagic flux and ATG8 induction in a TORC1-dependent and TORC1-independent manner, respectively. Thus, mitochondria modulate ATG8 induction via regulatory mechanisms other than TORC1.
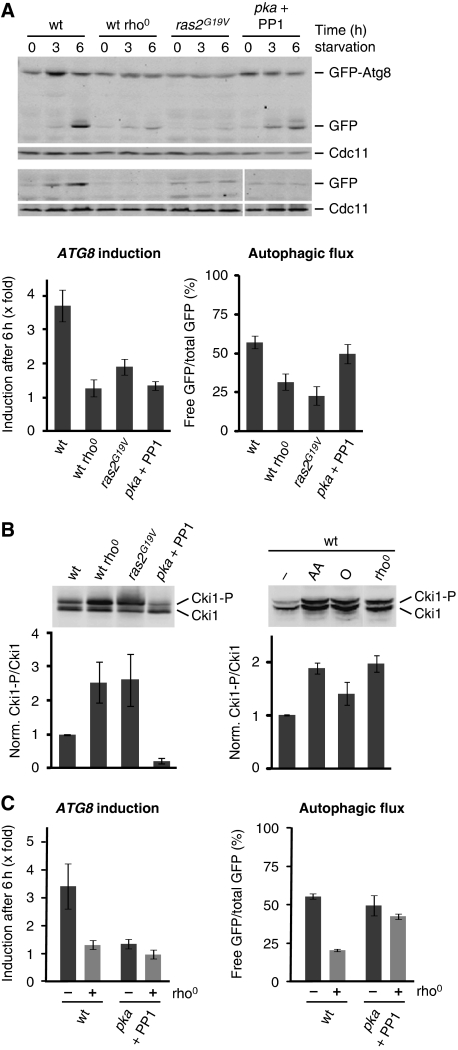
We tested whether mitochondria modulate the autophagic response via PKA by performing a comparative analysis of the role of PKA and mitochondrial function in the autophagic response. Specifically, we engineered cells that express a constitutively active variant of RAS2 (ras2G19V), which results in hyperactivation of PKA (Toda et al, 1985; Budovskaya et al, 2004). Additionally, we used cells that express variants of the three catalytic subunits of PKA (pka: tpk1M164G, tpk2M147G, tpk3M165G) instead of the wild-type alleles, which are sensitive to conditional inactivation by the inhibitor 1NM-PP1 (PP1) (Yorimitsu et al, 2007). We compared the autophagic response in these strains to wild-type and rho0 cells under amino-acid starvation. Consistent with previous reports, expression of the constitutively active ras2G19V mutant inhibited the autophagic response (Figure 5A) (Budovskaya et al, 2004). Remarkably, autophagic flux and ATG8 induction were impaired to a similar extent as observed for rho0 cells (Figure 5A), indicating that hyperactivation of PKA and mitochondrial respiratory deficiency lead to indistinguishable inhibition of the autophagy flux and ATG8 induction component of the autophagic response. Inhibition of PKA activity (pka + PP1) did not significantly influence autophagic flux, but, surprisingly, impaired ATG8 induction as observed in rho0 or ras2G19V-expressing cells (Figure 5A). Thus, some level of PKA activity is required for full ATG8 induction in wild-type cells under amino-acid starvation.
Figure 5.
Mitochondrial function controls autophagy by modulating PKA activity. (A) PKA-dependent regulation of the autophagic response under amino-acid starvation. Wild-type, rho0, pka, and ras2G19V-expressing cells harbouring prATG8-GFP-ATG8 (upper panels) or prATG8-GFP (lower panels) were exposed to amino-acid starvation medium supplemented with galactose. PKA activity in pka was inhibited by addition of 1NM-PP1 (PP1; 1 μg/ml). Samples were analysed as described in Figure 1A; autophagic flux was determined after 3 and 6 h. (B) In vivo activity of PKA. Wild-type, rho0, pka, and ras2G19V-expressing cells harbouring 6xMYC-cki12−200(S125/130A) (Cki1) were grown in galactose medium. When indicated, wild-type cells were grown in galactose medium in the presence of antimycin A (AA) or oligomycin (O) for 6 h. PKA-dependent phosphorylation of Cki1 was analysed by whole cell extraction and western blot analysis using a α-Myc antibody (upper panels). Ratio of phosphorylated (Cki1-P) and non-phosphorylated (Cki1) forms of Cki1 relative to wild-type cells (wt=1) (lower panels). (C) PKA inhibition restores autophagic flux, but not ATG8 induction in the presence of mitochondrial dysfunction. Wild-type, rho0, pka, and pka rho0 cells expressing prATG8-GFP-ATG8 were treated as described in (A). Samples were analysed as described in Figure 1A. The means and s.d. of four (n=4) independent experiments are indicated in (A–C).
These data raise the possibility that mitochondria regulate the autophagic response in a manner dependent on PKA activity. To test this, we measured PKA activity in vivo in wild-type, wild-type rho0, ras2G19V-expressing, and pka (+ PP1) cells, or in wild-type cells in the presence of antimycin A or oligomycin using a PKA substrate reporter, which was created from a native substrate protein, Cki1 (Deminoff et al, 2006). This PKA substrate reporter contains the first 200 amino acids of Cki1 and possesses mutations at two known protein kinase C sites (S125A, S130A) making its phosphorylation exclusively PKA dependent (S185). We detected PKA-dependent phosphorylation of the Cki1 reporter by a mobility shift in SDS–PAGE analysis and quantified the ratio of phosphorylated and unphosphorylated forms as a measure for the in vivo activity of PKA in cells during log-phase in synthetic galactose medium. As expected, we observed a decrease in the phosphorylated Cki1-P form upon inactivation of PKA (pka + PP1) compared with wild-type cells (Figure 5B). Remarkably, in rho0 cells and wild-type cells treated with antimycin A, we observed a two-fold increase in the Cki1-P/Cki1 ratio relative to wild-type cells, similar to what we detected in cells with hyperactive PKA (ras2G19V) (Figure 5B). Interestingly, PKA activity was only mildly increased in wild-type cells in the presence of oligomycin, consistent with its relatively small inhibitory effects on the autophagic response (Figures 2B and 5B). These data demonstrate that the in vivo activity of PKA is modulated by mitochondrial function and that mitochondrial respiratory deficiency acts as a positive regulator of PKA. In this context, it is interesting to note that the PKA pathway has been previously shown to regulate mitochondrial function (Chevtzoff et al, 2005; Chen and Powers, 2006). Thus, our findings suggest that mitochondria and PKA affect each other reciprocally, possibly to allow for adaptation of cell metabolism. Moreover, our data suggest that inhibition of ATG8 induction and autophagic flux in wild-type cells in the presence of antimycin A or rho0 cells is caused by an increase in PKA activity triggered by mitochondrial respiratory deficiency.
We tested whether inhibition of PKA activity reversed the defects in the autophagic response in cells with mitochondrial dysfunction by analysing rho0 cells harbouring analogue-sensitive pka alleles under amino-acid starvation in the presence of 1NM-PP1. As we previously observed, both components of the autophagic response were stimulated in wild-type cells (Figure 5C). The addition of 1NM-PP1 to analogue-sensitive wild-type cells (pka) inhibited ATG8 induction, but autophagic flux was stimulated in a manner similar to wild-type cells (Figure 5C). Importantly, in contrast to rho0 cells, a strong stimulation of autophagic flux was observed in the presence of 1NM-PP1 in analogue-sensitive pka rho0 cells (Figure 5C). These observations are consistent with our hypothesis that mitochondrial respiratory deficiency stimulates PKA, which in turn suppresses autophagic flux in rho0 cells. In contrast to autophagic flux, however, we did not observe stimulation in ATG8 induction upon addition of 1NM-PP1 to analogue-sensitive pka rho0 cells (Figure 5C). This observation is consistent with our previous finding that ATG8 induction requires some baseline level of PKA activity in wild-type cells (Figure 5A) and suggests that a similar PKA requirement exists in cells with mitochondrial dysfunction. Our data suggest that mitochondrial function controls the autophagic response by modulating PKA activity, which in turn regulates both arms of the autophagic response—autophagic flux and ATG8 induction—under amino-acid starvation.
In the autophagic flux response, mitochondrial respiratory deficiency impairs Atg1–Atg13 complex recruitment to the PAS
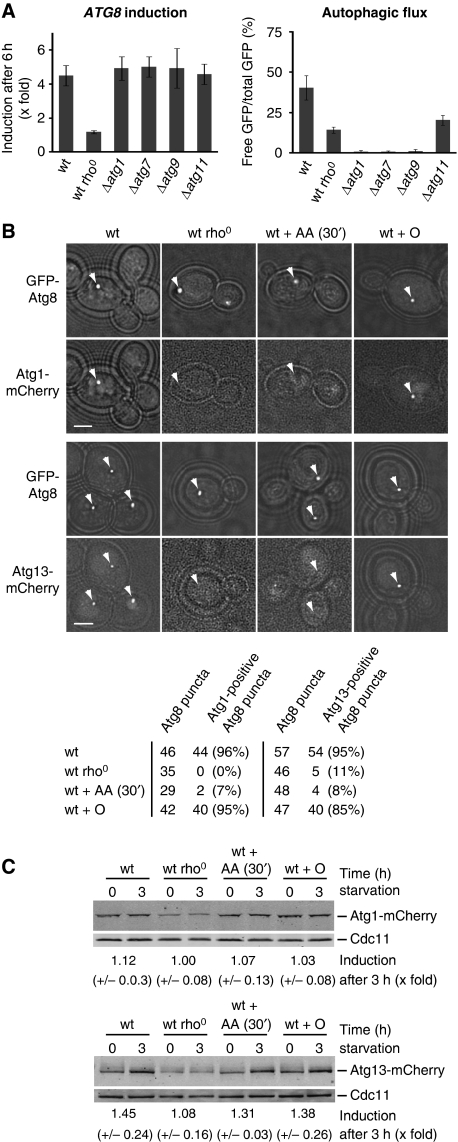
TORC1 and PKA regulate the Atg1–Atg13 complex, which in turn is a key regulator of the initial steps in autophagosome formation (Kamada et al, 2000; Cheong et al, 2008). First, we addressed whether ATG8 induction and autophagic flux depend on the Atg1–Atg13 complex and whether ATG8 induction and autophagic flux are interdependent events. Towards this goal, we analysed the autophagic response in Δatg1, Δatg7, and Δatg9 cells and Δatg11 cells, which lack essential factors for general and selective autophagy, respectively (Lang et al, 2000; Kim et al, 2001). Significantly, autophagic flux was completely blocked in Δatg1, Δatg7, or Δatg9 cells (Figure 6A). We also observed a partial inhibition in autophagic flux in Δatg11 cells during amino-acid starvation, indicating that Atg11 has a more general role during amino-acid starvation. However, ATG8 induction was unaffected in all tested ATG mutants (Figure 6A). Thus, during amino-acid starvation, the Atg1–Atg13 complex is required for autophagic flux regulation, but dispensable for ATG8 induction. In addition, ATG8 induction is not dependent on the autophagy machinery required for autophagic flux. These findings are consistent with the existence of two independent regulatory arms of the autophagic response pathway.
Figure 6.
Mitochondrial respiratory deficiency impairs PAS recruitment of the Atg1–Atg13 complex under amino-acid starvation. (A) ATG8 induction is independent of the Atg1–Atg13 complex and autophagic flux. Wild-type, rho0, Δatg1, Δatg7, Δatg9, and Δatg11 cells harbouring prATG8-GFP-ATG8 were analysed as described in Figure 1A. The means and s.d. of four (n=4) independent experiments are indicated. (B) Atg1 and Atg13 recruitment to the PAS depends on mitochondrial function. Wild-type and rho0 cells harbouring prATG8-GFP-ATG8 and prATG1-ATG1-mCherry (upper panels) or prATG13-ATG13-mCherry (lower panels) were exposed to amino-acid starvation medium supplemented with galactose for 3 h. Wild-type cells were treated with antimycin A (AA 30′) after 2.5 h of starvation for 30 min or with oligomycin (O) for 3 h of starvation. Arrowheads indicate the position of GFP-Atg8 puncta. Transmission and fluorescence light microscopy images were superimposed to visualize cellular boundaries. Scale bar represents 1.5 μm. (C) Steady-state levels of Atg1- and Atg13-mCherry during amino-acid starvation. Wild-type, rho0, and Δatg7 cells expressing prATG1-ATG1-mCherry (upper panels) or prATG13-ATG13-mCherry (lower panels) were exposed and treated as described in (B) and analysed by whole cell extraction and western blot analysis using α-dsRed and α-Cdc11 antibodies.
To determine the basis for mitochondrial regulation of autophagic flux, we exploited the observation that PKA-dependent phosphorylation of Atg1 and Atg13 regulates their localization to the PAS (Budovskaya et al, 2005; Stephan et al, 2009). Specifically, Atg1 and Atg13 are not recruited to the PAS in the presence of hyperactive PKA. Given that cells with mitochondrial respiratory deficiency display a strong increase in PKA in vivo activity (Figure 5B), we determined whether Atg1 and Atg13 localized to puntate structures labelled by GFP-Atg8, which are likely the PAS during amino-acid starvation under conditions of mitochondrial dysfunction. In wild-type cells, GFP-Atg8 and Atg1-mCherry or GFP-Atg8 and Atg13-mCherry co-localized in the majority of puncta under amino-acid starvation conditions, consistent with a stimulation of the autophagic response (Figure 6B). Although GFP-Atg8 puncta were present at similar frequency (50–75% of the cells; n⩾150) in rho0 or antimycin A-treated wild-type cells as compared with wild-type cells, there was a significant decrease in Atg1- or Atg13-mCherry-positive Atg8 puncta at these structures (Figure 6B). In contrast, Atg1- or Atg13-mCherry co-localized with GFP-Atg8 in the presence of oligomycin during amino-acid starvation to an extent similar to that observed in wild-type cells consistent with the smaller increase in PKA activity observed as compared with rho0 or antimycin A-treated wild-type cells (Figures 5B and 6B). Under all conditions, Atg1-mCherry was present at comparable steady-state levels in cells as monitored by western analysis (Figure 6C). The expression of Atg13-mCherry was induced ≈1.5-fold in wild-type cells (Figure 6C). This induction was inhibited in rho0 cells, but it was comparable with wild-type cells in antimycin A-treated or oligomycin-treated cells (Figure 6C). These findings suggest that mislocalization of Atg13 observed in response to mitochondrial dysfunction is a consequence of PKA regulation rather than protein levels. Thus, during amino-acid starvation, our data suggest that even short-term mitochondrial respiratory deficiency strongly impairs the recruitment of Atg1 or Atg13 to the PAS, consistent with our observations that mitochondrial respiratory deficiency increases PKA activity and impairs autophagic flux under these conditions. Taken together, these data are consistent with a model in which mitochondrial respiratory deficiency induces PKA activity and, thereby, suppresses the two arms of autophagy regulation: induction of critical ATG components like Atg8 and Atg14 and organization of the PAS required for autophagic flux.
Discussion
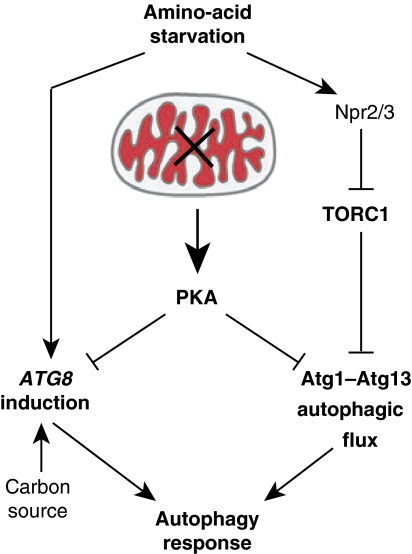
Our data demonstrate that mitochondrial function is a critical regulator of autophagy in S. cerevisiae and support the model summarized in Figure 7. Our data indicate that the autophagy response to amino-acid starvation is controlled by at least two components, autophagic flux and ATG gene induction, and are consistent with previously published data (Abeliovich et al, 2000). Interestingly, these two components are differentially regulated. Our data together with published work indicate that the autophagic flux component is controlled by the Atg1–Atg13 complex and is negatively regulated by independent phosphorylation by both TORC1 and PKA (Funakoshi et al, 1997; Matsuura et al, 1997; Kamada et al, 2000, 2010; Budovskaya et al, 2005; Stephan et al, 2009). The ATG gene induction component, specifically ATG8, modulates the magnitude of the autophagic response by controlling the size of the autophagic compartment (Abeliovich et al, 2000; Xie et al, 2008). In contrast to autophagic flux, our data indicate that ATG8 induction is negatively controlled by PKA in an Atg1- and TORC1-independent manner. Importantly, within this autophagy regulatory network, mitochondrial respiratory function is a critical modulator of PKA activity and regulates both the autophagic flux and ATG8 induction components of the autophagy pathway.
Figure 7.
Model for the role of mitochondrial function in autophagy regulation under amino-acid starvation. Amino-acid starvation induces the two regulatory arms of the autophagic response, ATG8 induction and autophagic flux. Autophagic flux is regulated in an Atg1, PKA, and TORC1-dependent manner, while ATG8 induction is regulated by PKA. Mitochondrial respiratory deficiency induces PKA activity and, thereby, suppresses both arms of the autophagic response.
TOR and PKA function together to coordinate nutrient availability with cell proliferation and growth in eukaryotic cells (Dechant and Peter, 2008; Zaman et al, 2008). PKA transitions cells from aerobic respiration to fermentative growth via the suppression of genes associated with mitochondrial activity (Chen and Powers, 2006; Dechant and Peter, 2008; Zaman et al, 2008). Our observation that mitochondrial dysfunction activates PKA in cells suggests that a reciprocal relationship exists between PKA activity and mitochondrial function. The physiological role of this regulatory circuit likely insures that cellular metabolism adapts to fermentation in response to respiratory deficiency. The circuit may also function to lower mitochondrial stress by suppressing the synthesis and import of new mitochondrial proteins so that the mitochondrial protein quality control systems can be devoted to the maintenance of existing proteins. Lowering stress would provide a window of time for recovery, similar to other stress responses, such as the unfolded protein response in the endoplasmic reticulum (Ron and Walter, 2007). In this context, the suppression of autophagy by mitochondrial dysfunction would be another facet in this putative stress pathway. Alternatively, the degradation of cellular components might be disadvantageous under conditions where ATP can be solely generated by glycolysis and PKA activation in response to mitochondrial dysfunction might insure adaptation of the autophagic response to the metabolic capacity of the cell.
One key question is how mitochondrial respiratory deficiency signals to PKA. Our data indicate that the generation of mitochondrial membrane potential by respiration is a critical factor for autophagy induction and might signal the functional state of mitochondria. Consistently, under these conditions, we find the strongest increase in cellular PKA activity. It is interesting that in both mammalian and yeast cells depolarized mitochondria can induce selective autophagic turnover (mitophagy), consistent with a quality control mechanism (Priault et al, 2005; Nowikovsky et al, 2007; Narendra et al, 2008; Twig et al, 2008). However, our data indicate that mitochondrial respiratory deficiency inhibits general and selective autophagy, including mitophagy under amino-acid starvation (Supplementary Figure S7). Hence, the consequences of mitochondrial dysfunction on autophagy and mitophagy regulation can substantially vary depending on the nature and severity of the defects, for example collapse of mitochondrial membrane potential versus respiratory deficiency, as well as growth conditions in eukaryotic cells. Furthermore, our findings suggest that the accumulation of respiratory-deficient mitochondria beyond a certain threshold decreases the autophagic and mitophagic capacity of the cell and initiates a negative feedback loop that ultimately results in cellular ageing or cell death and might contribute to or initiate age-related diseases. A similar principle has been proposed for the collapse of the proteostasis capacity during ageing in Caenorhabditis elegans (Ben-Zvi et al, 2009), suggesting a common interdependence of cell metabolism, stress responses, and quality control mechanisms. While this model remains speculative at this point, it is interesting to note that TORC1 inhibition can alleviate effects on autophagic flux in the presence of respiratory-deficient mitochondria in our system. This is consistent with numerous reports on beneficial effects of modulated TOR activity for life span and age-related diseases across species (Kapahi et al, 2010). It is interesting to note that in fly neurons during ageing, the expression of several essential autophagy genes including Atg8a declines (Simonsen et al, 2008). This raises the possibility that in neurons mitochondrial dysfunction might compromise ATG8 expression in ageing animal models in a similar manner to what we observe in yeast. Indeed, increased Atg8a expression can reverse neuronal damage and shortened life span, suggesting that Atg8a levels are rate limiting under these conditions (Simonsen et al, 2008). In this context, our findings, which indicate an interrelation of two common risk factors, mitochondrial dysfunction and impaired autophagy, may transform our understanding of the mechanism underlying ageing, cancerogenesis, and neurodegenerative diseases by raising the possibility of a common pathogenic mechanism.
Materials and methods
Cloning and plasmids
pRS416-prATG8-GFP-ATG8 has been described previously (Abeliovich et al, 2003). pRS426-prATG8-GFP was generated by cloning an EcoRI and HindIII flanked prATG8-GFP fragment from pRS416-prATG8-GFP-ATG8 into pRS426. pRS426-prATG14-GFP was generated by cloning prATG14 (−1000 to −1 bp 5′-region) flanked by EcoRI and PstI into pRS426 with PstI and XmaI flanked GFP. pRS314-prATG1-ATG1-mCherry and pRS314-prATG13-ATG13-mCherry were generated by cloning prATG1-ATG1 (−800 to −1 bp 5′-region plus ORF without stop codon) flanked by NotI and BamHI or prATG13-ATG13 (−1000 to −1 bp 5′-region plus ORF without stop codon) flanked by NotI and XmaI into pRS314 containing mCherry (Shaner et al, 2004), respectively. pRS314-prNPR1-NPR1-HA was generated by cloning prNPR1-NPR1 (−763 to −1 bp 5′-region plus ORF without stop codon) containing a single HA tag flanked by SacI and XmaI into pRS314. pCM184-ras2G19V was generated by cloning RAS2 flanked by BamHI and NotI into pCM184 (Gari et al, 1997), and mutation G19V was introduced by site-directed mutagenesis. pRS416-prOM45-OM45-GFP was generated by cloning prOM45-OM45 (−878 to −1 bp 5′-region plus ORF without stop codon) flanked by NotI and BamHI into pRS416 with BamHI and EcoRI flanked GFP. pRS423-prCUP-6xMYC-cki12−200(S125/130A) was generated as described in Deminoff et al (2006) and mutations S125/130A were introduced by mutagenesis PCR (kind gift from Dr Paul Herman).
Yeast strains and media
All strains used in this study are derivatives of W303 (leu2-3,112; ura3-1; his3-11,15; trp1-1; ade2-1; can1-100). Gene deletions were generated by PCR-based targeted homologous recombination replacing complete ORFs by kanMX6 (Δatg1, Δatg7, Δatg9), natMX (Δcbs1, Δmss51, Δpet111, Δpet122, or Δatp10), or HIS3MX6 (Δatg11, Δatg32, Δnpr2, Δvma2) cassettes (Longtine et al, 1998). The rho0 strains were generated by growth in YPD medium supplemented with ethidium bromide (25 μg/ml). Strains were grown to log-phase in synthetic galactose medium (0.7% yeast nitrogen base, 2% galactose, and 0.05% glucose) or YPlactate medium (1% yeast extract, 2% peptone, and 2% lactate) and exposed to amino-acid (0.1% yeast extract) or nitrogen (-N; 0.17% yeast nitrogen base without (NH4)2SO4 and amino acids) starvation medium supplemented with acetate (1%), galactose (2%), or glucose (2%) as indicated. Antimycin A (1 μg/ml), oligomycin (7.5 μg/ml), or rapamycin (400 ng/ml) was added when indicated. To suppress or allow expression, strains harbouring pCM184-ras2G19V were grown in the presence or absence (12 h) of doxycyclin (20 μg/ml), respectively. The 1NM-PP1-sensitive pka (tpk1M164G, tpk2M147G, and tpk3M165G) strain has been described in Yorimitsu et al (2007). To block PKA activity, pka strains were pre-incubated (60 min) and starved in the presence of 1NM-PP1 (1 μg/ml) in indicated media. Strains harbouring pRS423-prCUP-6xMYC-cki12−200(S125/130A) were grown in synthetic galactose medium as described in Deminoff et al (2006).
Whole cell extraction, western blot analysis, and quantification
At indicated time points, cells corresponding to 0.25 OD600 units were collected and lysed by alkaline whole cell extraction (0.255 M NaOH, 1% β-mercaptoethanol). Protein extracts were analysed by SDS–PAGE and immunoblotting (α-Atg13, polyclonal, Dr Klionsky; α-Cdc11, polyclonal, Novus Biologicals; α-dsRed, polyclonal, Clontech; α-GFP and α-HA, monoclonal, Covance; α-Myc, monoclonal, Sigma; α-Pgk1, monoclonal, Molecular Probes) and visualized with the appropriate secondary antibodies conjugated to IRDye (800CW; LI-COR Biosciences). Quantification was performed using the Odyssey Infrared Imaging System (LI-COR Biosciences).
Fluorescence microscopy
Cells were mounted in 0.5% low melt agarose in indicated starvation medium and viewed with a microscope (IX70 Deltavision; Olympus) using a × 60 1.4 NA objective (Olympus) and a 100-W mercury lamp (Applied Precision, LLC). Two- and three-dimensional light microscopy data were collected using an integrated, cooled charge-coupled device-based camera (MicroMax; Princeton) equipped with an interline chip (Sony). Three-dimensional data sets were processed using DeltaVision's iterative, constrained three-dimensional deconvolution method to remove out of focus light. Representative sections of deconvolved images were processed in SoftWorx (Applied Precision, LLC) and PHOTOSHOP (Adobe Systems, San Jose, CA) software unless otherwise indicated.
Measurement of cellular and mitochondrial ATP
At indicated time points, cells corresponding to 50 OD600 units were collected, washed, and resuspended in 1 ml NMIB buffer (0.6 M sorbitol; 5 mM MgCl2; 50 mM KCl; 100 mM KOAc; 20 mM HEPES pH 7.4), and frozen in liquid nitrogen as droplets. Cell breakage was performed using the SPEX freezer mill 6750 (3 cycles; rate 7 for 2 min). After cell debris was removed from thawed cell powder by centrifugation, the protein concentration of the supernatant (total cell) was determined by a standard Bradford assay and 10 μl were mixed with 90 μl TCA (5%) for ATP determination using the promega ENLITEN® ATP assay, according to the instructions of the manufacturer. Mitochondria were isolated from total cell supernatant by differential centrifugation and resuspended in 25 μl NMIB buffer. The protein concentration was determined by a standard Bradford assay and 10 μl were mixed with 90 μl TCA (5%) for ATP determination.
Measurement of mitochondrial membrane potential
After 3 h of amino-acid starvation, cells were exposed to 3,3′-dihexyloxacarbocyanine iodide (DiOC3(6)) (10 ng/ml final concentration) for 5 min at 23°C. Three-dimensional data sets of cells with identical exposure times (1 s) and intensity scales were collected. For analysis, pixel intensities for cross-sections of at least 10 mitochondrial tubules in 5 representative cells were determined using ImageJ software (NIH) for each condition.
Supplementary Material
Acknowledgments
We thank Dr Paul Herman and Dr Dan Klionsky for providing constructs, antibodies, and yeast strains and Dr Ted Powers as well as members of the Nunnari laboratory for critical discussion and comments. JN is supported by NIH Grant R01GM062942.
Author contributions: MG and JN designed the study and wrote the manuscript. MG performed all experiments and analysed the data.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abeliovich H, Dunn WA Jr, Kim J, Klionsky DJ (2000) Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol 151: 1025–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeliovich H, Zhang C, Dunn WA Jr, Shokat KM, Klionsky DJ (2003) Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol Biol Cell 14: 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman SH, Tzagoloff A (1990) ATP10, a yeast nuclear gene required for the assembly of the mitochondrial F1-F0 complex. J Biol Chem 265: 9952–9959 [PubMed] [Google Scholar]
- Banerjee R, Starkov AA, Beal MF, Thomas B (2009) Mitochondrial dysfunction in the limelight of Parkinson's disease pathogenesis. Biochim Biophys Acta 1792: 651–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zvi A, Miller EA, Morimoto RI (2009) Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci USA 106: 14914–14919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK (2005) An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc Natl Acad Sci USA 102: 13933–13938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK (2004) The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem 279: 20663–20671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso SM, Pereira CF, Moreira PI, Arduino DM, Esteves AR, Oliveira CR (2010) Mitochondrial control of autophagic lysosomal pathway in Alzheimer's disease. Exp Neurol 223: 294–298 [DOI] [PubMed] [Google Scholar]
- Cebollero E, Reggiori F (2009) Regulation of autophagy in yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1793: 1413–1421 [DOI] [PubMed] [Google Scholar]
- Chan EY, Kir S, Tooze SA (2007) siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem 282: 25464–25474 [DOI] [PubMed] [Google Scholar]
- Chan TF, Bertram PG, Ai W, Zheng XF (2001) Regulation of APG14 expression by the GATA-type transcription factor Gln3p. J Biol Chem 276: 6463–6467 [DOI] [PubMed] [Google Scholar]
- Chen JC, Powers T (2006) Coordinate regulation of multiple and distinct biosynthetic pathways by TOR and PKA kinases in S cerevisiae. Curr Genet 49: 281–293 [DOI] [PubMed] [Google Scholar]
- Chen LB (1988) Mitochondrial membrane potential in living cells. Annu Rev Cell Biol 4: 155–181 [DOI] [PubMed] [Google Scholar]
- Chen S, Tarsio M, Kane PM, Greenberg ML (2008) Cardiolipin mediates cross-talk between mitochondria and the vacuole. Mol Biol Cell 19: 5047–5058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Nair U, Geng J, Klionsky DJ (2008) The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell 19: 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevtzoff C, Vallortigara J, Averet N, Rigoulet M, Devin A (2005) The yeast cAMP protein kinase Tpk3p is involved in the regulation of mitochondrial enzymatic content during growth. Biochim Biophys Acta 1706: 117–125 [DOI] [PubMed] [Google Scholar]
- Costanzo MC, Fox TD (1988) Specific translational activation by nuclear gene products occurs in the 5′ untranslated leader of a yeast mitochondrial mRNA. Proc Natl Acad Sci USA 85: 2677–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechant R, Peter M (2008) Nutrient signals driving cell growth. Curr Opin Cell Biol 20: 678–687 [DOI] [PubMed] [Google Scholar]
- Decoster E, Simon M, Hatat D, Faye G (1990) The MSS51 gene product is required for the translation of the COX1 mRNA in yeast mitochondria. Mol Gen Genet 224: 111–118 [DOI] [PubMed] [Google Scholar]
- Deminoff SJ, Howard SC, Hester A, Warner S, Herman PK (2006) Using substrate-binding variants of the cAMP-dependent protein kinase to identify novel targets and a kinase domain important for substrate interactions in Saccharomyces cerevisiae. Genetics 173: 1909–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker N, Mor A, Journo D, Abeliovich H (2010) Induction of autophagic flux by amino acid deprivation is distinct from nitrogen starvation-induced macroautophagy. Autophagy 6: 879–890 [DOI] [PubMed] [Google Scholar]
- Funakoshi T, Matsuura A, Noda T, Ohsumi Y (1997) Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene 192: 207–213 [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Morselli E, Kepp O, Vitale I, Rigoni A, Vacchelli E, Michaud M, Zischka H, Castedo M, Kroemer G (2010) Mitochondrial gateways to cancer. Mol Aspects Med 31: 1–20 [DOI] [PubMed] [Google Scholar]
- Gander S, Bonenfant D, Altermatt P, Martin DE, Hauri S, Moes S, Hall MN, Jenoe P (2008) Identification of the rapamycin-sensitive phosphorylation sites within the Ser/Thr-rich domain of the yeast Npr1 protein kinase. Rapid Commun Mass Spectrom 22: 3743–3753 [DOI] [PubMed] [Google Scholar]
- Gari E, Piedrafita L, Aldea M, Herrero E (1997) A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13: 837–848 [DOI] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12: 119–131 [DOI] [PubMed] [Google Scholar]
- Graham LA, Powell B, Stevens TH (2000) Composition and assembly of the yeast vacuolar H(+)-ATPase complex. J Exp Biol 203: 61–70 [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441: 885–889 [DOI] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N (2008) FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 181: 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL (1999) Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci USA 96: 14866–14870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Klionsky DJ (2009) Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43: 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JM, Funes S (2005) Biogenesis of cytochrome oxidase-sophisticated assembly lines in the mitochondrial inner membrane. Gene 354: 43–52 [DOI] [PubMed] [Google Scholar]
- Huang J, Klionsky DJ (2007) Autophagy and human disease. Cell Cycle 6: 1837–1849 [DOI] [PubMed] [Google Scholar]
- Huang WP, Scott SV, Kim J, Klionsky DJ (2000) The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem 275: 5845–5851 [DOI] [PubMed] [Google Scholar]
- Johnson LV, Walsh ML, Bockus BJ, Chen LB (1981) Monitoring of relative mitochondrial membrane potential in living cells by fluorescence microscopy. J Cell Biol 88: 526–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y (2005) Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell 16: 2544–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y (2000) Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 150: 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Yoshino KI, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y (2010) Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol 30: 1049–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ (2008) Mitophagy in yeast occurs through a selective mechanism. J Biol Chem 283: 32386–32393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ (2009) Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell 17: 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, Thomas EL, Kockel L (2010) With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab 11: 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y (2008) Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell 19: 2039–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA Jr, Klionsky DJ (2001) Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol 153: 381–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y (1999) Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 147: 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissova I, Deffieu M, Manon S, Camougrand N (2004) Uth1p is involved in the autophagic degradation of mitochondria. J Biol Chem 279: 39068–39074 [DOI] [PubMed] [Google Scholar]
- Kissova I, Salin B, Schaeffer J, Bhatia S, Manon S, Camougrand N (2007) Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy 3: 329–336 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441: 880–884 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert AJ, Brand MD (2009) Reactive oxygen species production by mitochondria. Methods Mol Biol 554: 165–181 [DOI] [PubMed] [Google Scholar]
- Lang T, Reiche S, Straub M, Bredschneider M, Thumm M (2000) Autophagy and the cvt pathway both depend on AUT9. J Bacteriol 182: 2125–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G, Uchiyama Y, Westaway D, Cuervo AM, Nixon RA (2010) Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141: 1146–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Ludovico P, Sansonetty F, Corte-Real M (2001) Assessment of mitochondrial membrane potential in yeast cell populations by flow cytometry. Microbiology 147: 3335–3343 [DOI] [PubMed] [Google Scholar]
- Matsuura A, Tsukada M, Wada Y, Ohsumi Y (1997) Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192: 245–250 [DOI] [PubMed] [Google Scholar]
- Meijer AJ, Codogno P (2009) Autophagy: regulation and role in disease. Crit Rev Clin Lab Sci 46: 210–240 [DOI] [PubMed] [Google Scholar]
- Morselli E, Galluzzi L, Kepp O, Vicencio JM, Criollo A, Maiuri MC, Kroemer G (2009) Anti- and pro-tumor functions of autophagy. Biochim Biophys Acta 1793: 1524–1532 [DOI] [PubMed] [Google Scholar]
- Nakamura N, Matsuura A, Wada Y, Ohsumi Y (1997) Acidification of vacuoles is required for autophagic degradation in the yeast, Saccharomyces cerevisiae. J Biochem 121: 338–344 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10: 458–467 [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183: 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ (2010) PINK1 is selectively stabilized on impaired mitochondria to activate parkin. PLoS Biol 8: e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neklesa TK, Davis RW (2009) A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex. PLoS Genet 5: e1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y (1998) Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem 273: 3963–3966 [DOI] [PubMed] [Google Scholar]
- Nowikovsky K, Reipert S, Devenish RJ, Schweyen RJ (2007) Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ 14: 1647–1656 [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y (2009) Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell 17: 87–97 [DOI] [PubMed] [Google Scholar]
- Petit P, Glab N, Marie D, Kieffer H, Metezeau P (1996) Discrimination of respiratory dysfunction in yeast mutants by confocal microscopy, image, and flow cytometry. Cytometry 23: 28–38 [DOI] [PubMed] [Google Scholar]
- Poutre CG, Fox TD (1987) PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics 115: 637–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priault M, Salin B, Schaeffer J, Vallette FM, di Rago JP, Martinou JC (2005) Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ 12: 1613–1621 [DOI] [PubMed] [Google Scholar]
- Rodel G (1997) Translational activator proteins required for cytochrome b synthesis in Saccharomyces cerevisiae. Curr Genet 31: 375–379 [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8: 519–529 [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z (2007) Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 26: 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Beck T, Koller A, Kunz J, Hall MN (1998) The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J 17: 6924–6931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ (2004) Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem 279: 29889–29894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD (2008) Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4: 176–184 [DOI] [PubMed] [Google Scholar]
- Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK (2009) The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci USA 106: 17049–17054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen DF, Narendra DP, Tanaka A, Manfredi G, Youle RJ (2010) Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc Natl Acad Sci USA 107: 11835–11840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y (2007) Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12: 209–218 [DOI] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y (1992) Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol 119: 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal R, Winter G, Ecker N, Klionsky DJ, Abeliovich H (2007) Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J Biol Chem 282: 5617–5624 [DOI] [PubMed] [Google Scholar]
- Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E (1999) Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol Biol Cell 10: 1367–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Uno I, Ishikawa T, Powers S, Kataoka T, Broek D, Cameron S, Broach J, Matsumoto K, Wigler M (1985) In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40: 27–36 [DOI] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333: 169–174 [DOI] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS (2008) Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 27: 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A, Barrientos A, Neupert W, Herrmann JM (2004) Atp10p assists assembly of Atp6p into the F0 unit of the yeast mitochondrial ATPase. J Biol Chem 279: 19775–19780 [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, Magrane J, Moore DJ, Dawson VL, Grailhe R, Dawson TM, Li C, Tieu K, Przedborski S (2010) PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA 107: 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Cuervo AM (2010) Autophagy gone awry in neurodegenerative diseases. Nat Neurosci 13: 805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ (2007) Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Xie Z, Nair U, Klionsky DJ (2008) Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 19: 3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Geng J, Yen WL, Wang K, Klionsky DJ (2010) Positive or negative roles of different cyclin-dependent kinase Pho85-cyclin complexes orchestrate induction of autophagy in Saccharomyces cerevisiae. Mol Cell 38: 250–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, He C, Wang K, Klionsky DJ (2009) Tap42-associated protein phosphatase type 2A negatively regulates induction of autophagy. Autophagy 5: 616–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Zaman S, Broach JR, Klionsky DJ (2007) Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell 18: 4180–4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR (2008) How Saccharomyces responds to nutrients. Annu Rev Genet 42: 27–81 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.