Abstract
The PITX3 bicoid-type homeodomain transcription factor plays an important role in lens development in vertebrates. PITX3 deficiency results in a spectrum of phenotypes from isolated cataracts to microphthalmia in humans, and lens degeneration in mice and zebrafish. While identification of downstream targets of PITX3 is vital for understanding the mechanisms of normal ocular development and human disease, these targets remain largely unknown. To isolate genes that are directly regulated by PITX3, we performed a search for genomic sequences that contain evolutionarily conserved bicoid/PITX3 binding sites and are located in the proximity of known genes. Two bicoid sites that are conserved from zebrafish to human were identified within the human promoter of the major intrinsic protein of lens fiber, MIP/AQP0. MIP/AQP0 deficiency was previously shown to be associated with lens defects in humans and mice. We demonstrate by both chromatin immunoprecipitation and electrophoretic mobility shift assay that PITX3 binds to MIP/AQP0 promoter region in vivo and is able to interact with both bicoid sites in vitro. In addition, we show that wild-type PITX3 is able to activate the MIP/AQP0 promoter via interaction with the proximal bicoid site in cotransfection experiments and that the introduction of mutations disrupting binding to this site abolishes this activation. Furthermore, mutant forms of PITX3 fail to produce the same levels of transactivation as wild-type when cotransfected with the MIP/AQP0 reporter. Finally, knockdown of pitx3 in zebrafish affects formation of a DNA-protein complex associated with mip1 promoter sequences; and examination of expression in pitx3 morphant and control zebrafish revealed a delay in and reduction of mip1 expression in pitx3-deficient embryos. Therefore, our data suggest that PITX3 is involved in direct regulation of MIP/AQP0 expression and that the alteration of MIP/AQP0 expression is likely to contribute to the lens phenotype in cataract patients with PITX3 mutations.
Introduction
The PITX3 bicoid-related homeodomain transcription factor represents an important regulator of lens development in vertebrates. Mutations in PITX3 result in congenital cataracts, anterior segment mesenchymal dysgenesis (ASMD), Peter's anomaly, and microphthalmia in humans [1]–[6]. Deletions within the Pitx3 promoter region in mice produce the aphakia phenotype, which is characterized by small eyes lacking a lens [7], [8]. In lower vertebrates (zebrafish and frog), pitx3 was shown to be essential to normal lens and retina formation [9]–[13]. Knockdown of pitx3 protein in zebrafish embryos via translational morpholino results in small eyes, lens degeneration, misshapen head and reduced jaw and fins [9], [10], [12]. In vertebrates, expression of Pitx3/pitx3 is first detected in the lens placode and then the lens vesicle; early expression is observed in the lens epithelial cells and primary fibers while later expression is restricted to the equator regions of the developing lens [1], [14].
Despite its vital importance for eye development, little is currently known about the ocular function of PITX3/Pitx3 and its downstream targets. Expression of several genes/proteins was found to be altered in the lenses of Pitx3-deficient mice. Some early reports demonstrated that expression of β- and γ -crystallins is completely absent at developmental stages 10–18 days as well as in newborn aphakia mice [15]–[17]. Two recent publications provided additional data on this matter; Ho and colleagues detected precocious activation of both β- and γ-crystallins in the eyes of 10.5–11.5-dpc Pitx3–knockout mice [18] while Medina-Martinez and coauthors reported deregulation of crystallin expression in aphakia mice with α- and β- crystallin expression being reduced at both transcript and protein levels and γ –crystallin expression being downregulated at the protein level [19]. In addition to crystallins, expression of the transcription factors Foxe3 [18]–[20] and Prox1 [19] as well as the cell cycle regulator p57KIP2 [19] were found to be affected in Pitx3-deficient animals, which seems more likely to be related to the overall abnormal lens development in aphakia mice rather than direct involvement of Pitx3 in transcriptional regulation of these genes [19].
PITX3 belongs to the PITX family of bicoid-type homeodomain-containing proteins that regulate expression of other genes during development and, possibly, in adult organisms. Other members of this family were shown to be involved in developmental disorders such as idiopathic clubfoot [PITX1; 21] and Axenfeld-Rieger syndrome [PITX2; 22]. PITX factors are known to interact with bicoid-type DNA sequences and to regulate downstream gene expression through these interactions [23]–[28]. PITX factors are primarily known as activators of transcription, though they may also act as repressors [29], [30]. Several transcriptional targets of PITX homeoproteins have been identified and bicoid sequences located in the regulatory regions of these downstream genes were shown to mediate these interactions; two or more bicoid sites were found in some promoters [25], [26], [28], [31], [32], although a single bicoid element was demonstrated to be sufficient in several other cases [24], [33]–[35]. Interspecies conservation of bicoid sequences has been reported for some promoters [26], [32]. Preservation of regulatory sequences is frequently observed for developmental genes which demonstrate a conserved expression pattern; therefore, identification of regulatory sequences represents a useful tool in uncovering genetic pathways [36], [37].
In order to isolate downstream targets of the PITX3 homeodomain transcription factor we performed a search for evolutionarily conserved non-coding sequences containing bicoid sites and located in proximity to known genes, therefore potentially interacting with PITX3 to regulate expression of that gene. As a result, we identified two bicoid sites located in the promoter of Major Intrinsic Protein of lens fiber (MIP) or Aquaporin 0 (AQP0) that are conserved between human, mouse, zebrafish and several other species. We further demonstrated that PITX3 is able to specifically interact with the identified sequences both in vitro and in vivo and to transactivate gene expression as a result of this interaction. In addition to this, expression of mip1 was found to be altered in pitx3 deficient zebrafish morphants. Our data suggest that PITX3 is involved in direct regulation of MIP/AQP0 expression and provide new insight into the PITX3 pathway as well as mechanisms of lens development.
Materials and Methods
Ethics statement
The study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee at the Medical College of Wisconsin (protocol number AUA00000352).
In silico analysis
ECR Browser web-based tool (http://ecrbrowser.dcode.org) was used to identify conserved paired bicoid sites in the promoters/intronic regions of genes with known expression/function. Paired comparison of human and mouse genomes was performed using the following parameters: presence of two conserved bicoid sites with distance between the sites not to exceed 650-bp. Secondary analysis of identified regions for sequence conservation was performed using the UCSC Genome Brower multiple alignment module (http://genome.ucsc.edu) as well as the BLAST tool (http://blast.ncbi.nlm.nih.gov), including examination of the corresponding genes in lower vertebrates when available.
Cell culture
Human lens epithelial cells (B3) and human embryonic kidney cells (293HEK) were obtained from ATCC (Manassas, VA). B3 cells were cultured in MEM medium (Invitrogen; Carlsbad, CA) supplemented with heat-inactivated 20% fetal calf serum (FBS), glutamine, sodium pyruvate to a final concentration of 1 mM, non-essential amino acids and antibiotic-antimycotic (Invitrogen; Carlsbad, CA). 293HEK cells were maintained in DMEM medium containing 10% FBS, glutamine and antibiotic-antimycotic solution.
Electrophoretic Mobility Shift Assay (EMSA)
Nuclear extracts were prepared from B3 cells transiently transfected with PITX3-pcDNA3.1 vector with CelLytic NuCLEAR extraction kit (Sigma, St. Louis, MO). Cells were harvested after 48 hours with a cell scraper and the compact cellular pellet was re-suspended in 5 volumes of Hypotonic lysis buffer (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl) with Protease inhibitor cocktail (Sigma; St. Louis, MO). After 15 minutes of incubation on ice, Igepal CA-630 was added to a final concentration 0.6%, then the cells were vortexed and spun down for 30 seconds at 10000 g. Crude nuclear pellet was extracted with about 2/3 of the original packed cell volume of Extraction buffer (20 mM HEPES, pH 7.9, 1.5 mM MgCl2, 0.42 M NaCl, 0.2 mM EDTA, 1 mM DTT and 25% Glycerol) in the presence of protease inhibitors for 30 minutes on ice. 32-mer 5′-GGAGAAAGGCTTCTAATCCCTGGGAACTAAAG oligonucleotide spanning region −533/−502 from transcriptional start site (tss) of MIP/AQP0 promoter, 32-mer 5′-CTGCCCCTCCCAGGGATTAAGAGTCCTCTATA corresponding to the promoter sequence −71/−40 and their complement oligonucletides as well as both sets of oligonucleotides with TAATCC (GGATTA) bicoid sites replaced by TAATTT (AAATTA) (see above) were labeled with Biotin 3′ End DNA Labeling kit (PIERCE) and annealed. Electrophoretic mobility shift assays (EMSA) were performed with LightShift Chemiluminescent EMSA kit (Pierce; Rockford, IL) in accordance with the manufacturer's protocol and using 50 ng/µl of Poly(dI-dC), 20 fmol of labeled DNA and 2 µl of nuclear extracts. After 20 minutes of incubation at room temperature, reactions were either diluted with 5× Loading buffer or further incubated for 30 minutes in presence of 1 µg of polyclonal Pitx3 antibody for supershift assay. Binding reactions and free probe were run on 5% native polyacrylamide gel in 0.5× TBE buffer.
For EMSA experiments performed using whole zebrafish embryo nuclear extracts, the 32-mer and its compliment corresponding to the region from −44 to −76 of zebrafish mip1 promoter were utilized: 5′-CAA TTC AGC CAA AGG ATT ACA GTG TCA CAG AG. In addition to this, both sets of oligonucleotides were made with TAATCC (GGATTA) bicoid sites replaced by TAATTT (AAATTA) to be used as a control for bicoid site binding specificity. Nuclear extracts were generated from sixty 48-hpf zebrafish pitx3 morphant or wild-type embryos that demonstrated normal body length and morphology; the preparation was carried out as described above except for that the embryos were first grinded with glass tissue homogenizer equipped with type B pestle in hypotonic detergent-less lysis buffer to assist nuclei release. Binding reaction was performed in the presence of 2.5% glycerol, 5 mM MgCl2 and 0.05% NP-40 in addition to the buffer composition described above; 3 µl of extract was used in each binding reaction. Five embryos from each group were analyzed for pitx3 transcript presence to verify the degree of morpholino-mediated knockdown.
Chromatin immunoprecipitation (ChIP)
ChIP was performed with ChIP-IT enzymatic kit or ChIP-IT Express enzymatic kit (Active Motif; Carlsbad CA) according to manufacturer recommendations.
B3 human lens epithelial (HLE) cells were grown in 100 mm tissue culture dish to 90–95% confluence and utilized for ChIP assays; experiments were performed using native untransfected cells as well as cells transfected with PITX3 expression plasmids. Cells were transfected with 7.5–10 µg of pcDNA3.1_PITX3_FLAG or empty pcDNA3.1 plasmid and cross-linked after 48 hours with 1% formaldehyde for 10 minutes at room temperature with agitation. Following this, the monolayers were washed with 125 mM of glycine and lysed. The nuclear pellet was resuspended in digestion buffer and DNA was sheared with Enzymatic shearing cocktail for 10 minutes at 37°C in a water bath. The resulting fragments ranged between 200- and 1000-bp in size. The quality of chromatin was verified in a control experiment of immunoprecipitation with the Polymerase II antibody followed by PCR with primers specific for the GADPH promoter. Only those chromatin preparations that demonstrated significant enrichment in these control experiments were used in further analysis. Immunoprecipitation was performed with 2 µg of Pitx3, FLAG or control IgG antibody overnight in a cold room and, after washing and de-crosslinking, the precipitated DNA was analyzed by PCR. Goat polyclonal PITX3 (N-20) and normal goat IgG were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) and FLAG-M2 mouse monoclonal antibody from Sigma (St. Louis, MO). For PCR amplification of MIP/AQP0 promoter, the following primers were utilized: set 1 (spanning region −110/+95) forward, 5′-GCTGTGAAGGGGTTAAGAGG-3′ and reverse 5′-GAGGGTGGCAAAGAACTCAG-3′ and set 2 (spanning region −473/−275) forward, 5′-CTGAACCCCACTCCTTACCA-3′ and reverse, 5′- TCTGCCCTTCTGTGTGTGTC-3′. For control experiments, the following primers were used: GAPDH forward 5′- TACTAGCGGTTTTACGGGCG-3′ and reverse 5′-TCGAACAGGAGGAGCAGAGAGCGA-3′, product = 166 bp (provided as positive control as part of ChIP-IT Express Enzymatic kit (Active Motif; Carlsbad, CA); forward 5′-ATGGTTGCCACTGGGGATCT-3′ and reverse 5′-TGCCAAAGCCTAGGGGAAGA-3′, product = 174 bp (provided as negative control as part of ChIP-IT Express Enzymatic kit, Active Motif, Carlsbad CA). The PCR reactions were repeated at least three times using precipitated DNA from independent chromatin immunoprecipitation experiments. Quantification of ChIP PCR products was performed by densitometry using the ImageJ program developed by Dr. Rasband, NIH (http://rsbweb.nih.gov/ij/). The measurements obtained for ChIP PCR results were normalized by input DNA and expressed as percent of its value. Data from at least three independent experiments were combined to calculate mean and standard deviation. Statistical significance was determined using the homoscedastic Student's t-test with two-tailed distribution.
Expression and reporter plasmids
PITX3 wild type, PITX3-WT, and mutant expression constructs, PITX3-K111E, PITX3-S13N and PITX3- G219f, were previously described [27]. To produce the MIP656-bcd1,2 reporter plasmid, a 656-bp fragment containing 597-bp of upstream and 59-bp of downstream sequence from the transcriptional start site (tss) of MIP/AQP0 was amplified by PCR and cloned into the pCRII-TOPO vector (Invitrogen; Carlsbad, CA) and then subcloned into basic pGL3 luciferase reporter vector (Promega, Madison, WI). The transcriptional start site (tss) of MIP/AQP0 was designated based on the ENST00000257979 entry in Ensemble Database. Site-directed mutagenesis was performed with QuikChange II Site-Directed mutagenesis kit (Stratagene, La Jolla, CA), oligonucleotide 5′-CTCAGCCTGCCCCTCCCAGAAATTAAGAGTCCTCTATAAA-3′ and its complement for the proximal bicoid site and oligonucleotide 5′-CTAGCCAATGGGAGAAAGGCTTCTAATTTCTGGGAACTAAAGAATT-3′ and its compliment for the distal site on MIP/AQP0 promoter. Therefore, in both bicoid sites the consensus recognition sequence TAATCC was replaced by TAATTT and three additional constructs were produced: MIP656-bcd1 (carrying mutant bcd2 site), MIP656-bcd2 (carrying mutant bcd1 site) and MIP656-bcd0 (carrying mutations in both bicoid sites). All constructs were verified by sequencing.
Reporter assays
Human embryonic kidney cells (293HEK) were plated in 24-well plates and transfected using Lipofectamine 2000 (Invitrogen; Carlsbad, CA) according to the manufacturer's protocol. Equimolar amounts of basic pGL3 reporter plasmid (100 ng) and MIP656 wild-type and mutant reporters (114 ng) were used. Each cotransfection included 300 ng of effector (PITX3 wild type and mutant expression constructs) and 60 ng of β-galactosidase in pcDNA3.1 vector (internal control for efficiency of transfection); the total DNA amount was kept the same in all transfections by adding empty pcDNA3.1 vector when needed. Cells were harvested after 48 hours; luciferase and β-galactosidase activities were determined using Luciferase assay and Enzyme Assay systems (Promega, Madison, WI), respectively. Every experiment was performed at least three times in triplicate. Student's paired t-Test with a one-tailed distribution was utilized to determine the statistical significance of any differences in activity level.
Zebrafish care and morpholino injections
Zebrafish (Danio rerio) were maintained on a 14-hour light/ 10-hour dark cycle. The embryos were obtained by natural spawning and maintained at 28.5°C. The pitx3 morpholino, 5′-AGGTTAAAATCCATCACCTCTACCG-3′, that was previously reported [9] or control morpholino (Gene Tools, Philomath, OR) were suspended at 250 µM in injection buffer [0.1% (w/v) phenol red (Sigma) in 0.3× Danieau buffer (17 mM NaCl, 2 mM KCl, 0.12 mM MgSO4, 1.8 mM Ca(NO3)2 and 1.5 mM HEPES, pH 7.6) and 19.2 ng was injected into zebrafish embryos immediately after fertilization at the 1–2 cell stage. Microinjections were performed using the Nanoject II injector (Drummond Scientific, Broomall, PA). Embryos were incubated at 28.5°C in 0.2 mM 1-phenyl-2-thiourea (PTU) to inhibit pigment formation and anesthetized with 0.05% Tricane before imaging. The developmental stage was determined by time (hours post fertilization (hpf)) and by morphological criteria [38]. Nikon SMZ 1500 and Zeiss M2 Discovery microscopes were utilized for embryo imaging.
RNA isolation, RT-PCR and in situ hybridization
For RNA isolation, the embryos were homogenized in TRI reagent (Sigma) in the presence of glycogen and processed using a standard extraction protocol. The cDNA was generated using equal amounts of RNA for every sample and SuperScript III First-Strand Synthesis system (Invitrogen, Carlsbad, CA) according to manufacturer recommendations. Semi-quantitative PCR was performed using gene-specific oligonucleotides for mip1, exon_1F, 5′- CTCCCAGATGTCCCTGTTTC-3′, and exon_2R, 5′- CATACTGATGCCAGGCTGAA-3′, (PCR product = 148 bp); for pitx3, exon_1F, 5′-CTCCACTAGACCGGGATTCA-3′, and exon_3R, 5′- AAAGGTGGCTTCCAGTTCCT-3′ (PCR product = 276 bp); and for β-actin, exon2F, 5′-GAGAAGATCTGGCATCACAC-3′ and exon_3R, 5′-ATCAGGTAGTCTGTCAGGTC-3′ (PCR product = 323 bp). The PCR conditions were as follows: initial denaturation at 94°C for 3 min followed by 23–32 cycles of 94°C for 20 seconds, 59°C for 30 seconds and 72°C for 30 seconds and final extension at 72°C for 7 min. The number of cycles was optimized to maintain PCR reaction in linear range.
To construct an antisense riboprobe for in situ hybridization experiments, a 368-bp fragment specific to zebrafish mip1 transcript was generated using the following primers, forward, 5′-CTGCAGGACATGCTCATCAC-3′ and reverse, 5′-GGCTGCAAAAAGTCAACAGA-3′, and inserted into pCRII-TOPO plasmid vector. An antisense RNA probe was generated using DIG RNA Labeling Kit (Roche Applied Science, Indianapolis, IN) following manufacturer recommendation; in situ hybridization was performed as previously described [39].
Results
Genome search for regulatory regions containing conserved PITX3 binding sites identifies MIP/AQP0 promoter
Examination of the ECR Browser web-based tool for clusters of PITX3 binding sites conserved between different species yielded a total of 976 genomic regions: 511 sequences were found inside of intergenic regions, 454 elements were located within genes (309 in intronic, 90 in coding and 55 in untranslated regions), and only 11 were positioned within 1500 bp from a transcriptional start site. The eleven identified promoter regions were subjected to a secondary analysis of sequence conservation that included examination of the corresponding genes in lower vertebrates when available. One sequence demonstrated the strongest level of conservation of bicoid sites across multiple species- the promoter region of the gene encoding for the major intrinsic protein of lens fiber or aquaporin 0 (MIP/AQP0).
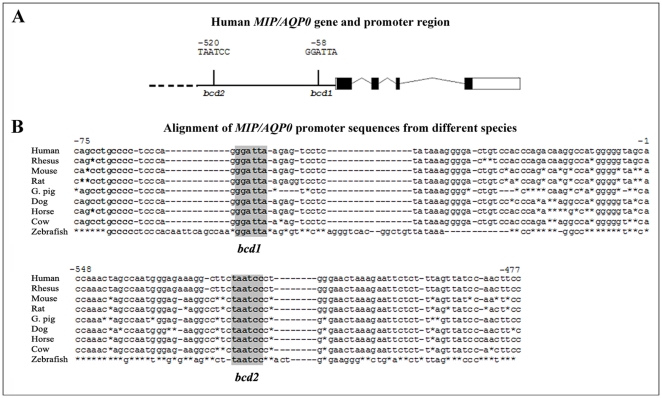
The MIP/AQP0 promoter region contains two bicoid sites separated by 456 base pairs at positions −58 (bcd1) and −520 (bcd2) from the transcriptional start site. Alignment of MIP/AQP0 promoters from different species demonstrates high conservation of both bicoid sites in nine mammalian/vertebrate species from human to zebrafish (Figure 1). Zebrafish (Danio rerio) has two orthologs of the human MIP/AQP0 gene designated mip1 and mip2 [40]. The promoter sequence/structure of zebrafish mip1 appears to be more similar to mammalian species showing conservation for both bicoid sites (positions −59 (bcd1) and −442 (bcd2)) and surrounding sequence (Figure 1).
Figure 1. Human MIP/AQP0 genomic region and bicoid elements.
A. Schematic representation of the MIP/AQP0 gene and promoter region; bcd1 and bcd2 sites are indicated. B. Multiple species alignment of genomic sequences surrounding the bcd1 and bcd2 sites (highlighted in grey). GenBank accession numbers are as follows: NT_029419.12 (Homo sapiens); NC_007868.1 (rhesus, Macaca mulatta), NT_039500.7 (mouse, Mus musculus), NC_005106.2 (rat, Rattus Norvegicus), NC_006592.2 (dog, Canis lupus familiaris), AAKN02014837.1 G.Pig, Cavia porcellus), NC_009149.2 (horse, Equus caballus), NC_007303.4 (cow, Bos taurus), NC_007134.4 (zebrafish, Danio rerio).
Conservation of the bicoid sites in MIP promoters of different species points to the potential importance of these sequences in the regulation of MIP/AQP0 expression.
The conserved bicoid sequences within the MIP/AQP0 promoter are capable of binding PITX3
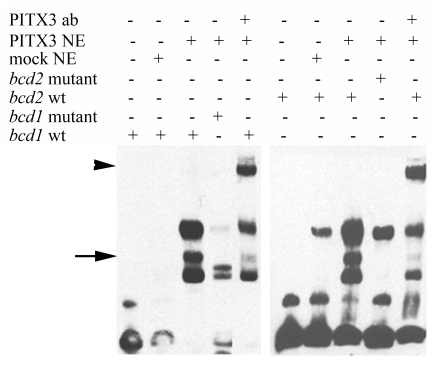
We first performed electrophoretic mobility shift assay (EMSA) to examine whether these putative bicoid sequences are able to bind PITX3 in vitro. Nuclear extracts were isolated from human lens epithelial cells transiently transfected with either a PITX3 expression plasmid or an empty pcDNA vector and incubated with labeled oligonucleotides containing the TAATCC motif and 13-bp of flanking sequences on either side of each bicoid site. The samples derived from PITX3-enriched nuclear extracts produced clearly visible shifts with both probes which were not observed with samples prepared from mock-transfected cells (Figure 2). The PITX3-DNA complexes were further verified by addition of PITX3 polyclonal antibody, which resulted in reduction of the intensities of the shifted bands and formation of supershifts (Figure 2). In addition to this, the specificity of binding was confirmed by EMSA analysis using modified oligonucleotides carrying a 2-nt mutation within the bicoid sites: the TAATCC sequence was replaced with TAATTT in both probes to abolish PITX3 binding [27]. Mutations in the bicoid sites resulted in the disappearance of protein-DNA complexes, confirming that these bands are the product of specific PITX3-DNA interactions (Figure 2).
Figure 2. Electrophoretic mobility shift assays (EMSA) demonstrate interaction between PITX3 and bcd1 and bcd2 sites.
EMSA performed with bcd1 and bcd2 oligonucleotides. DNA-PITX3 complexes are indicated with a full arrow; supershifts are shown with an arrowhead. ab = antibody, NE = Nuclear extracts, wt = wild type.
These results demonstrate that PITX3 is capable of binding to both bicoid sites located in the human MIP/AQP0 promoter in vitro.
PITX3 interacts with MIP/AQP0 promoter in human lens epithelial cells in vivo
To investigate whether PITX3 interacts with the MIP/AQP0 promoter in vivo, we performed chromatin immunoprecipitation (ChIP) assays. Native untransfected human lens epithelial (HLE) cells or HLE cells following transfection with pcDNA3.1_PITX3_FLAG expression plasmid or pcDNA3.1 empty vector were used in these experiments.
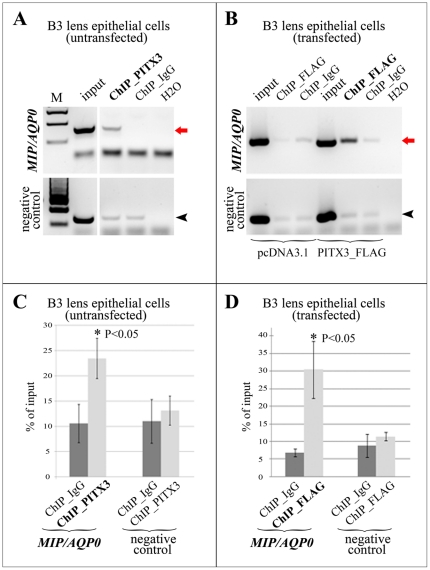
Immunoprecipitations with PITX3 antibody that used nuclear extracts from native untransfected HLE cells resulted in enrichment of MIP/AQP0 promoter sequences in the precipitated DNA in comparison to ChIP samples produced with control antibody (IgG) (Figure 3A). Immunoprecipitation with FLAG antibodies that used nuclear extracts from pcDNA3.1_PITX3_FLAG transfected cells resulted in enrichment of MIP/AQP0 promoter sequences in the precipitated DNA in comparison to ChIP samples produced with control antibody (IgG)/same nuclear extracts as well as precipitations that utilized the same antibody (FLAG) but employed mock-transfected cells (Figure 3B). This enrichment for MIP/AQP0 promoter sequences in the precipitated DNA was demonstrated by semi-quantitative PCR using specific MIP/AQP0 and control primers (described in Materials and Methods) and calculated to be ∼2.2 times in experiments performed in native untransfected HLE cells and ∼4.5 times in assays that used transfected HLE cells; the observed differences were found to be statistically significant with P<0.05 based on t-test (Figure 3C and D). The chromatin immunoprecipitation and PCR-based enrichment analysis was repeated eight times using independently transfected cells with consistent results.
Figure 3. MIP/AQP0 region demonstrates enrichment in chromatin immunoprecipitation experiments with PITX3 or FLAG antibody.
A. Endogenous PITX3 is bound to the proximal MIP/AQP0 promoter in human lens epithelial (HLE) cell cultures. Chromatin immunoprecipitation assays were performed using untransfected HLE cells and human PITX3 or IgG (control) antibodies. The samples were analyzed by semi-quantitative PCR using MIP/AQP0 proximal promoter- specific primers and negative control primers. Please note robust amplification of MIP/AQP0 promoter region from ChIP sample precipitated with PITX3 but not with control IgG antibody (red arrow) and equal levels of DNA amplification for negative control region in both samples (black arrowhead). B. PITX3_FLAG is bound to proximal MIP/AQP0 promoter in HLE cell cultures following transfection with PITX3-FLAG expression plasmid. HLE cells were transfected with either PITX3-FLAG expression plasmid or control pcDNA3.1 expression vector. ChIP assays were performed with FLAG-M2 or control IgG antibody. The ChIP samples were analyzed by semi-quantitative PCR as described in A. Please note enrichment of MIP/AQP0 promoter region in ChIP sample obtained from PITX3-FLAG transfected cells and precipitated with FLAG antibody in comparison to FLAG-precipitated ChIP sample obtained from pcDNA3.1 transfected cells as well as IgG-precipitated ChIP sample obtained using either PITX3-FLAG or pcDNA3.1 transfected cells (red arrow). In addition to this, amplification of negative control region demonstrated similar levels across all samples (black arrowhead). C and D. Statistical analysis of multiple semi-quantitative PCR/ChIP experiments performed in untransfected (C) and transfected HLE cells (D) as described in A and B, correspondingly. Presence of MIP/AQP0 promoter or negative control region DNA in various ChIP samples was evaluated by semi-quantitative PCR followed by densitometric analysis and expressed as a percentage of input values; mean and standard deviation for at least 3 independent experiments were calculated and analyzed by Student's t test. Please note statistically significant enrichment for MIP/AQP0 promoter region DNA precipitated with PITX3 (C) or FLAG (D) antibody in comparison to control IgG-precipitated chromatin in HLE untransfected (C) or transfected (D) cells. IgG = normal mouse IgG; PITX3 = PITX3 polyclonal antibody; FLAG = anti-FLAG monoclonal antibody.
These experiments demonstrated the specific association of PITX3 with the MIP/AQP0 promoter region in vivo.
Mutations in bicoid sites located in the MIP/AQP0 promoter affect the activity of the promoter
To examine whether the conserved bicoid sites within the MIP/AQP0 promoter are involved in regulation of its activity, we created several reporter constructs: MIP656-bcd1,2, which contained a 656-bp fragment of the MIP/AQP0 wild-type promoter encompassing both bicoid sites and nucleotides from positions −597 to +59 in relation to the MIP/AQP0 transcriptional start site inserted into a basic pGL3 plasmid containing the luciferase reporter gene; MIP656-bcd1, which contained a mutation in the bcd2 site that changed the 5′-TAATCC-3′ sequence to 5′-TAATTT-3′, thus abolishing its interaction with wild-type PITX3 [27]; MIP656-bcd2, which contained a similar mutation in the bcd1 site changing the 5′-GGATTA-3′ sequence into 5′-AAATTA-3′; and MIP656-bcd0, which included both of the above described mutations.
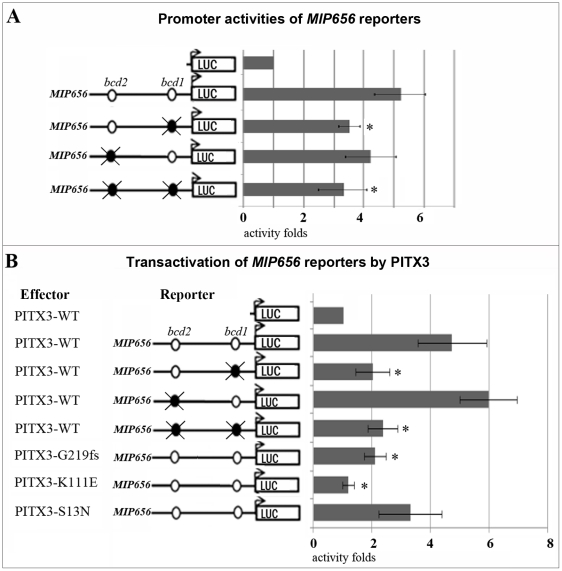
Reporter assays demonstrated a 5.2-fold upregulation of luciferase expression in the presence of the MIP656-bcd1,2 promoter fragment in comparison to the empty vector in human embryonic kidney cells (Figure 4A). Mutations in either the bcd1 or bcd2 sites resulted in a decrease in MIP/AQP0 promoter activity compared to the wild-type promoter: to 3.5-fold (67% of MIP656-bcd1,2 activity; P<0.001) when the bcd1 site was mutated, to 4.3-fold (83%; P = 0.014) when the bcd2 site was abolished and to 3.4-fold (65%; P<0.001) when both sites were disrupted.
Figure 4. MIP/AQP0 is activated by PITX3 via interaction with the proximal bicoid site, bcd1.
A. Promoter activities of the MIP656 reporters in human embryonic kidney cells B. Transactivation of MIP656 reporters by PITX3 and its mutants in human embryonic kidney cells. Constructs and positions of bicoid sites are indicated on the left side. Wild-type bcd1 or bcd2 sites are depicted as open circles. Mutations (TAATCC to TAATTT substitutions) in bcd1 or bcd2 sites are depicted as dark circles with a strike-through. Student's paired t-Test with a one-tailed distribution was utilized to compare values. Experiments marked with asterisk (*) demonstrated a significant difference (P<0.001) in comparison to experiments performed with MIP656 wild-type promoter (A) or MIP656 wild-type promoter with PITX3-WT (B).
Based upon these results, both bicoid sites appear to be involved in regulation of MIP/AQP0 expression with the proximal site, bcd1, playing a more significant role in its activation.
PITX3 is capable of transactivating the MIP/AQP0 promoter via the bcd1 bicoid site
In order to investigate the effect of PITX3 on the transcriptional activity of the MIP/AQP0 promoter, we performed cotransfection assays using the above described MIP656-bcd1,2, MIP656-bcd1, MIP656-bcd2 and MIP656-bcd0 reporter constructs and a PITX3 expression plasmid.
Cotransfection of the PITX3-WT expression plasmid with the MIP656-bcd1,2 luciferase reporter into human embryonic kidney cells resulted in a ∼4.8-fold normalized activation in comparison to the cotransfection of the PITX3-WT plasmid with the promoter-less reporter (Figure 4B). In contrast, cotransfection of the MIP656-bcd1,2 luciferase reporter with an expression plasmid carrying PITX3 mutants produced a ∼2-fold increase over the same control for the G219f s mutant (42% of wild-type activity; P<0.001), ∼3.1-fold for S13N (65%; P = 0.015) and no transactivation was observed for the K111E mutant. These results are consistent with the previously reported data on the residual activities of the corresponding mutant PITX3 forms [27].
We next examined the two bicoid sites present in the MIP/AQP0 promoter for their role in this observed transactivation. The PITX3-WT expression plasmid and mutant MIP/AQP0 promoter constructs, MIP656-bcd1 (carrying mutant bcd2 site), MIP656-bcd2 (carrying mutant bcd1 site) and MIP656-bcd0 (carrying mutations in both bicoid sites) were cotransfected into human embryonic kidney cells and the resultant luciferase activities were compared to values observed in experiments involving cotransfection of wild-type MIP/AQP0 promoter (MIP656-bcd1,2) and PITX3-WT. The transactivation of the MIP656-bcd1,2 promoter by PITX3-WT decreased to ∼2-fold (42% of PITX3 induced wild-type promoter transactivation) when the mutation in the proximal bicoid site, bcd1, was introduced; increased to ∼6-fold (125%; P = 0.03) when the distal bicoid site, bcd2, was mutated; and produced ∼2.2-folds (46%; P<0.001) when both bicoid sites were disrupted (Figure 4B).
These data suggest that PITX3 is involved in activation of the MIP/AQP0 promoter via its proximal bicoid site, bcd1.
Knockdown of pitx3 affects formation of a DNA-protein complex associated with mip1 promoter sequences
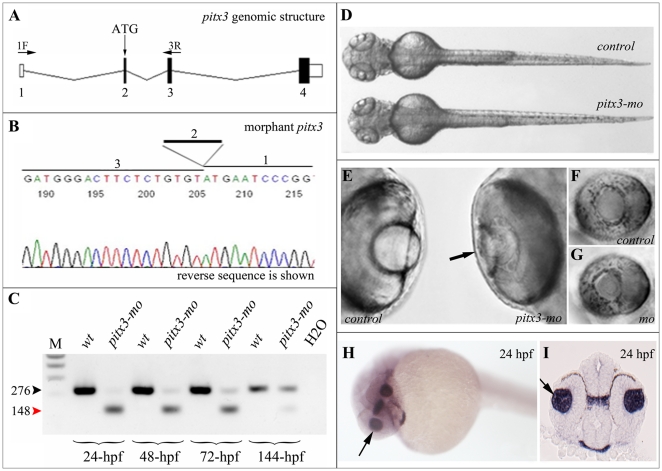
In order to efficiently disrupt pitx3 gene expression in zebrafish and to be able to tightly monitor residual activity/knockdown level, we tested several splicing morpholino that were designed against pitx3 intron-exon junctions. Unfortunately, none of these morpholinos produced the desired outcome, resulting in either no effect on pitx3 splicing or highly abnormal phenotype due to toxicity/non-specific defects. Then we tested the previously reported translational morpholinos [9], [12] and discovered that the antisense morpholino reported by Dutta and coauthors [9] and designed against the sequence containing the translation initiation codon located in exon 2 of pitx3 results in abnormal splicing of the pitx3 transcript due to exon 2 skipping (Figure 5). This pitx3 morpholino [9] matches the nucleotide sequence at positions +8 to +32 of exon 2 and therefore is located only 7-nt upstream of the intron 1/exon 2 acceptor site. In our experiments, we found this morpholino to be highly efficient in blocking normal splicing with the abnormal 148-bp product lacking exon 2 generated in pitx3-mo injected embryos versus the normal 276-bp product containing exon 2 seen in control-mo injected embryos (Figure 5B, C). The first potential initiation codon (for methionine) in the pitx3-mo transcript is located at position 28 of the pitx3 homeodomain and, as a result, the translation of this transcript will produce an abnormal protein lacking the N-terminal region and 45% of its homeodomain and, therefore, predicted to be nonfunctional.
Figure 5. Injection of pitx3 morpholino results in abnormal splicing of pitx3 transcript and small lens phenotype.
A. Schematic drawing of pitx3 gene; position of initiation codon (ATG) and RT-PCR primers (1F and 3R) are indicated, exons are numbered. B. Sequencing of the pitx3-mo transcript generated with primers located in the first and third pitx3 exons demonstrates absence of exon 2 in the resultant product. C. RT-PCR results obtained with pitx3 1F/3R primers using RNA extracted from embryos injected with control or pitx3 morpholino. Please note a strong decrease in normal 276-bp product (black arrowhead) and presence of abnormal 148-bp product (red arrowhead) in pitx3-mo samples; hpf- hours post fertilization of analyzed embryos. D–G. Morphological phenotypes of zebrafish pitx3-mo embryos. In comparison to control embryos, a smaller head can be observed in pitx3 morphants (72-hpf embryo is shown; D) as well as an obvious reduction in lens size (black arrow) at later stages (96-hpf embryos are shown; E–G). H and I. Expression of pitx3 in the developing lens in 24-hpf embryos. Please note robust expression in the lens vesicle (arrows) as demonstrated by both whole mount (H) and section (I) in situ.
An abnormal phenotype was detected in ∼95% of pitx3-mo injected embryos; early lethality (before 20-hpf) was observed in ∼20% of pitx3-mo and control-mo injected embryos. The pitx3-mo displayed a misshapen smaller head, jaw abnormalities and reduction in eye size due to progressive lens degeneration and retinal defects leading to a complete loss of lens by 7-dpf consistent with the previous reports [9], [12] (Figure 5D–G). Robust expression of pitx3 is seen in lens vesicle at 24-hpf and it continues to be highly expressed during all stages of lens development [9]–[13; Figure 5H and I]. In addition to the strong lens expression, pitx3 transcripts are also detected in the developing brain, craniofacial region and trunk musculature as previously described [9], [12], [13], [41], [42].
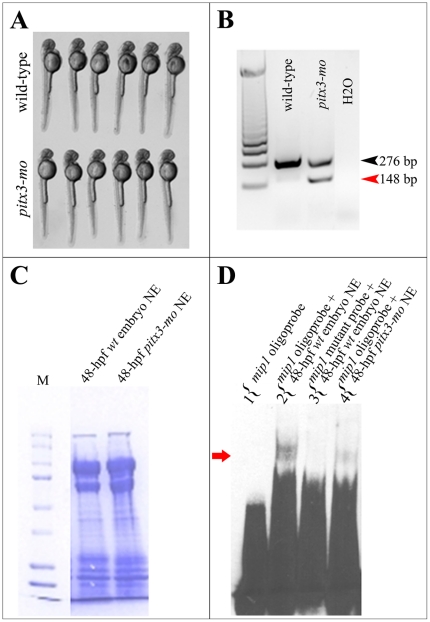
Since we demonstrated above that PITX3 is capable of binding human MIP/AQP0 promoter in vivo, further experiments were performed to establish if knockdown of pitx3 would affect formation of protein-mip1 promoter complexes in zebrafish. We injected zebrafish embryos with above described pitx3 morpholino oligonucleotides that result in abnormal splicing of pitx3 transcript. Embryos were harvested at 48-hpf and nuclear extracts from wild-type embryos and pitx3 morphants were tested for their ability to bind a biotinylated DNA fragment containing the proximal bicoid site and corresponding to zebrafish −44/−76 mip1 promoter region (Figure 6). The experiments were performed using nuclear protein extracts isolated from the upper trunk/head region of the 48-hpf wild-type and pitx3 morphant embryos that displayed normal body length and morphology (Figure 6A) and demonstrated a normal presence (wild-type) or a significant reduction (pitx3-mo) in normal pitx3 transcript based on RT-PCR analysis performed using RNA extracted from the lower trunk region of the same embryos (Figure 6B). For positive control, aliquots of nuclear extracts from wild-type and pitx3 morphants were analyzed on 10% polyacrylamide gel followed by Coomassie Blue R-250 staining to assure equal protein concentration in both samples (Figure 6C). Two apparent slow-migrating complexes were formed that were evident at the top of the gel when EMSA was performed with nuclear extracts obtained from wild-type embryos. The formation of these complexes was abolished by introduction of a mutation into the bicoid site contained within the −44/−76 mip1 promoter, which suggests that pitx3 is directly involved in DNA-binding of this fragment (Figure 6D). In addition to this, the slow-migrating complexes were significantly diminished when nuclear extract obtained from pitx3 morphants were utilized (Figure 6D). Therefore the observed reduction in the intensity of the slow-migrating DNA-protein complexes correlates well with the residual amount of normal pitx3 transcript in morphants in this experiment.
Figure 6. Formation of high molecular weight mip1 promoter- protein complexes is dependent on pitx3 presence.
A. 48-hpf wild-type and pitx3 morphant embryos showing normal body length and morphology that were selected for EMSA experiments. B. Results of RT-PCR performed with RNA extracted from the pooled tail tissues from wild-type and pitx3 morphant embryos shown in C. A sharp reduction in normal pitx3 transcript (black arrowhead) and the presence of abnormally spliced product (red arrowhead) are evident in pitx3 morphant embryos. C. Coomassie Blue R-250 stained polyacrilamide gel demonstrating equal protein concentration in nuclear extracts obtained from wild-type (lane 1) and pitx3 morphant (lane 2) embryos that were used in EMSA experiments shown in A. D. Electrophoretic mobility shift assays (EMSA) show formation of a DNA-protein complex when an oligonucleotide corresponding to the −44/−76 region of zebrafish mip1 promoter and nuclear extracts from 48-hpf wild-type zebrafish embryos are used. Please note a presence of a specific slow migrating complex, which is formed by wild-type mip1 probe and proteins extracted from nuclei of 48-hpf wild-type zebrafish embryos (lane 2), absence of this complex in lane 3 when the same nuclear extracts were combined with a mutant mip1 probe where the pitx3-binding bicoid site GGATTA was replaced by AAATTA, and sharp reduction of this complex in lane 4 containing a combination of a wild-type mip1 probe and nuclear extracts obtained from pitx3 morphants (red arrow).
These data support our previous findings which demonstrate that pitx3 is a part of large complex occupying the mip1 promoter in the developing zebrafish embryo.
Knockdown of pitx3 affects mip1 expression in zebrafish embryos
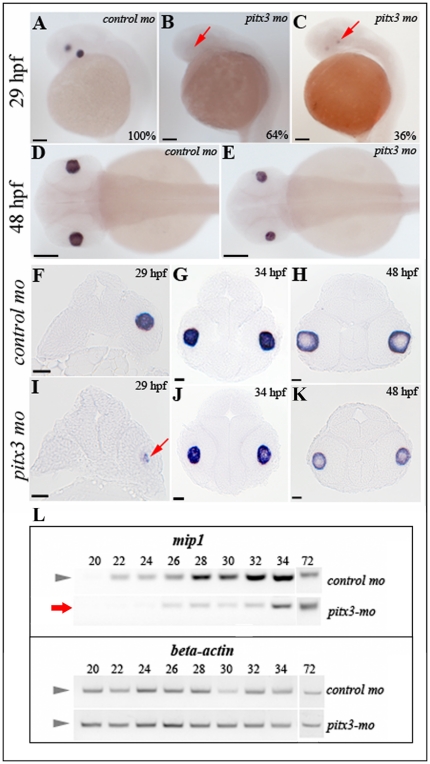
Examination of mip1 expression by in situ hybridization identified a specific and robust expression pattern in 100% of control-mo injected embryos (15/15), while a complete absence (9/14 or 64.3%) or a very low level (5/14 or 35.7%) of mip1 expression was seen in pitx3-mo embryos at 29-hpf (Figure 7A–C, F, I). mip1 expression is clearly observed in both control and pitx3-mo injected embryos at later stages but appears to be somewhat reduced in pitx3 morphants (Figure 7 D, E, G, H, J, K).
Figure 7. Analysis of mip1 expression in pitx3-mo and control embryos via in situ hybridization and RT-PCR.
A, D, F-H. Normal mip1 expression in control-injected embryos at 29-, 34- and 48-hpf. B, C, E, I–K. Altered mip1 expression is observed in pitx3 morphants at 29-hpf with 64% of embryos demonstrating a complete absence of mip1 expression (B) and the remaining larvae showing markedly reduced mip1 expression (C and I). Reduced mip1 expression is also observed in 34- and 48-hpf embryos (E, J, K). Red arrows show sites of expected mip1 expression. Scale bars: A–E: 100 µM; F–L: 20 µM. L. Results of semi-quantitative RT-PCR showing reduced expression of mip1 in pitx3 morphants at early stages of development (red arrow).
Semi-quantitative RT-PCR analysis of mip1 expression in the pitx3 morphants and control-injected embryos confirmed a specific delay and decrease in mip1 expression in pitx3 morphants. The expression was initiated at ∼22-hpf in control injected embryos consistent with the start of lens fiber cell differentiation. The expression increased at later stages with the highest levels being detected in 48-hpf embryos and decreased levels by 72-hpf. In the pitx3-mo embryos, the first expression was observed in 26-hpf embryos with expression levels being noticeably lower in comparison to the control-mo injected larvae. The mip1 expression in 28–34-hpf pitx3 morphants continued to be reduced in comparison to control-injected larvae and but reached similar expression levels by 72-hpf (Figure 7L). Examination of beta-actin (loading control) demonstrated similar levels of expression between pitx3-mo and control-mo injected embryos (Figure 7L).
These experiments revealed a specific delay and decrease in expression of an important lens factor, mip1, in response to pitx3 deficiency.
Discussion
PITX3 is a homeodomain transcription factor that is essential to normal eye development in vertebrates. Yet, its direct downstream targets and mechanism of action are poorly understood. In this manuscript, we present identification of the first direct target of PITX3 during lens development, the major intrinsic protein of lens fibers, MIP/AQP0.
The MIP/AQP0 promoter was identified via scanning of the human genome for regions containing conserved clusters of bicoid sequences and located in the proximity of known genes. Since the sequence, expression and function of PITX3 are conserved in vertebrates and bicoid sites are known to mediate its interaction with DNA, the conserved presence of these elements in a gene's promoter/regulatory region suggests that it may be regulated by PITX3. In addition to this, MIP/AQP0 represents a logical downstream target of PITX3 because of its known role in lens development/function.
MIP/AQP0 is one of the most abundant proteins found in lens fibers where it acts as a water channel and adhesion molecule [43]–[46]. Mouse Pitx3 and Mip/Aqp0 display overlapping expression patterns during eye development as both genes are expressed in the developing primary and secondary lens fibers, with continued expression in adult organisms [1], [14], [47], [48]; expression of zebrafish pitx3 precedes mip1 in the developing lens [9]–[13; this manuscript] consistent with its proposed role in activation of mip1 expression. Mutations in both PITX3 and MIP/AQP0 are implicated in congenital cataracts in humans [1]–[6], [49] and result in lens phenotypes in mice [7], [8], [18], [50]–[52]. Gene expression patterns as well as phenotypic abnormalities observed in mutant animals suggest an earlier appearance of Pitx3 in comparison to Mip/Aqp0, which would also be consistent with Mip/Aqo0 being a downstream target of Pitx3. Expression of the Mip/Aqp0 transcript and protein is first detectable at mouse embryonic stage E11.25 in the ventro-temporal half of the lens vesicle concurrent with the initial stages of primary fiber cell differentiation and continues to be restricted to the lens differentiating primary and secondary fiber cells throughout adulthood [47], [48].
Transcriptional regulation of the Mip/Aqp0 expression pattern is not yet well understood with several potential players discussed in the literature. Previous studies have shown that the human MIP/AQP0 5′ flanking sequence −253/+42 is sufficient for promoter activity in embryonic chicken lens epithelia primary cultures but is inactive in non-lens cells, suggesting that this region contains regulatory sequences required for the lens-specific expression of MIP/AQP0 [53]. Ohtaka-Maruyama and colleagues reported that MIP/AQP0 promoter fragments are protected in a DNase I footprinting assay performed with purified AP-2 and Sp1 and, therefore, the promoter is likely to interact with these factors [54]. Transgenic mice expressing AP2-α in lens fiber cells under the control of the αA-crystallin promoter were found to have reduced amounts of Mip/Aqp0 in all fiber cells while expanded Mip/Aqp0 expression was detected in the lens stalk of AP2-α−/− mice [55], [56]. Based on these observations and earlier reports [54], West-Mays and colleagues concluded that Ap2-α acts as a negative regulator of Mip/Aqp0 expression and may, directly or indirectly, be responsible for its tight spatial/ temporal control [56]. In addition to these reports, Mip expression was shown to be triggered by treatment with FGF-2 and to accompany ERK1/2 and JNK activation in rat lens epithelia explants; treatment with specific ERK1/2 or JNK inhibitors resulted in abrogation of Mip/Aqp0 expression in response to FGF-2 [57].
In this manuscript, we present evidence that the two evolutionarily conserved sequences containing bicoid sites within the MIP/AQP0 promoter are capable of binding specifically to PITX3. Moreover, through chromatin immunoprecipitation assays we demonstrated that PITX3 is bound to the MIP/AQP0 promoter in vivo in human lens epithelial cells. Analysis of the functional significance of this binding using luciferase reporter assays demonstrated that wild-type PITX3 is able to transactivate the MIP/AQP0 promoter while mutant PITX3 forms showed reduced or absent transactivation ability. Further functional analysis utilizing site-specific mutations revealed the importance of the proximal bicoid site for the observed transactivation. The obliteration of the proximal bicoid site resulted in a statistically significant reduction of MIP/AQP0 promoter activity as well as a decreased level of transactivation by PITX3. Finally, analyses performed in zebrafish embryos suggested that pitx3 is bound to the mip1 promoter sequences during embryonic development and that mip1 expression is altered in zebrafish pitx3 morphants. At later stages of development (48–72-hpf), the expression of mip1 in pitx3 morphants appears to be largely unaffected, suggesting that regulation of mip1 activity at these stages may be mainly controlled by other, pitx3 pathway-independent, factors; pitx3 may also contribute to the recovery of mip1 expression since increasing amounts of normal pitx3 transcript can be observed in zebrafish embryos starting at 48-hpf due to weakening of the effects of morpholino injections (Figure 5C). Identification/development of permanent pitx3 mutant lines is needed to allow more careful evaluation of the relationship between these factors.
Mutations in the human MIP/AQP0 gene were shown to underlie various dominant forms of cataracts [49], [58]–[65]. To the best of our knowledge, only two of the reported MIP/AQP0 mutations were explored for functional defects and a dominant-negative mechanism was suggested [58]. The dominant nature of MIP/AQP0 mutations may also be explained by haploinsufficiency which would suggest that lens development is highly sensitive to dosage/timely expression of MIP/AQP0. The later possibility is further supported by the phenotype reported in the mouse carrying a null allele of Aqp0 [54]. Deletion of mouse Aqp0 was shown to result in cataracts at 3 weeks of age and at 24 weeks of age in homozygous and heterozygous mice, respectively. In heterozygous animals, the lens osmotic water permeability value was reduced to around 46% and the lens focusing power was significantly decreased in comparison to wild-type [52]. These findings demonstrated that a loss of one Aqp0 allele, which presumably leads to reduced Aqp0 expression, can be associated with lens abnormalities. Therefore, since MIP/AQP0 represents an apparent transcriptional target of PITX3, the alteration of the MIP/AQP0 expression in patients affected with PITX3 mutations is likely to contribute to the lens phenotype observed in these individuals.
Further studies of the MIP/AQP0 promoter will not only yield important insight into the transcriptional regulation of this critical lens differentiation factor but will also provide better understanding of the function of PITX3 and its interacting partners and allow for more specific identification of additional downstream targets of this ocular factor. Also, genetic screening of both PITX3 and MIP/AQP0 in human patients affected with ocular conditions may lead to an identification of synergistic or compensatory mutations/variants that may help to explain the considerable intra- and interfamilial phenotypic variability associated with mutations in either gene.
Acknowledgments
The authors would like to thank Rebecca C. Tyler and Gary Gardner for assistance with experiments involving DNA sequencing and zebrafish maintenance, and Linda M. Reis for critical reading of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funding was provided by grant EY013606 from the National Eye Institute and by support from the Children's Research Institute Foundation at Children's Hospital of Wisconsin to EVS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Semina EV, Ferrell RE, Mintz-Hittner HA, Bitoun P, Alward WL, et al. A novel homeobox gene PITX3 is mutated in families with autosomal-dominant cataracts and ASMD. Nat Genet. 1998;19(2):167–170. doi: 10.1038/527. [DOI] [PubMed] [Google Scholar]
- 2.Berry V, Yang Z, Addison PK, Francis PJ, Ionides A, et al. Recurrent 17 bp duplication in PITX3 is primarily associated with posterior polar cataract (CPP4). J Med Genet. 2004;41(8):e109. doi: 10.1136/jmg.2004.020289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bidinost C, Matsumoto M, Chung D, Salem N, Zhang K, et al. Heterozygous and homozygous mutations in PITX3 in a large Lebanese family with posterior polar cataracts and neurodevelopmental abnormalities. Invest Ophthalmol Vis Sci. 2006;47(4):1274–1280. doi: 10.1167/iovs.05-1095. [DOI] [PubMed] [Google Scholar]
- 4.Finzi S, Li Y, Mitchell TN, Farr A, Maumenee IH, et al. Posterior polar cataract: genetic analysis of a large family. Ophthalmic Genet. 2005;26(3):125–130. doi: 10.1080/13816810500229124. [DOI] [PubMed] [Google Scholar]
- 5.Burdon KP, McKay JD, Wirth MG, Russell-Eggit IM, Bhatti S, et al. The PITX3 gene in posterior polar congenital cataract in Australia. Mol Vis. 2006;12:367–371. [PubMed] [Google Scholar]
- 6.Summers KM, Withers SJ, Gole GA, Piras S, Taylor PJ. Anterior segment mesenchymal dysgenesis in a large Australian family is associated with the recurrent 17 bp duplication in PITX3. Mol Vis. 2008;14:2010–2015. [PMC free article] [PubMed] [Google Scholar]
- 7.Semina EV, Murray JC, Reiter R, Hrstka RF, Graw J. Deletion in the promoter region and altered expression of Pitx3 homeobox gene in aphakia mice. Hum Mol Genet. 2000;9(11):1575–1585. doi: 10.1093/hmg/9.11.1575. [DOI] [PubMed] [Google Scholar]
- 8.Rieger DK, Reichenberger E, McLean W, Sidow A, Olsen BR. A double-deletion mutation in the Pitx3 gene causes arrested lens development in aphakia mice. Genomics. 2001;72(1):61–72. doi: 10.1006/geno.2000.6464. [DOI] [PubMed] [Google Scholar]
- 9.Dutta S, Dietrich JE, Aspöck G, Burdine RD, Schier A, et al. Pitx3 defines an equivalence domain for lens and anterior pituitary placode. Development. 2005;132(7):1579–1590. doi: 10.1242/dev.01723. [DOI] [PubMed] [Google Scholar]
- 10.Khosrowshahian F, Wolanski M, Chang WY, Fujiki K, Jacobs L, et al. Lens and retina formation require expression of Pitx3 in Xenopus pre-lens ectoderm. Dev Dyn. 2005;234(3):577–589. doi: 10.1002/dvdy.20540. [DOI] [PubMed] [Google Scholar]
- 11.Pommereit D, Pieler T, Hollemann TX. Pitx3: a member of the Rieg/Pitx gene family expressed during pituitary and lens formation in Xenopus laevis. Mech Dev. 2001;102(1–2):255–257. doi: 10.1016/s0925-4773(01)00305-7. [DOI] [PubMed] [Google Scholar]
- 12.Shi X, Bosenko DV, Zinkevich NS, Foley S, Hyde DR, et al. Zebrafish pitx3 is necessary for normal lens and retinal development. Mech Dev. 2005;122(4):513–527. doi: 10.1016/j.mod.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 13.Zilinski CA, Shah R, Lane ME, Jamrich M. Modulation of zebrafish pitx3 expression in the primordia of the pituitary, lens, olfactory epithelium and cranial ganglia by hedgehog and nodal signaling. Genesis. 2005;41(1):33–40. doi: 10.1002/gene.20094. [DOI] [PubMed] [Google Scholar]
- 14.Semina EV, Reiter RS, Murray JC. Isolation of a new homeobox gene belonging to the Pitx/Rieg family: expression during lens development and mapping to the aphakia region on mouse chromosome 19. Hum Mol Genet. 1997;6(12):2109–2116. doi: 10.1093/hmg/6.12.2109. [DOI] [PubMed] [Google Scholar]
- 15.Malinina NA, Koniukhov BV. Action of mutant genes on crystallin synthesis in the developing mouse lens. III. The aphakia gene. Ontogenez. 1981;12(6):589–595. [PubMed] [Google Scholar]
- 16.Webster EH, Jr, Zwaan J, Cooper PJ. Abnormal accumulation of sulphated materials in lens tissue of mice with the aphakia mutation. Embryol Exp Morphol. 1986;92:85–101. [PubMed] [Google Scholar]
- 17.Zwaan J. Immunofluorescent studies on aphakia, a mutation of a gene involved in the control of lens differentiation in the mouse embryo. Dev Biol. 1975;44(2):306–312. doi: 10.1016/0012-1606(75)90401-7. [DOI] [PubMed] [Google Scholar]
- 18.Ho HY, Chang KH, Nichols J, Li M. Homeodomain protein Pitx3 maintains the mitotic activity of lens epithelial cells. Mech Dev. 2009;126(1–2):18–29. doi: 10.1016/j.mod.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Medina-Martinez O, Shah R, Jamrich M. Pitx3 controls multiple aspects of lens development. Dev Dyn. 2009;238(9):2193–2201. doi: 10.1002/dvdy.21924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi X, Luo Y, Howley S, Dzialo A, Foley S, et al. Zebrafish foxe3: roles in ocular lens morphogenesis through interaction with pitx3. Mech Dev. 2006;123(10):761–782. doi: 10.1016/j.mod.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Gurnett CA, Alaee F, Kruse LM, Desruisseau DM, Hecht JT, et al. Asymmetric lower-limb malformations in individuals with homeobox PITX1 gene mutation. Am J Hum Genet. 2008;83(5):616–622. doi: 10.1016/j.ajhg.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, et al. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14(4):392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- 23.Amendt BA, Sutherland LB, Semina EV, Russo AF. The molecular basis of Rieger syndrome. Analysis of Pitx2 homeodomain protein activities. J Biol Chem. 1998;273(32):20066–20072. doi: 10.1074/jbc.273.32.20066. [DOI] [PubMed] [Google Scholar]
- 24.Lebel M, Gauthier Y, Moreau A, Drouin J. Pitx3 activates mouse tyrosine hydroxylase promoter via a high-affinity binding site. J Neurochem. 2001;77(2):558–567. doi: 10.1046/j.1471-4159.2001.00257.x. [DOI] [PubMed] [Google Scholar]
- 25.Quentien MH, Manfroid I, Moncet D, Gunz G, Muller M, et al. Pitx factors are involved in basal and hormone-regulated activity of the human prolactin promoter. J Biol Chem. 2002;277(46):44408–44416. doi: 10.1074/jbc.M207824200. [DOI] [PubMed] [Google Scholar]
- 26.Hjalt TA, Amendt BA, Murray JC. PITX2 regulates procollagen lysyl hydroxylase (PLOD) gene expression: implications for the pathology of Rieger syndrome. J Cell Biol. 2001;152(3):545–552. doi: 10.1083/jcb.152.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakazume S, Sorokina E, Iwamoto Y, Semina EV. Functional analysis of human mutations in homeodomain transcription factor PITX3. BMC Mol Biol. 2007;8:84. doi: 10.1186/1471-2199-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vadlamudi U, Espinoza HM, Ganga M, Martin DM, Liu X, et al. PITX2, beta-catenin and LEF-1 interact to synergistically regulate the LEF-1 promoter. J Cell Sci. 2005;118(Pt 6):1129–1137. doi: 10.1242/jcs.01706. [DOI] [PubMed] [Google Scholar]
- 29.Island ML, Mesplede T, Darracq N, Bandu MT, Christeff N, et al. Repression by homeoprotein pitx1 of virus-induced interferon a promoters is mediated by physical interaction and trans repression of IRF3 and IRF7. Mol Cell Biol. 2002;22(20):7120–7133. doi: 10.1128/MCB.22.20.7120-7133.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, et al. Identification of a Wnt/Dvl/beta-Catenin→Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111(5):673–685. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- 31.Amen M, Liu X, Vadlamudi U, Elizondo G, Diamond E, et al. PITX2 and beta-catenin interactions regulate Lef-1 isoform expression. Mol Cell Biol. 2007;27(21):7560–7573. doi: 10.1128/MCB.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamba P, Khivansara V, D'Alessio AC, Santos MM, Bernard DJ. Paired-like homeodomain transcription factors 1 and 2 regulate follicle-stimulating hormone beta-subunit transcription through a conserved cis-element. Endocrinology. 2008;149(6):3095–3108. doi: 10.1210/en.2007-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quirk CC, Lozada KL, Keri RA, Nilson JH. A single Pitx1 binding site is essential for activity of the LHbeta promoter in transgenic mice. Mol Endocrinol. 2001;15(5):734–746. doi: 10.1210/mend.15.5.0628. [DOI] [PubMed] [Google Scholar]
- 34.Ganga M, Espinoza HM, Cox CJ, Morton L, Hjalt TA, et al. PITX2 isoform-specific regulation of atrial natriuretic factor expression: synergism and repression with Nkx2.5. J Biol Chem. 2003;278(25):22437–22445. doi: 10.1074/jbc.M210163200. [DOI] [PubMed] [Google Scholar]
- 35.Suszko MI, Lo DJ, Suh H, Camper SA, Woodruff TK. Regulation of the rat follicle-stimulating hormone beta-subunit promoter by activin. Mol Endocrinol. 2002;17(3):318–332. doi: 10.1210/me.2002-0081. [DOI] [PubMed] [Google Scholar]
- 36.Elgar G, Vavouri T. Tuning in to the signals: noncoding sequence conservation in vertebrate genomes. Trends Genet. 2008;24(7):344–352. doi: 10.1016/j.tig.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 37.Woolfe A, Elgar G. Organization of conserved elements near key developmental regulators in vertebrate genomes. Adv Genet. 2008;61:307–338. doi: 10.1016/S0065-2660(07)00012-0. [DOI] [PubMed] [Google Scholar]
- 38.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 39.Zinkevich NS, Bosenko DV, Link BA, Semina EV. laminin alpha 1 gene is essential for normal lens development in zebrafish. BMC Dev Biol. 2006;6:13. doi: 10.1186/1471-213X-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vihtelic TS, Fadool JM, Gao J, Thornton KA, Hyde DR, et al. Expressed sequence tag analysis of zebrafish eye tissues for NEIBank. Mol Vis. 2005;11:1083–1100. [PubMed] [Google Scholar]
- 41.Qiu HY, Guo C, Cheng XW, Huang Y, Xiong ZQ, Ding YQ. Pitx3-CreER mice showing restricted Cre expression in developing ocular lens and skeletal muscles. Genesis. 2008;46(6):324–328. doi: 10.1002/dvg.20399. [DOI] [PubMed] [Google Scholar]
- 42.L'Honoré A, Coulon V, Marcil A, Lebel M, Lafrance-Vanasse J, et al. Sequential expression and redundancy of Pitx2 and Pitx3 genes during muscle development. Dev Biol. 2007;307(2):421–433. doi: 10.1016/j.ydbio.2007.04.034. [DOI] [PubMed] [Google Scholar]
- 43.Chepelinsky AB. Structural Function of MIP/Aquaporin 0 in the Eye Lens; Genetic Defects Lead to Congenital Inherited Cataracts. Handb Exp Pharmacol. 2009;(190):265–297. doi: 10.1007/978-3-540-79885-9_14. [DOI] [PubMed] [Google Scholar]
- 44.Dunia I, Recouvreur M, Nicolas P, Kumar N, Bloemendal H, et al. Assembly of connexins and MP26 in lens fiber plasma membranes studied by SDS-fracture immunolabeling. J Cell Sci. 1998;111(Pt 15):2109–2120. doi: 10.1242/jcs.111.15.2109. [DOI] [PubMed] [Google Scholar]
- 45.Mulders SM, Preston GM, Deen PM, Guggino WB, van Os CH, et al. Water channel properties of major intrinsic protein of lens. J Biol Chem. 1995;270(15):9010–9016. doi: 10.1074/jbc.270.15.9010. [DOI] [PubMed] [Google Scholar]
- 46.Pisano MM, Chepelinsky AB. Genomic cloning, complete nucleotide sequence, and structure of the human gene encoding the major intrinsic protein (MIP) of the lens. Genomics. 1991;11(4):981–990. doi: 10.1016/0888-7543(91)90023-8. [DOI] [PubMed] [Google Scholar]
- 47.Varadaraj K, Kumari SS, Mathias RT. Functional expression of aquaporins in embryonic, postnatal, and adult mouse lenses. Dev Dyn. 2007;236(5):1319–1328. doi: 10.1002/dvdy.21125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou L, Chen T, Church RL. Temporal expression of three mouse lens fiber cell membrane protein genes during early development. Mol Vis. 2002;8:143–148. [PubMed] [Google Scholar]
- 49.Berry V, Francis P, Kaushal S, Moore A, Bhattacharya S. Missense mutations in MIP underlie autosomal dominant ‘polymorphic’ and lamellar cataracts linked to 12q. Nat Genet. 2000;25(1):15–17. doi: 10.1038/75538. [DOI] [PubMed] [Google Scholar]
- 50.Sidjanin DJ, Parker-Wilson DM, Neuhäuser-Klaus A, Pretsch W, Favor J, et al. A 76-bp deletion in the Mip gene causes autosomal dominant cataract in Hfi mice. Genomics. 2001;74(3):313–319. doi: 10.1006/geno.2001.6509. [DOI] [PubMed] [Google Scholar]
- 51.Shiels A, Bassnett S. Mutations in the founder of the MIP gene family underlie cataract development in the mouse. Nat Genet. 1996;12(2):212–215. doi: 10.1038/ng0296-212. [DOI] [PubMed] [Google Scholar]
- 52.Shiels A, Bassnett S, Varadaraj K, Mathias R, Al-Ghoul K, et al. Optical dysfunction of the crystalline lens in aquaporin-0-deficient mice. Physiol Genomics. 2001;7(2):179–186. doi: 10.1152/physiolgenomics.00078.2001. [DOI] [PubMed] [Google Scholar]
- 53.Wang XY, Ohtaka-Maruyama C, Pisano MM, Jaworski CJ, Chepelinsky AB. Isolation and characterization of the 5′-flanking sequence of the human ocular lens MIP gene. Gene. 1995;167(1–2):321–325. doi: 10.1016/0378-1119(95)00637-0. [DOI] [PubMed] [Google Scholar]
- 54.Ohtaka-Maruyama C, Wang X, Ge H, Chepelinsky AB. Overlapping Sp1 and AP2 binding sites in a promoter element of the lens-specific MIP gene. Nucleic Acids Res. 1998;26(2):407–414. doi: 10.1093/nar/26.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.West-Mays JA, Zhang J, Nottoli T, Hagopian-Donaldson S, Libby D, et al. AP-2alpha transcription factor is required for early morphogenesis of the lens vesicle. Dev Biol. 1999;206(1):46–62. doi: 10.1006/dbio.1998.9132. [DOI] [PubMed] [Google Scholar]
- 56.West-Mays JA, Coyle BM, Piatigorsky J, Papagiotas S, Libby D. Ectopic expression of AP-2alpha transcription factor in the lens disrupts fiber cell differentiation. Dev Biol. 2002;245(1):13–27. doi: 10.1006/dbio.2002.0624. [DOI] [PubMed] [Google Scholar]
- 57.Golestaneh N, Fan J, Fariss RN, Lo WK, Zelenka PS, et al. Lens major intrinsic protein (MIP)/aquaporin 0 expression in rat lens epithelia explants requires fibroblast growth factor-induced ERK and JNK signaling. J Biol Chem. 2004;279(30):31813–31822. doi: 10.1074/jbc.M403473200. [DOI] [PubMed] [Google Scholar]
- 58.Francis P, Chung JJ, Yasui M, Berry V, Moore A, et al. Functional impairment of lens aquaporin in two families with dominantly inherited cataracts. Hum Mol Genet. 2000;9(15):2329–2334. doi: 10.1093/oxfordjournals.hmg.a018925. [DOI] [PubMed] [Google Scholar]
- 59.Francis P, Berry V, Bhattacharya S, Moore A. Congenital progressive polymorphic cataract caused by a mutation in the major intrinsic protein of the lens, MIP (AQP0). Br J Ophthalmol. 2000;84(12):1376–1379. doi: 10.1136/bjo.84.12.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Geyer DD, Spence MA, Johannes M, Flodman P, Clancy KP, et al. Novel single-base deletional mutation in major intrinsic protein (MIP) in autosomal dominant cataract. Am J Ophthalmol. 2006;141(4):761–763. doi: 10.1016/j.ajo.2005.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gu F, Zhai H, Li D, Zhao L, Li C, et al. A novel mutation in major intrinsic protein of the lens gene (MIP) underlies autosomal dominant cataract in a chinese family. Mol Vis. 2007;13:1651–1656. [PubMed] [Google Scholar]
- 62.Jiang J, Jin C, Wang W, Tang X, Shentu X, et al. Identification of a novel splice-site mutation in MIP in a chinese congenital cataract family. Mol Vis. 2009;15:38–44. [PMC free article] [PubMed] [Google Scholar]
- 63.Lin H, Hejtmancik JF, Qi Y. A substitution of arginine to lysine at the COOH-terminus of MIP caused a different binocular phenotype in a congenital cataract family. Mol Vis. 2007;13:1822–1827. [PubMed] [Google Scholar]
- 64.Wang KJ, Li SS, Yun B, Ma WX, Jiang TG, et al. A novel mutation in MIP associated with congenital nuclear cataract in a chinese family. Mol Vis. 2011;17:70–77. [PMC free article] [PubMed] [Google Scholar]
- 65.Wang W, Jiang J, Zhu Y, Li J, Jin C, et al. A novel mutation in the major intrinsic protein (MIP) associated with autosomal dominant congenital cataracts in a Chinese family. Mol Vis. 2010;16:534–539. [PMC free article] [PubMed] [Google Scholar]