Abstract
Transmissible spongiform encephalopathies, or prion diseases, are fatal degenerative disorders of the central nervous system that affect humans and animals. Prions are nonconventional infectious agents whose replication depends on the host prion protein (PrP). Transmission of prions to cultured cells has proved to be a particularly difficult task, and with a few exceptions, their experimental propagation relies on inoculation to laboratory animals. Here, we report on the development of a permanent cell line supporting propagation of natural sheep scrapie. This model was obtained by stable expression of a tetracycline-regulatable ovine PrP gene in a rabbit epithelial cell line. After exposure to scrapie agent, cultures were repeatedly found to accumulate high levels of abnormal PrP (PrPres). Cell extracts induced a scrapie-like disease in transgenic mice overexpressing ovine PrP. These cultures remained healthy and stably infected upon subpassaging. Such data show that (i) cultivated cells from a nonneuronal origin can efficiently replicate prions; and (ii) species barrier can be crossed ex vivo through the expression of a relevant PrP gene. This approach led to the ex vivo propagation of a natural transmissible spongiform encephalopathy agent (i.e., without previous experimental adaptation to rodents) and might be applied to human or bovine prions.
Transmissible spongiform encephalopathies (TSEs) are fatal infectious neurodegenerative diseases of the central nervous system (1, 2). These naturally occurring human and animal disorders include Creutzfeldt–Jakob disease, scrapie in sheep and goats, and the more recently observed bovine spongiform encephalopathy in cattle. In most cases, these diseases are associated with the accumulation of an abnormal pathological isoform of the host-derived prion protein (PrP).
PrP is a glycoprotein located at the cell surface, where it is bound by a glycosyl-phosphatidylinositol anchor (3). It is present in a variety of tissues (4, 5) and is mainly expressed in the central nervous system, particularly in the neurons (6). Although the precise physiological function of PrP remains to be established, a wealth of experimental data demonstrates its essential role in the susceptibility and in the pathogenesis of TSEs. Ablation of the Prnp gene renders mice unable to replicate murine-adapted scrapie strains (7), and increasing PrP overexpression levels in transgenic mice generally reduces the incubation time (8, 9). Allelic forms of PrP have been linked to the disease susceptibility in several species including mice (10), sheep (11), and humans (12, 13). The genetic linkage between familial TSEs and mutations in the human Prnp gene (14) also exemplifies the crucial role of PrP in TSEs. In mice, expression of PrP homologous to that of the infecting species can lower the transmission barrier among species (15, 16).
The accumulation of an abnormal isoform (PrPres), characterized by an increased β-sheet content (17) and by the acquisition of partial resistance to proteinase K (PK) proteolysis (18), is the hallmark of the agent replication. The conversion of PrP to abnormal PrP appears to be catalyzed by PrPres itself, probably by acting as a template or a seed to allow further conversion of PrP to PrPres (1–3). According to the protein-only hypothesis (19), the TSE infectious agent is PrPres itself or a precursor of it. However, although PrP and its conversion are recognized as key events in TSE replication and pathogenesis, formal proof that abnormal PrP is the actual transmissible agent has yet to be obtained.
Only a few cell culture models permissive to prion replication are available to date (3). Mouse neuroblastoma N2a, the more intensively used cell line to date (20), and other infectable rodent cell cultures including PC12 rat pheochromocytoma cells (21) and the GT-1 hypothalamic neuronal cell line (22) have provided some valuable insights into the biogenesis of PrPres in infected cells (3). Additionally, these and the persistently infected SMB murine cell line (23), have been used to screen potential drugs for their ability to inhibit PrPres accumulation (24–27). Recently, apoptosis was demonstrated in infected GT-1 cells (22), suggesting that this model could be relevant to study neurodegenerative changes in TSE. However, a common feature of the above cell lines is to support propagation of TSE strains experimentally adapted to rodents only. So far, despite repeated attempts (28), relevant cell culture models for strains from naturally occurring TSE diseases such as sheep scrapie, bovine spongiform encephalopathy, and Creutzfeldt–Jakob disease are lacking, although propagation of human prions was reported on one occasion (29).
In this paper, we report the characteristics of a cell model in which a prototypical, naturally occurring animal TSE, sheep scrapie, can actively replicate.
Materials and Methods
Vector Construction and Transfection of RK13 Rabbit Cell Line.
The complete coding sequence of the VRQ allele (Val-136, Arg-154, and Gln-171) of ovine PrP was PCR-cloned in the pTRE plasmid (CLONTECH). The PrP ORF was verified by DNA sequencing, and the resulting plasmid was transfected by the Lipofectamine method (GIBCO/BRL) into rabbit kidney epithelial cells (30). Stable transfectants were selected in the presence of puromycin (1 μg/ml) and one (Rov9) was amplified for further study. Rov9 cells were grown at 37°C in 6% CO2 in MEM supplemented with 10% FBS and were usually split at a one-fourth dilution every week.
Immunocytochemistry and Immunoblot Analysis.
Immunofluorescence analysis on living Rov9 cells was performed at 4°C, with 4F2 anti-PrP mAbs (31). Fixed cells (10 min at room temperature in PBS containing 4% paraformaldehyde and 4% sucrose) were permeabilized (3 min with 0.1% Triton X-100), incubated sequentially with 4F2 and anti-mouse IgG alkaline phosphatase-conjugated antibodies. Bound antibodies were visualized with Fast Red TR/Naphthol AS-MX (Sigma).
For immunoblot analysis of inoculated cell cultures, proteins from cell lysates were either methanol-precipitated or digested with PK for 2 h at 37°C (2 μg of PK for 500 μg of protein, i.e., 4–6 μg of PK per ml of cell lysate). Pefabloc (4 mM) was added and aggregated PK-resistant PrP was collected by centrifugation at 13,000 rpm for 20 min at room temperature. Pellets from methanol precipitation and PK-treated lysates were resuspended in sample buffer, subjected to SDS/PAGE electrophoresis, and transferred to nitrocellulose membranes. PrP was visualized either with l42 mAbs (32) or 4F2, which does not recognize NH2-terminally truncated abnormal PrP in PK-treated cell lysates. Western blots were revealed with an enhanced chemiluminescence detection system (ECL, Amersham Pharmacia).
Preparation of Inocula.
PG127 (PG127/98, Veterinary Laboratory Agency, U.K.) and LA404 [954044, Institut National de la Recherche Agronomique (INRA), Jouy-en-Josas, France] isolates are from VRQ-genotyped sheep affected by natural scrapie. Infected sheep brains were homogenized at 10% (wt/vol) in a sterile 5% glucose solution. These two sheep isolates also have been transmitted to Prnp0/0 transgenic mice expressing the VRQ allele of ovine PrP (TgOv mice, see below for description). The resulting material was used as an alternative source of inoculum, as specified. Extracts from inoculated Rov9 cultures were prepared by scraping cells into PBS, pelleting them by centrifugation, and resuspending the cell pellets in a sterile 5% glucose solution. After four freezing-thawing cycles, suspensions were sonicated for 1–2 min in a cup-horn apparatus before being inoculated to cells or transgenic mice.
Isolation of PrPres From Infected Brains.
Brain homogenates (typically 200 μl of 10%) were digested for 1 h with 10 μg/ml PK, and the reactions were stopped with 4 mM Pefabloc. After addition of 10% sarcosyl and 10 mM Tris⋅HCl (pH 7.4), samples were incubated for 15 min at room temperature. They were then centrifuged at 245,000 × g for 30–45 min at 20°C on 10% sucrose cushions. Pelleted material was resuspended in sample buffer and analyzed by immunoblotting.
Ex Vivo Infection of Rov9 Cells.
All of the brain homogenates used for cell inoculation were heated at 80°C for 20 min and sonicated for 1–2 min. Confluent Rov9 monolayers (grown in single wells of 12-well plates for 2 days in the presence of 1 μg/ml doxycycline (dox) were overlaid with 500 μl of culture medium containing 2.5% (wt/vol) of brain homogenate. Six hours later, 500 μl of culture medium was added and the cultures were incubated for 2 days. The supernant was then removed, and the cultures were rinsed once with PBS and left for 2.5 days in regular culture medium before being split into two 25-cm2 flasks. One week later, one flask was used for subcultivation [one passage at a one-fourth dilution per week, dox (1 μg/ml) was maintained during the whole experiment], and the other was rinsed once with cold PBS and lysed for 10 min at 4°C in Triton/DOC lysis buffer (50 mM Tris⋅HCl, pH 7.4/0.5% Triton X-100/0.5% sodium deoxycholate). Lysates were clarified (2,000 rpm, 1 min) and stored at −20°C.
In Situ PrPres Detection.
Paraffin inclusions of trypsinized, infected and mock-infected Rov9 cells were performed by using a cell block preparation system (Cytoblock, Shandon, Pittsburgh) and then treated as described (33). In brief, sections (2 μm) were mounted and dried overnight at 56°C before being deparaffinized and rehydrated. Slides were first incubated in 98% formic acid for 30 min, followed by 5 min of PK treatment (5 μg/ml) at 37°C, and then autoclaved for 30 min at 121°C in 10 mM citrate buffer (pH 6.1) and incubated with 20% normal goat serum for 20 min. PrP immunolabeling was carried by using 8G8 mAbs (31). Biotinylated antibodies were applied as secondary antibodies, and the streptavidin-biotinylated peroxidase complex was used for amplification. Revelation was performed by using diaminobenzidin. Nucleus counterstain was achieved with Mayer's hematoxylin.
Mouse Bioassay.
The in vivo infectivity assays were performed on TgOv hemizygous for the ovine Prnp gene (VRQ allele) and nullizygous for the mouse Prnp gene (Prnp0/0). Such mice were reported by some of us to be more susceptible to natural sheep scrapie than conventional mice (J.L.V., D.V., and H.L., unpublished results). The tg301 line used in these experiments carries a large DNA fragment isolated from an ovine bacterial artificial chromosome library, and the expression levels of ovine PrP were ≈8-fold those observed in sheep brain. Animals (≈6 wk old) were infected intracerebrally with 20 μl of inoculum. Inoculated mice were examined for neurological dysfunction every 2 days and then daily once clinical signs of scrapie were detected. Most of the diseased animals were killed when the death was imminent, i.e., within 1 wk after the onset of symptoms in this model. The brain of each diseased animal was taken and examined for the presence of PrPres by immunoblotting. Some brains were subjected to histologic examination so as to confirm the diagnosis of scrapie (data not shown).
Results
Inducible Expression of Ovine PrP in Rov9 Cells.
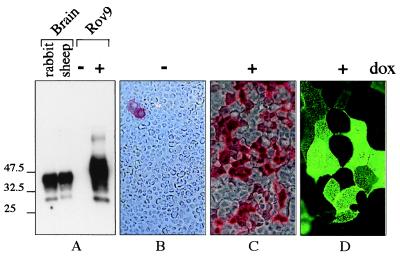
The susceptibility of sheep to scrapie is strongly determined by Prnp, the host gene for PrP. The V136R154Q171 allele of PrP (where V, R, and Q stand for valine, arginine, and glutamine, respectively) confers high susceptibility and short incubation time to sheep naturally exposed to scrapie whereas the A136R154R171 allele (A for alanine) is associated with an absolute clinical resistance to the disease (11). In a search for cellular models infectable by sheep scrapie agent, we have used the tetracycline-inducible (tet-on) system (34) to achieve regulated, high-level expression of the ovine PrP (VRQ allele). After transfection of several, including ovine brain-derived (35), cell lines, a strong inducible expression of ovine PrP was obtained in most of the clones derived from a rabbit kidney epithelial cell line (RK13). Data obtained with a representative clone (Rov9) are presented in this paper. The dox-induced Rov9 cells synthesized highly glycosylated PrP at levels close to those seen in sheep brain (Fig. 1A). No PrP could be detected in unstimulated Rov9 cells (Fig. 1A), indicating that expression of endogenous, rabbit PrP was very low in these cells. Up to 50% of the cells within induced Rov9 cell monolayers synthesized PrP at a high level (Fig. 1 B and C) and expressed it at the outer membrane (Fig. 1D).
Figure 1.
Inducible expression of ovine PrP in Rov9 cells as analyzed by Western blotting (A) or immunostaining (B–D). (A) Equal amounts of methanol-precipitated proteins (10 μg) from Rov9 cells after treatment with (+) or without (−) 1 μg/ml dox were analyzed by Western blotting with 4F2 mAbs. Ten micrograms of proteins from rabbit or sheep brain was included for comparison. The positions of molecular size marker proteins are indicated (in kDa). (B–D) Immunostaining of ovine PrP in fixed (B and C) or living (D) Rov9 cells. Dox-treated Rov9 cells (1 μg/ml, C and D) or untreated control Rov9 cells (B) were fixed and permeabilized (B and C) or left unfixed and unpermeabilized (D) before labeling with 4F2 mAbs. Alkaline phosphatase-conjugated IgG (B and C) or fluorescein-conjugated IgG (D) were used as second antibodies. (Original magnification: B and C, ×100; D, ×500.)
PrPres Detection in Rov9 Cells Inoculated With Sheep Scrapie Agent.
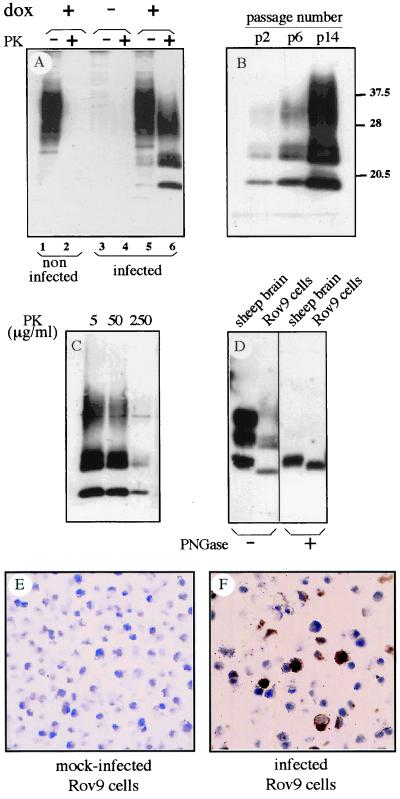
Rov9 cells were inoculated with an isolate issued from a naturally scrapie-affected sheep homozygous for the VRQ allele (PG127, Veterinary Laboratory Agency, U.K.). After incubation with infectious brain homogenate, Rov9 cell monolayers were rinsed and serially passaged. The cultures were then checked periodically for abnormal protease-resistant PrP (PrPres), the only known molecular marker of prion propagation. PrPres was readily detected in inoculated, induced Rov9 cells (Fig. 2A). However, PrPres was not observed in inoculated Rov9 cells expressing low levels of PrP (not induced) or in uninoculated, induced Rov9 cultures. PrPres appeared to accumulate at increasing levels through serial passages of the infected cultures (Fig. 2B), reaching a maximum at passages 14–18 postinoculation (p.i.). The level of PK resistance of Rov9-generated abnormal PrP was tested (Fig. 2C), and it was found to be high enough to compare to that from infected tissues. Interestingly, sheep brain- and Rov9-derived PrPres showed distinct glycosylation profile and electrophoretic mobility (Fig. 2D). This finding is in agreement with the observation that the molecular characteristics of PrPres, initially regarded as strain-specific (36), also can depend on the tissue producing the abnormal PrP (37, 38). Altogether, these data led to the conclusion that PrPres was actually produced de novo by infected Rov9 cells and did not originate from residual inoculum.
Figure 2.
Detection of PrPres in Rov9 cells inoculated with a natural sheep isolate (PG127) by Western blotting of PK-digested cell lysates (A-D) and in situ immunostaining (E and F). (A) Detection of abnormal PrP in Rov9 cells, 1 passage p.i. Dox-treated Rov9 cells (lanes 5 and 6) or untreated Rov9 cells (lanes 3 and 4) were infected by PG127 sheep brain homogenate. Control dox-treated Rov9 cells were left uninfected (lanes 1 and 2). After inoculation, cultures were passaged once, lysed, and analyzed for the presence of PrPres isolated from PK-digested cell lysates (lanes 2, 4, and 6; 200 μg of proteins). Total PrP was obtained by methanol precipitation of undigested cell lysates (lanes 1, 3, and 5; 20 μg of proteins). (B) PrPres accumulation in serially passaged, infected Rov9 cultures. PK-treated cell lysates from inoculated Rov9 cells were analyzed 2, 6, and 14 passages p.i. The levels of normal PrP at the different passages p.i. were found to be similar, based on Western blot analysis with 4F2 (not shown). The positions of molecular size marker proteins are indicated (in kDa). (C). Level of PK resistance of Rov9-generated PrPres. Cell lysate from infected Rov9 cells was digested with 5, 50, or 250 μg/ml of PK for 2 h before Western blot analysis. (D) Comparison of brain sheep- and Rov9-derived PrPres. PrPres was isolated from scrapie-infected sheep brain homogenate (isolate PG127, 2 mg of brain equivalent) and infected Rov9 cultures (passage 20 p.i., 100 μg of proteins). Aliquots were deglycosylated by PNGase F treatment (+). Note the higher mobility of the unglycosylated band of cell-derived PrPres compared to that of brain-derived PrPres. 142 mAbs were used to detect PrP in A–D. (E and F) Mock-infected (E) and infected (F) Rov9 cultures (at passage 18 p.i.) were paraffin-embedded. PK-treated slices were immunolabeled for PrPres by using 8G8 mAbs as described in Materials and Methods. (Original magnification = ×400.)
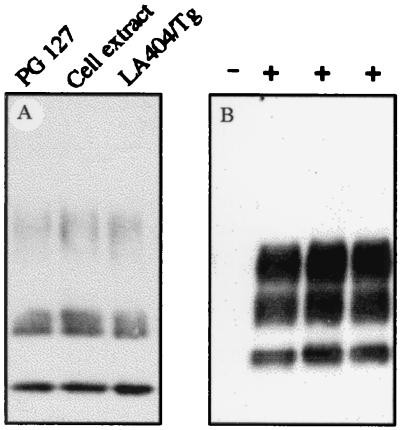
The proportion of PrPres-producing cells was estimated by using in situ detection of abnormal PrP. Although no labeling was seen in mock-infected cells (Fig. 2E), intense PrPres deposits were observed in at least 30% of the cells of the infected cultures (Fig. 2F). Notwithstanding, this substantial intracellular accumulation of PrPres in a large fraction of the cell population, uncloned infected cultures have been passaged for months, with no obvious loss of viability, alteration of cell morphology, or decline of PrPres accumulation. Infected Rov9 cells fully retained their infected status upon storage in liquid nitrogen (data not shown). The PG127 sheep isolate, passaged once in “ovinized” transgenic mice (TgOv mice, expressing the VRQ allele, see Materials and Methods), also was used as an alternative source of infectious agent. The infection of Rov9 cells proved to be highly reproducible because all of the experiments done so far were successful (n = 21, including inocula from either sheep or TgOv mice). The observed permissiveness of Rov9 cells to infection was not restricted to a single scrapie isolate. LA404 (from INRA) is an isolate from sheep homozygous for the VRQ allele, with biological properties clearly different from PG127 when transmitted to TgOv mice (J.L.V., D.V., and H.L., unpublished results). The LA404 isolate passaged once in TgOv mice also proved to be infectious for Rov9 cells, based on detection of newly accumulated PrPres (Fig. 3A). Whatever the inocula used for infection was Rov9-derived PrPres showed similar electrophoretic patterns (Fig. 3A).
Figure 3.
Detection of PrPres in infected Rov9 cells (A) and TgOv mice inoculated with infected Rov9 cells (B). (A) Transmission of two scrapie isolates and transmission from infected Rov9 cells to uninfected Rov9 cultures. Dox-treated Rov9 cells were inoculated with either PG127 sheep brain homogenate, extracts prepared from 5 × 106 PG127-infected Rov9 cells or with LA404 isolate passaged once in TgOv mice. In the latter case, the inoculum was left 7 days in contact with the cells. Analysis of PK-digested cell lysates was performed for 6 passages or 12 passages (for LA404/Tg) p.i. (B) Presence of PrPres in TgOv mice challenged with infected Rov9 cells. TgOv mice (n = 5) were inoculated intracerebrally with infected Rov9 cells (24 passages p.i.). PrPres was isolated from 2 mg of brain equivalent of terminally ill animals, and three of them are shown after immunoblotting analysis (+). A sample from uninoculated TgOv mice also was included (−). Antibodies l42 were used in A and B.
Sheep Scrapie Replication in Inoculated Rov9 Cells.
To assess whether infectivity was associated with PrPres-producing Rov9 cells, fresh cultures were inoculated with cell extracts from infected Rov9 cultures. The detection of newly accumulated abnormal PrP indicated that the PrPres-containing cell extracts induced PrP conversion in recipient cells (Fig. 3A). Next, inoculated cultures were bioassayed in TgOv mice (see Materials and Methods), which are more susceptible than conventional mice to sheep scrapie (J.L.V., D.V., and H.L., unpublished results). Extracts from inoculated Rov9 cultures (either PrPres positive or negative, depending on the dox-dependent level of ovine PrP cell expression) and from inoculated parental RK13 cells were bioassayed. All mice challenged with PrPres-positive Rov9 cultures at passages 6 and 24 p.i. died after acute, typical neurological disorders (Table 1) and showed brain accumulation of PrPres (Fig. 3B). Importantly, there was no detectable infectivity associated with inoculated cells expressing no ovine PrP (parental RK13 culture, Table 1), again excluding any significant effect of residual inoculum. Altogether these findings led us to conclude that a truly infectious, TSE-engendering agent was propagated in Rov9 cells.
Table 1.
Bioassay of ovine scrapie-infected Rov9 cells in transgenic mice expressing the VRQ allele of ovine PrP (TgOv mice)
| Inoculum* | Survival time (n/n0)† |
|---|---|
| Brain homogenate 10% | 59 ± 1.2 (6/6) |
| Brain homogenate 0.1% | 71 ± 1.8 (6/6) |
| Infected RK13 cells p8 | >240 (0/6) |
| Infected Rov9 (+dox) p6 | 65 ± 1.9 (6/6) |
| Infected Rov9 (+dox) p24 | 62 ± 0.9 (5/5) |
| Infected Rov9 (+dox) p24 × 10−4 | 83 ± 1.5 (5/5) |
| Infected Rov9 (−dox) p6 | 92 ± 3 (6/6) |
| Mock-infected Rov9 (+dox) p12 | >180 (0/5) |
Infected dox-induced (+dox), infected untreated (−dox), and mock-infected (+dox) Rov9 cells were tested at the indicated passage (p) p.i. The inoculated parental RK13 cells, expressing no ovine PrP, were assayed at 8 passages p.i. The brain homogenate (PG127 sheep isolate passaged once in TgOv mice) used to infect the cultures was bioassayed in parallel. Each animal was inoculated intracerebrally with 20 μl of either one of the following materials: cell extract from infected Rov9 and RK13 cultures, undiluted (3 × 106 cells per mice) or 104-fold diluted (3 × 102 cells per mice); or brain homogenate at the indicated dilution.
Mean days to death ± SEM; n: number of terminally ill animals; n0: number of animals inoculated.
The efficiency of scrapie agent replication depended on the level of ovine PrP expression in Rov9 cells. The incubation time of TgOv mice challenged with unstimulated, inoculated Rov9 cells (in which ovine PrP is not detectable by immunoblotting) was increased by 27 days when compared to animals inoculated with dox-treated cultures [infected Rov9 (−dox)p6 vs. infected Rov9 (+dox)p6, Table 1]. This result represented a 50% increase of the incubation time. Although infectious titer associated with the infected Rov9 cultures has not yet been determined by end-point titration, these data indicate that unstimulated, infected Rov9 cells had infectivity levels several orders of magnitude (at least 104-fold) lower than those observed in infected Rov9 cells expressing high levels of PrP. The presence of very few cells expressing ovine PrP in the absence of dox (see Fig. 1B) that can presumably replicate the agent is the more likely explanation for the presence of low levels of infectivity in uninduced, inoculated Rov9 cells.
Discussion
In this study, we have demonstrated the efficient propagation of ovine prions in cultured cells from a different species. These findings are unique in several respects. Prions from a natural TSE (i.e., without previous experimental adaptation to rodents) were propagated without resorting to animal inoculation, and thus permitting their study at the cellular level. Moreover, we have provided evidence that expression of ovine PrP in an otherwise refractory cell line may be sufficient to allow the species barrier to be crossed ex vivo. Whether a similar rationale could apply to the development of the urgently needed bovine and human cellular models is currently being examined. Among the perspectives offered by such models is their use as a rapid bioassay for prions. Experiments are in progress to address the sensitivity of Rov9 cells to sheep scrapie.
In transgenic mice and TSE-infected cultures, expression of an additional and distinct PrP can slow down the replication and/or the propagation of the scrapie agent (7, 39). This has been proposed to be due to the binding of the heterologous PrP to PrPres and the subsequent inhibition of further conversion of PrP in the abnormal isoform, as revealed by cell-free conversion analysis (40). As expression of endogenous rabbit PrP was found undetectable in Rov9 cells, one possible factor that may have contributed for the crossing of the species barrier and the efficient cell transmission of sheep scrapie to Rov9 could be the high ratio of ovine PrP vs. endogenous rabbit PrP.
The influence of PrP expression levels on the susceptibility to TSE is a well documented phenomenon, as evidenced by PrP overexpression in transgenic mice that often results in a marked reduction of the incubation time (8, 9). In a murine TSE cell culture model, it has been proposed that transmission of TSE agents might be improved by heightening of the PrP expression level through transfection (41). By using the cell line described here, the ability to induce elevated levels of Prnp gene expression through dox stimulation has allowed us to formally demonstrate this point. Indeed, the PrP expression level was found to have a critical effect on the transmission rate (100%) of sheep scrapie to Rov9 cells (as assessed by the biochemical detection of abnormal PrP) and on the efficiency of infectivity propagation (thousands-fold higher in the presence of dox). The availability of cell clones combining permissivity and regulatable prion protein expression may provide a unique opportunity to further elucidate the events underlying the establishment and/or maintenance of the infected status.
Our data raise the question of whether PrP overexpression per se would confer susceptibility to any given cell type. This possibility seems unlikely in view of the published data. Indeed, ectopic PrP expression in transgenic mice has resulted in mouse tissues, which do not replicate prions despite high levels of PrP (42). Moreover, marked differences in susceptibility to infection were recently observed among N2a sublines (43). These were apparently unrelated to PrP levels, suggesting the implication of additional factors for efficient prion replication. Our observation, together with a 25-yr-old report describing the infection of murine fibroblasts (44), supports the view that such cofactors are unlikely to be restricted to neuronal or lymphoreticular cells, which are so far the only recognized targets for prion replication. It is tempting to speculate that some features shared by neurons and epithelial cells might be involved in their ability to replicate prions. The nervous system is developmentally derived from an epithelium and both neurons and epithelial cells are polarized (45). Similar mechanisms are responsible for the polarized sorting of at least some proteins in neurons and epithelial cells (46). In epithelial cells, most of the glycosyl–phosphatidylinositol-anchored proteins are usually sorted in a polarized manner (47). Although the precise localization of PrP on epithelial cell surface has not been determined yet, evidence has been found for the presence of PrP in specific membrane domains in cultured neurons (48). Further studies should aim at determining the sorting of PrP in Rov9 cells.
The finding that active prion replication can take place in cells of an epithelial type is of particular interest. After peripheral challenge, and before reaching the central nervous system, the spread of the agent throughout the body critically depends on the expression of PrP (49). Cells of the reticuloendothelial system have been identified as one link of the chain leading to neuroinvasion (50). However, a number of other tissues, including epithelia (51, 52), express PrP and therefore might be involved in the spread of the agent. Epithelial cells are present in several organs (placenta, digestive tract, and skin) implicated in TSE pathogenesis and transmission (53–55). This, together with the present demonstration that cultivated epithelial cells support prion replication, should lead to a careful assessment of a possible involvement of epithelia in TSEs.
In several species, including humans, it is well established that PrP polymorphism affects susceptibility to TSE (10–13), although the mechanisms involved are still unknown. In sheep, the notion that PrP derived from certain alleles might be more efficiently converted to abnormal PrP than others recently has been supported by cell-free conversion experiments in which susceptible PrP alleles showed a greater propensity to be converted than the resistant ones (56). One current limitation of the assay is that the relation between any newly converted PrPres and infectivity cannot be established. In contrast, infectivity produced in Rov9 cells can readily be quantified through bioassay into TgOv mice. Hence, we believe that this ex vivo assay should allow a more accurate assessment of the influence of PrP allelic variants on the transmission of prion diseases. More generally, engineering of permissive cells with regulatable genes encoding PrPs of a specified sequence may represent a promising strategy to further explore, at a cellular level, important aspects of TSE diseases, including that of interspecific transmission.
Acknowledgments
We thank M. Dawson (Central Veterinary Laboratory/Veterinary Laboratory Agency, U.K.) and J. M. Elsen, and F. Eychenne (Institut National de la Recherche Agronomique/Station d'Amélioration Génétique des Animaux, France) for the kind gift of sheep isolates PG127 and LA404, respectively. We also acknowledge M. H. Groshup for l42 antibodies and J. Grassi (Service de Pharmacologie et d'Immunologie/Commissariat á l'Energie Atomique) for providing 4F2 and 8G8 antibodies. We are grateful to the members of the Service de Neurovirologie (Commissariat à l'Energie Atomique, Fontenay aux Roses, France) for teaching us some basic, “TSE-specific” skills. We thank C. La Bonnardière, J. A. Gingrich, and L. V. Ronco for critical reading of the manuscript and M. Nezonde for the artwork. This work was partially supported by grants from the French government (CI-ESST) and from the European Union (Biotech. PL976064).
Abbreviations
- TSE
transmissible spongiform encephalopathy
- PrP
prion protein
- PrPres
abnormal PrP
- PK
proteinase K
- p.i.
postinoculation
- dox
doxycycline
- VRQ
Val-136, Arg-154, Gln-171
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Prusiner S B. Trends Biochem Sci. 1996;21:482–487. doi: 10.1016/s0968-0004(96)10063-3. [DOI] [PubMed] [Google Scholar]
- 2.Weissmann C. FEBS Lett. 1996;389:3–11. doi: 10.1016/0014-5793(96)00610-2. [DOI] [PubMed] [Google Scholar]
- 3.Harris D A. Clin Microbiol Rev. 1999;12:429–444. doi: 10.1128/cmr.12.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendheim P E, Brown H R, Rudelli R D, Scala L J, Goller N L, Wen G Y, Kascsak R J, Cashman N R, Bolton DC. Neurology. 1992;42:149–156. doi: 10.1212/wnl.42.1.149. [DOI] [PubMed] [Google Scholar]
- 5.Manson J, West J D, Thomson V, McBride P, Kaufman M H, Hope J. Development (Cambridge, UK) 1992;115:117–122. doi: 10.1242/dev.115.1.117. [DOI] [PubMed] [Google Scholar]
- 6.Kretzschmar H A, Prusiner S B, Stowring L E, DeArmond S J. Am J Pathol. 1986;122:1–5. [PMC free article] [PubMed] [Google Scholar]
- 7.Büeler H, Aguzzi A, Sailer A, Greiner R-A, Autenreid P, Aguet M, Weissmann C. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 8.Prusiner S B, Scott M, Foster D, Pan K-M, Groth D, Mirenda C, Torchia M, Yang S-L, Serban D, Carlson G A, et al. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 9.Fisher M, Rülicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C. EMBO J. 1996;15:1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 10.Moore R C, Hope J, McBride P A, McConnell I, Selfridge J, Melton D W, Manson J C. Nat Genet. 1998;18:118–125. doi: 10.1038/ng0298-118. [DOI] [PubMed] [Google Scholar]
- 11.Hunter N. Trends Microbiol. 1997;5:331–334. doi: 10.1016/s0966-842x(97)01081-0. [DOI] [PubMed] [Google Scholar]
- 12.Collinge J, Palmer M S, Dryden A J. Lancet. 1991;337:1441–1442. doi: 10.1016/0140-6736(91)93128-v. [DOI] [PubMed] [Google Scholar]
- 13.Palmer M S, Dryden A J, Hughes J T, Collinge J. Nature (London) 1991;352:340–342. doi: 10.1038/352340a0. [DOI] [PubMed] [Google Scholar]
- 14.Goldfarb L G, Brown P, Cervenakova L, Gajdusek D C. Philos Trans R Soc London B. 1994;343:379–384. doi: 10.1098/rstb.1994.0032. [DOI] [PubMed] [Google Scholar]
- 15.Scott M, Foster D, Mirenda C, Serban D, Coufal F, Wälchli M, Torchia M, Groth D, Carlson G, DeArmond S J, et al. Cell. 1989;59:847–857. doi: 10.1016/0092-8674(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 16.Westaway D. In: Prion Diseases. Baker H, Ridley R M, editors. Totowa, NJ: Humana; 1996. pp. 251–263. [Google Scholar]
- 17.Caughey B W, Dong A, Bhat K S, Ernst D, Hayes S F, Caughey W S. Biochemistry. 1991;30:7672–7680. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- 18.Bolton D C, McKinley M P, Prusiner S B. Science. 1982;218:1309–1311. doi: 10.1126/science.6815801. [DOI] [PubMed] [Google Scholar]
- 19.Prusiner S B. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 20.Race R E, Fadness L H, Chesebro B. J Gen Virol. 1987;68:1391–1399. doi: 10.1099/0022-1317-68-5-1391. [DOI] [PubMed] [Google Scholar]
- 21.Rubenstein R, Carp R I, Callahan S M. J Gen Virol. 1984;65:2191–2198. doi: 10.1099/0022-1317-65-12-2191. [DOI] [PubMed] [Google Scholar]
- 22.Schätzl H M, Laszlo L, Holtzman D M, Tatzelt J, DeArmond S J, Weiner R I, Mobley W C, Prusiner S B. J Virol. 1997;71:8821–8831. doi: 10.1128/jvi.71.11.8821-8831.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clarke M C, Haig D A. Nature (London) 1970;225:100–101. doi: 10.1038/225100a0. [DOI] [PubMed] [Google Scholar]
- 24.Caughey W S, Raymond L D, Horiuchi M, Caughey B. Proc Natl Acad Sci USA. 1998;95:12117–12122. doi: 10.1073/pnas.95.21.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Supattapone S, Nguyen H O, Cohen F E, Prusiner S B, Scott M R. Proc Natl Acad Sci USA. 1999;96:14529–14534. doi: 10.1073/pnas.96.25.14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudyk H, Vasiljevic S, Hennion R M, Birkett C R, Hope J, Gilbert I H. J Gen Virol. 2000;81:1155–1164. doi: 10.1099/0022-1317-81-4-1155. [DOI] [PubMed] [Google Scholar]
- 27.Perrier V, Wallace A C, Kaneko K, Safar J, Prusiner S B, Cohen F E. Proc Natl Acad Sci USA. 2000;97:6073–6078. doi: 10.1073/pnas.97.11.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Race R. Curr Top Microbiol Immunol. 1991;172:181–193. doi: 10.1007/978-3-642-76540-7_12. [DOI] [PubMed] [Google Scholar]
- 29.Ladogana A, Liu Q, Xi Y G, Pocchiari M. Lancet. 1995;345:594–595. doi: 10.1016/s0140-6736(95)90508-1. [DOI] [PubMed] [Google Scholar]
- 30.Christofinis G J, Beale A J. J Pathol Bacteriol. 1968;95:377–381. doi: 10.1002/path.1700950204. [DOI] [PubMed] [Google Scholar]
- 31.Krasemann S, Groschup M H, Harmeyer S, Hunsmann G, Bodemer W. Mol Med. 1996;2:725–734. [PMC free article] [PubMed] [Google Scholar]
- 32.Vorberg I, Buschmann A, Harmeyer S, Saalmüller A, Pfaff E, Groshup M H. Virology. 1999;255:26–31. doi: 10.1006/viro.1998.9561. [DOI] [PubMed] [Google Scholar]
- 33.Andreoletti O, Berthon P, Marc D, Sarradin P, Grosclaude J, Schelcher F, Elsen J-M, Lantier F. J Gen Virol. 2001;81:3115–3126. doi: 10.1099/0022-1317-81-12-3115. [DOI] [PubMed] [Google Scholar]
- 34.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vilette D, Madelaine M F, Laude H. In Vitro Cell Dev Biol Anim. 2000;36:45–49. doi: 10.1290/1071-2690(2000)036<0045:EOACLF>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 36.Collinge J, Sidle K C, Meads J, Ironside J, Hill A F. Nature (London) 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- 37.Rubenstein R, Merz P A, Kascsak R J, Scalici C L, Papini M C, Carp R I, Kimberlin R H. J Infect Dis. 1991;164:29–35. doi: 10.1093/infdis/164.1.29. [DOI] [PubMed] [Google Scholar]
- 38.Hill A F, Butterworth R J, Joiner S, Jackson G, Rossor M N, Thomas D J, Frosh A, Tolley N, Bell J E, Spencer M, et al. Lancet. 1999;353:183–189. doi: 10.1016/s0140-6736(98)12075-5. [DOI] [PubMed] [Google Scholar]
- 39.Priola S A, Caughey B, Race R E, Chesebro B. J Virol. 1994;68:4873–4878. doi: 10.1128/jvi.68.8.4873-4878.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horiuchi M, Priola S A, Chabry J, Caughey B. Proc Natl Acad Sci USA. 2000;97:5836–5841. doi: 10.1073/pnas.110523897. . (First Published May 16, 2000; 10.1073/pnas.110523897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishida N, Harris D A, Vilette D, Laude H, Frobert Y, Grassi J, Casanova D, Milhavet O, Lehmann S. J Virol. 2000;74:320–325. doi: 10.1128/jvi.74.1.320-325.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raeber A J, Sailer A, Hegyi I, Klein M A, Rulicke T, Fisher M, Brandner S, Aguzzi A, Weissmann C. Proc Natl Acad Sci USA. 1999;96:3987–3992. doi: 10.1073/pnas.96.7.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bosque P J, Prusiner S B. J Virol. 2000;74:4377–4386. doi: 10.1128/jvi.74.9.4377-4386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clarke M C, Millson G C. Nature (London) 1976;261:144–145. doi: 10.1038/261144a0. [DOI] [PubMed] [Google Scholar]
- 45.Colman D R. Neuron. 1999;23:649–651. doi: 10.1016/s0896-6273(01)80024-6. [DOI] [PubMed] [Google Scholar]
- 46.Dotti C G, Simons K. Cell. 1990;62:63–72. doi: 10.1016/0092-8674(90)90240-f. [DOI] [PubMed] [Google Scholar]
- 47.Lisanti M P, Rodriguez-Boulan E. Trends Biochem Sci. 1990;15:113–118. doi: 10.1016/0968-0004(90)90195-h. [DOI] [PubMed] [Google Scholar]
- 48.Madore N, Smith K L, Graham C H, Jen A, Brady K, Hall S, Morris R. EMBO J. 1999;18:6917–6926. doi: 10.1093/emboj/18.24.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blattler T, Brandner S, Raeber A J, Klein M A, Voigtlander T, Weissmann C, Aguzzi A. Nature (London) 1997;389:69–73. doi: 10.1038/37981. [DOI] [PubMed] [Google Scholar]
- 50.Kimberlin R H, Walker C A. J Comp Pathol. 1979;89:551–562. doi: 10.1016/0021-9975(79)90046-x. [DOI] [PubMed] [Google Scholar]
- 51.Pammer J, Suchy A, Rendl M, Tschachler E. Lancet. 1999;354:1702–1703. doi: 10.1016/S0140-6736(99)02800-7. [DOI] [PubMed] [Google Scholar]
- 52.Lemaire-Vieille C, Schulze T, Podevin-Dimster V, Follet J, Bailly Y, Blanquet-Grossard F, Decavel J-P, Heinen E, Cesbron J-Y. Proc Natl Acad Sci USA. 2000;97:5422–5427. doi: 10.1073/pnas.080081197. . (First Published May 2, 2000; 10.1073/pnas.080081197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor D M, McConnell I, Fraser H. J Gen Virol. 1996;77:1595–1599. doi: 10.1099/0022-1317-77-7-1595. [DOI] [PubMed] [Google Scholar]
- 54.Pattison I H, Hoare M N, Jebbett J N, Watson W A. Vet Rec. 1972;90:465–468. doi: 10.1136/vr.90.17.465. [DOI] [PubMed] [Google Scholar]
- 55.Bons N, Mestre-Frances N, Belli P, Cathala F, Gajdusek D C, Brown P. Proc Natl Acad Sci USA. 1999;96:4046–4051. doi: 10.1073/pnas.96.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bossers A, de Vries R, Smits M A. J Virol. 2000;74:1407–1414. doi: 10.1128/jvi.74.3.1407-1414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]