Abstract
Antibacterial polymers have potential as pharmaceuticals and as coatings for implantation devices. The design of these materials will be optimized when we have a complete understanding of the structural features that impart activity toward target organisms and those that are benign with respect to the mammalian host. In this work, four series of polymers in which cationic and hydrophobic groups were distributed along the backbone were tested against six different bacterial species (both Gram positive and Gram negative) and for host cytotoxicities (red blood cell lysis). The most effective of the polymers studied are regularly spaced, featuring a 6–8 carbon stretch along the backbone between side chains that present positively charged groups. They cause potassium efflux, disorder the bacterial cytoplasmic membrane, and disrupt the membrane potential. These polymers, available from alternating ring opening metathesis polymerization (AROMP), offer proof of principle for the importance of regular spacing in antibacterial polymers and for the synthesis of additional functional materials based on regularly spaced scaffolds.
In the more than 80 years since Fleming discovered penicillin, humans have expended a great deal of effort in the search for, optimization of, and testing of new antibiotics. During the same period, new pathogens have appeared and antibiotic-resistant strains have evolved. The war against the microbes is far from over (1).
Of particular concern in the western world is the incidence of hospital-acquired infections and the drug resistance of many of the causative phenotypes (2, 3). The appearance of methicillin-resistant Staphylococcus aureus (4) and resistant strains of pneumonia and tuberculosis (5, 6) as community-acquired infections have served to focus public attention on this growing health problem, leading The Infectious Diseases Society of America to call for renewed efforts to develop antimicrobial therapies (7).
One attractive avenue for new antibiotic development is the exploitation or development of “host-defense” antimicrobial peptides (AMPs). Eukaryotes produce these small peptides (about 12 to 80 amino acid residues) as part of their innate immune response against pathogen infection (8–10). Some AMPs are preorganized so that they are amphipathic; i.e. cationic residues are segregated from hydrophobic residues onto opposing faces of the peptide (11). Others, it appears, are induced to adopt an amphipathic topology by contact with cell membranes (or, in laboratory experiments, with micellar surfaces).
The inherent or induced amphipathic conformation of an AMP facilitates binding to and insertion into lipid bilayers. Subsequent disruption of the cytoplasmic membrane (8) can lead to bacterial death. Alternatively, some antimicrobial peptides are thought to cause cell death by additional mechanisms (11) including membrane depolarization, binding to cytoplasmic components (12, 13), and inhibition of cell wall synthesis (14). Regardless of the killing mechanism, the ability to interact with the bacterial membrane is requisite for activity.
Development of resistance to AMPs is infrequent because the mechanisms of action are varied and not easily eluded. Although bacteria can develop resistance to toxicity, for example by increases in protease levels, such evolutionary processes are relatively slow (15, 16) and, at least in the cases studied; they lead to organisms that are not robust.
Mimicry of antimicrobial peptides is an attractive approach to the development of synthetic antibiotic compounds that may be used as coatings or as pharmaceuticals (17). Synthetic mimics may be designed to have improved chemical and biochemical stability, for example, by increased resistance to proteases. The first generation of mimics, based on synthetic peptides, β-peptides, and peptoids constructed by step-wise chain elongation have been examined for applications as antibacterials and imaging agents (18).
A significant advantage of synthetic polymers is that they can be efficiently and inexpensively prepared in a single chain propagation. The use of polymeric mimics based on hydrocarbon backbones generated by ring opening metathesis polymerization (ROMP) (19) is particularly appealing because of the efficiency with which they may be produced in large-scale, one-pot reactions and the level of functionality that may be introduced. The potential of some of these second generation polymers as sensors (20) and as lung surfactants (21) has already been demonstrated.
Although it is now clear that a number of immunodefensive AMPs operate primarily by mechanisms other than amphiphilic membrane disruption, strategies for inventing antimicrobial polymers that interact with membranes are still valid. However, the prediction of secondary structure of hydrocarbon-based polymers, unlike that of small polypeptides and foldamers, is not well understood. The fact that the regio- and stereochemistry in polymers is generally not controlled further complicates matters.
We recognized that the regular polymers available from alternating ring-opening polymerization (AROMP) (22) could provide hydrocarbon polymers with a linear polymer backbone and regularly arrayed cationic and lipophilic residues. The hydrophilicity of the polymers could be tuned by modifying the substituents on the cyclobutene and/or cyclohexene monomers. Furthermore, polymers that present only one functional group (homopolymers) would be available from ROMP for direct comparison of the functional group spacing requirements.
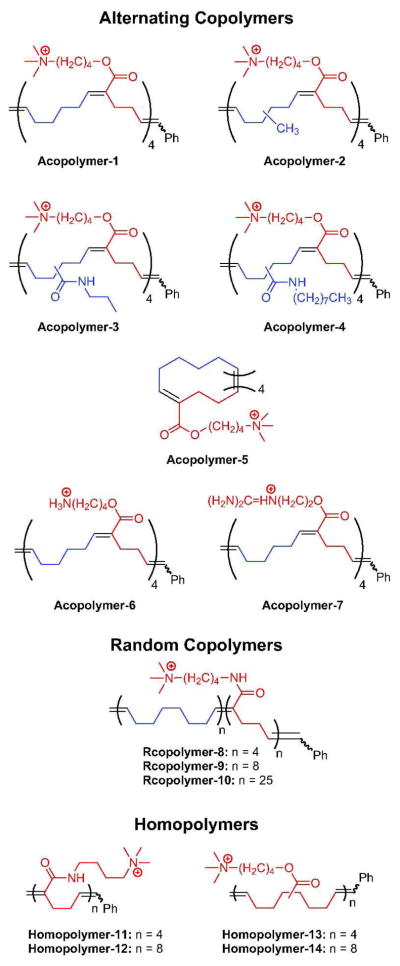
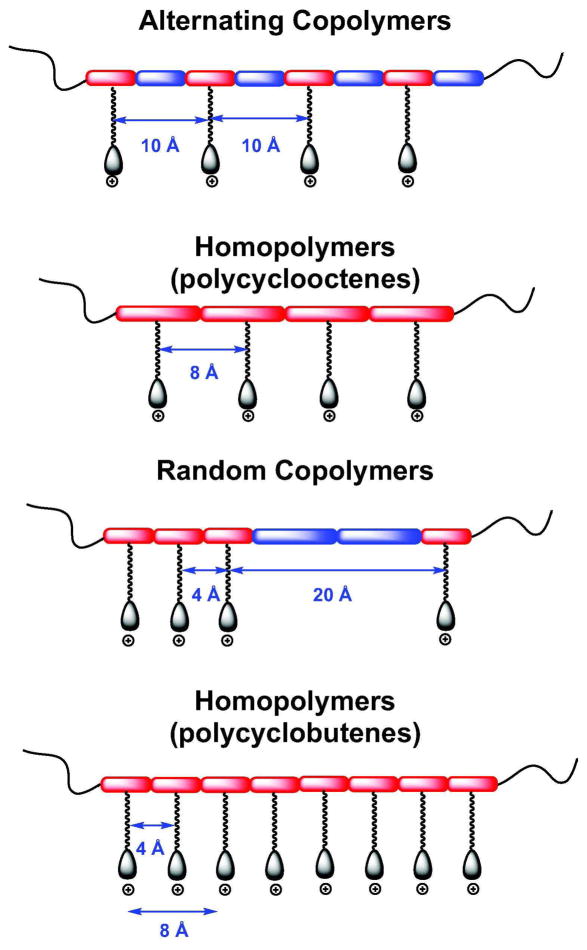
Here we report the synthesis and antibacterial evaluation of a series of alternating copolymers, random copolymers and homopolymers in which cationic side chains are interspersed with lipophilic side chains or with unsubstituted backbone regions in (Figure 1). In the regularly spaced polymers, the cationic side chains are spaced at 4, 8, and 10 carbon intervals along the backbone. Experiments with additional related polymers tested the significance of the nature of the charged group and the distance between the charge-bearing side chains.
Figure 1.
Structures of amphiphilic polymers. The reported n is based on experimentally determined number-average molecular weights (Table S5).
Effective antibacterial peptides and synthetic mimics typically have 2–9 cationic charges (23, 24). However, an increasing charge density can contribute to hemolysis (23). We prepared polymers containing four to eight, and in one case up to 25, cationic groups for initial evaluation. Screened against six species of Gram-positive and Gram-negative bacteria, they showed good in vitro antibacterial activity against all strains, causing membrane depolarization, lysis, and leakage of cellular contents as has been observed with other synthetic polymer mimics (24). Moreover, they had low host cell cytotoxicities as measured by erythrocyte lysis. In comparison to random copolymers of similar backbone, size, and functionality but different or variable spacing, polymers with regular 8 to 10 Å backbone spacing are more effective antibacterial agents.
RESULTS AND DISCUSSION
Synthesis of Antibacterial Polymers
The polymers tested (alternating copolymers, designated Acopolymers; random copolymers, designated Rcopolymers; and Homopolymers, Figure 1) bear trialkylammonium, ammonium, and guanidinium substitutents. All were obtained by schemes based on ROMP.
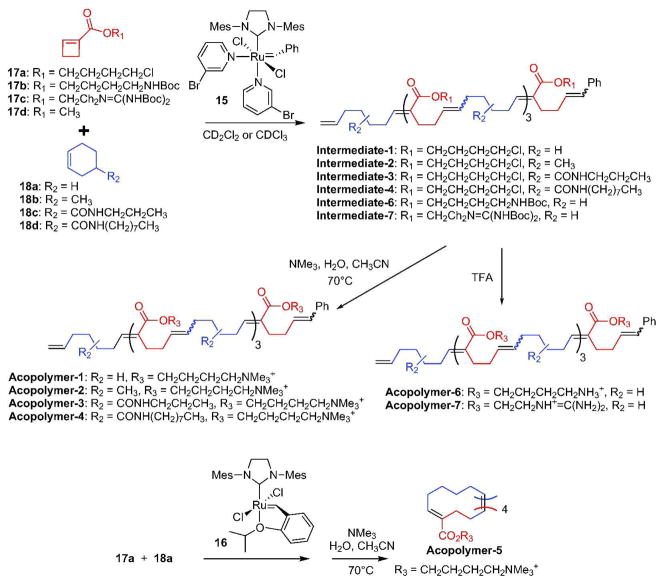
Polymers-1, -2, -3, -4, -8, -9, -10, -11, -12, -13, and -14 were prepared from the corresponding chloro-substituted polymers (see below). The cyclic Acopolymer-5 was obtained from the corresponding cyclic chloro-substituted polymer that was prepared with catalyst 16 as described in the literature (25).
Copolymers Intermediates-1, -2, -3, and -4 were prepared from cyclobutene ester 17a and cyclohexenes 18a-18d by AROMP methodology with high monomer conversion yields (90–96%) as shown in Scheme 1 and Song et al. (22, 26) (Table S1, Figure S1). Nucleophilic substitution with trimethylamine provided the corresponding quaternary ammonium cations. 1H-NMR spectroscopic analysis revealed that the degree of substitution was greater than 99% (Figure S2).
Scheme 1.
Synthetic routes for alternating copolymers. The reported n is based on experimentally determined number-average molecular weights (Table S5).
Acopolymer-6 was obtained by AROMP of cyclobutene ester 17b and cyclohexene (18a) followed by deprotection of the primary amine group in Intermediate-6 with trifluoroacetic acid. Likewise, Acopolymer-7 was obtained by AROMP of cyclobutene ester 17c with cyclohexene and deprotection of the guanidine group in Intermediate-7 (Scheme 1, Figure S3).
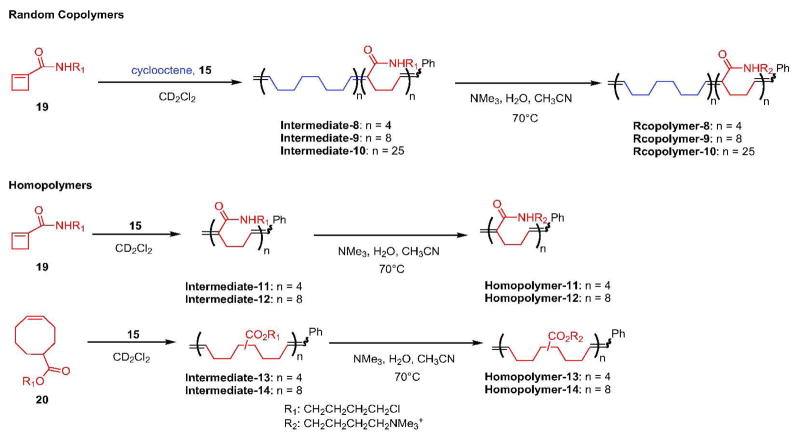
Random copolymers Intermediate-8, -9, and -10 were prepared from 1-cyclobutene amide 19 and cyclooctene by ROMP (Scheme 2, Table S2) with greater than 99% conversion. The random nature of their polymer backbones was confirmed by 1H-NMR spectroscopy (Figure S4) and 1H-1H gCOSY spectroscopy (Figure S5). In the 1H-1H gCOSY spectrum of Intermediate-10, couplings between a trisubstituted olefin proton and its adjacent CH2 protons or between a disubstituted olefin proton and its neighboring CH2 protons corresponding to the AA, BB and AB repeats were observed. The multiple couplings confirm the random polymer backbone structure of Intermediate-10 (Scheme 2).
Scheme 2.
Synthetic routes for random copolymers and homopolymers. The reported n is based on experimentally determined number-average molecular weights (Table S5).
Homopolymers Intermediates-11 and -12 were synthesized by ROMP of 1-substituted cyclobutene amide 19 in greater than 90% conversion (Table S3) (26, 27). The stereo- and regioregularity of the polymer backbones were confirmed by 1H-NMR analysis (Figure S6).
Homopolymers Intermediates-13 and -14 were prepared by ROMP of 5-substituted cyclooctene 20. Within 4 h, more than 99% conversion was achieved (Table S4). The structures of Intermediates-13 and -14 were confirmed by 1H-NMR analysis (Figure S7).
Molecular Weight Characterization
The molecular weights and polydispersity indices (PDI) of the intermediate polymers were characterized by gel permeation chromatography (GPC) with polystyrene standards (Table S5). The alternating polymers exhibited lower molecular weights than calculated on the basis of degree of polymerization, and their PDIs were high (1.3–2.4) (22). We attribute these properties to back-biting during the AROMP. The random copolymers (Rcopolymers) and Homopolymers exhibited average molecular weights close to the values expected.
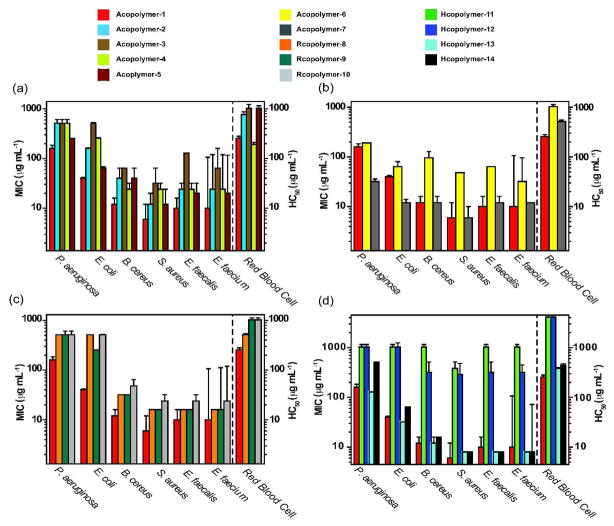
Antibacterial Activities and Selectivities
Medicinally useful polymers must be both potent and selective. We tested our synthetic polymers against both Gram positive and Gram negative bacterial strains and we also tested them for mammalian toxicity. We measured the minimum inhibitory concentration (MIC) as the lowest concentration required to completely inhibit bacterial growth. Figure 2 and Table S6 show the antibacterial activities of Polymers 1–14 against Pseudomonas aeruginosa ATCC 27853 (P. aeruginosa), Escherichia coli ATCC 25922 (E. coli), Bacillus cereus ATCC 10987 (B. cereus), Staphylococcus aureus ATCC 25923 (S. aureus), Enterococcus faecalis ATCC 19433 (E. faecalis), and Enterococcus faecium ATCC 19434 (E. faecium) (Figure 2). In addition, the polymer concentrations at which 50% lysis of sheep red blood cells occurred (HC50) were determined. The selectivity of each polymer is defined as the ratio of the HC50 to the MIC.
Figure 2.
MIC and HC50 data for amphiphilic polymers. a) Alternating copolymers with varying hydrophobic spacers: Acopolymers-1–5. b) Alternating copolymers with different cationic groups: Acopolymer-1, -6; and -7. c) Acopolymer-1 compared to random copolymers Rcopolymers-8–10. d) Acopolymer-1 compared to homopolymers: Homopolymers-11–14. The MIC data are the average of results from two independent preparations of each polymer performed in triplicate. Error bar: standard error of the mean (SEM).
As a measure of the relative hydrophobicities of Polymers 1–5 and 8–14, we calculated the logP values of the smallest repeating unit containing a cationic group with a fragment approach (CLogP) (28). We calculated the difference in CLogP relative to Acopolymer-1 for the ring-opened AB dyad for each copolymer and that of the ring-opened A monomer for each homopolymer. The CLogPs reported in Table S6 are relative to Acopolymer-1 that was arbitrarily set to 0.
First, we adjusted the lipophilicity of the alternating polymers by utilizing cyclohexene monomers that bear hydrophobic groups, i.e., methyl, n-propyl carboxamide or n-octyl carboxamide, at the 4-carbon. Varying the hydrophobicity from Acopolymer-1 to Acopolymer-4 did not increase the antibacterial activity (Figure 2A). The more hydrophobic polymers Acopolymers-2 and -4 had lower antibacterial activities than those of Acopolymer-1. Acopolymer-3 also had lower antibacterial activities than Acopolymer-1 despite their similar estimated hydrophobicities. Moreover, increasing hydrophobicity did not increase erythrocyte lysis as has been observed with other mimics (29). Thus, of the four polymers examined, Acopolymer-1 has the best microstructural level hydrophobicity to effectively kill bacteria. Its effectiveness was equivalent to ~10-fold better (on a molar or weight basis) than the naturally occurring magainin antimicrobial peptide when tested against the same strains (30). Acopolymer-5 exhibited much lower antibacterial activities than Acopolymer-1. The cyclic polymers formed by our method are estimated to be small, i.e., n = 3–4, on the basis of mass spectra and gel permeation chromatography measurements (25). Therefore, the curvature in this polymer is quite high, and the conformational flexibility is low. These features appear to preclude antibacterial activity.
We also examined the efficacy of additional cationic groups by testing Acopolymers-6, and -7 for antibacterial activity. Acopolymers-1 and -7 were effective against all six bacteria; both were potent against S. aureus and E. faecalis and E. faecium. Acopolymer-6 was less effective than either Acopolymer-1 or -7. In a neutral buffer solution, the primary amine groups in Acopolymer-6 are not completely protonated, whereas Acopolymers-1 and -7 are entirely positively charged at physiologic pH. Thus, these polymers behave like many other antibacterial peptide mimics in which positive charges enable attachment to anionic bacterial membranes. The selectivities of Acopolymers-1 and -7 for bacteria were as high as 40–80 in the case of S. aureus (Figure 2B, Table S6). However, they showed little to no selectivity for Pseudomonas aeruginosa which has a low permeability outer membrane compared to other bacteria as well as active efflux pumps which eliminate molecules that penetrate the cell membrane (31). Although Acopolymer-7 scored slightly better than Acopolymer-1 in the antibacterial tests, we chose the trimethyl ammonium rather than guanidinium moiety for the study of additional analogs, because it is easily introduced.
The antibacterial activities of the random copolymers (Rcopolymers-8, -9, and -10) against E. coli, B. cereus, and S. aureus were consistently (p < 0.05) 2 to 6-fold lower than those of alternating copolymers Acopolymers-1 and -7 (Figure 2C). Thus it appears that the precise spatial separation of the quaternary ammonium groups is critical for activity in these strains. Local regions within the polymer backbone may achieve optimal cation spacing and hydrophobicity, for example upon formation of an AB dyad. The AB dyad hydrophobicities of Rcopolymers-8, -9, and -10 correspond closely to that of the Acopolymer-1 dyad (Table S6). In contrast, an Rcopolymer AA dyad results in a closer spacing of cations, a lower hydrophobicity, and potentially, a lower antibacterial efficacy (see below). The requirement for the spacing and hydrophobicity of the AB dyad is consistent with the high antibacterial efficiencies (generally within an order of magnitude of Acopolymer-1 for similar strains) observed with rigid aromatic backbone arylamide foldamers that precisely space cationic group linkers 10 Å apart (32). Likewise, longer spans in oligomeric acyl lysine (OAK) peptides are more optimal than shorter spans (33).
Homopolymers-11, -12, -13, and -14 were prepared in order to explore further the correlation between spacer distance and antibacterial activity. Acopolymer-1 has an approximately 10-Å backbone distance between charged side chains. Homopolymers-11 and -12 have a cation backbone spacing of ~4 Å and Homopolymers-13 and -14 have a cation backbone spacing of ~8 Å. These distances are approximate given the number of degrees of freedom and the variable alkene stereochemistry in the all-carbon backbones of our polymers.
Neither Homopolymer-11 nor -12 had any antibacterial activity (Figure 2D), suggesting that their charge density is too high for efficient bacterial killing. Homopolymers-13 and -14 had identical activities (Figure 2D) and selectivities; increasing the average total number of cationic charges from 4 to 8 did not improve efficacy on a weight basis. The activities and selectivities of Homopolymers-13 and -14 were comparable to those of Acopolymer-1. We conclude that the backbone distance between cation side chains required for antibacterial activity is greater than 4 Å and that a range of 8 to 10 Å is the minimum spacing required. Moreover, the spacer must be hydrophobic as increasing the length of Homopolymer-11 to form Homopolymer-12, that is using the cation monomer as a spacer, did not improve efficacy. This hydrophobic distance requirement is consistent with the cation spacing (> 5 Å) in the naturally occurring AMPs. Gellman and coworkers have observed an analogous effect of charge density in random β-peptide copolymers (34).
Mechanism of Polymer Action
To further establish the relationship between structure and antibacterial properties, we selected Acopolymer-1, Rcopolymer-8, Homopolymer-11, and Homopolymer-13 for additional investigation. On average, each of these polymers has 4 cations presented along polymer backbones that range in length from 25 to 30 Å. Therefore, study of their mechanisms of action reports on the influence of spacing along nearly identical backbones (Figure 3).
Figure 3.
Structure-activity relationship for amphiphilic polymers. Polymers containing longer spacing exhibit better antibacterial activities.
Depolarization assay
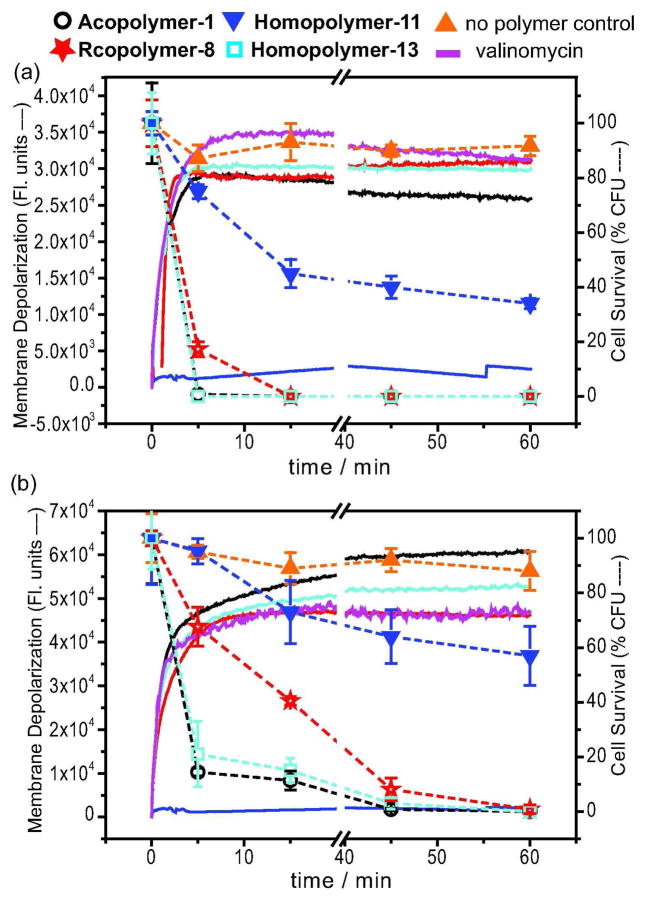
A potential gradient (Δ⇁) of approximately 140 mV at neutral pH is present across a typical bacterial membrane. Disruption of the cytoplasmic membrane can depolarize the membrane potential gradient and contribute to bacterial death (35–38). We assessed whether the bacterial plasma membranes were depolarized by mimic polymers using a membrane potential sensitive dye diSC35 (3,3'-dipropylthiadicarbocyanine iodide). DiSC35 intercalates into the cytoplasmic membrane of energized cells under the effect of Δ⇁, and its fluorescence is quenched. Upon disruption of the potential, diSC35 is displaced into the buffer solution, and an increase in fluorescence is observed. Coincident with the membrane depolarization assay, we measured cell viability by counting remaining colony forming units (CFU) to assess the relationship between disruption of membrane potential and loss of viability.
We compared the effect of adding Acopolymer-1 (MIC = 40 μg mL−1 & 6 μg mL−1), Rcopolymer-8 (MIC > 256μg mL−1 & 16 μg mL−1), and Homopolymer-13 (MIC = 32 μg mL−1 & 8μg mL−1) to E. coli and S. aureus at fixed concentrations of 16 μg mL−1 and 4 μg mL−1, respectively. Maximal release of diSC35 after the addition of Acopolymer-1, Rcopolymer-8, and Homopolymer-13 to both E. coli and S. aureus was observed within 10 min. This timescale is similar to that associated with the rate of depolarization induced by the antibiotic valinomycin (Figure 4).
Figure 4.
Relationship between cytoplasmic membrane depolarization, as assessed by the diSC35 assay, and cell viability, as measured by the counting of CFU at the same time as the membrane depolarization assay for a) E. coli and b) S. aureus. Solid curves represent data obtained from the diSC35 assay, and dashed curves represent data from the cell viability assay. Acopolymer-1 (16 μg mL−1 for E. coli; 4 μg/mL for S. aureus); Rcopolymer-8 (16 μg mL−1 for E. coli; 4 μg/mL for S. aureus); Homopolymer-11 (16 μg mL−1 for E. coli: 4 μg/mL for S. aureus); Homopolymer-13 (16 μg mL−1 for E. coli; 4 μg mL−1 for S. aureus); The negative control was buffer with no polymer. The positive control for the diSC35 assay was valinomycin (10 μg mL−1). The data shown are the average of triplicate measurements for two independent replicates. Error bar: standard error of the mean (SEM).
After 15 minutes, E. coli cells treated with 16 μg mL−1 Acopolymer-1, Rcopolymer-8, or Homopolymer-13 were no longer viable. Cell viability decreases in parallel with membrane depolarization for E. coli suggesting that cell killing is closely correlated with membrane potential disruption. Loss of S. aureus viability upon addition of 4 μg mL−1 Acopolymer-1, Rcopolymer-8, or Homopolymer-13 was slower than observed for E. coli. After 15 minutes, cell viability was only reduced to 11%, 41% and 15% for Acopolymer-1, Rcopolymer-8, and Homopolymer-13 respectively. The incomplete decrease in cell viability for S. aureus demonstrates that membrane depolarization is not sufficient to kill the cells and suggests that other slower mechanisms, e.g., pore formation or intracellular binding, may be involved in triggering cell death. Acopolymer-1 killed S. aureus more rapidly than Rcopolymer-8 (Figure 4B), and the faster killing kinetics of Acopolymer-1 correlate with its lower MIC.
Over the course of one hour very little (<5%) membrane depolarization occurred upon addition of Homopolymer-11 to either E. coli (16 μg mL−1) or S. aureus (4 μg mL−1), as expected on the basis of its high MIC (Figure 4). Furthermore, approximately 40–70% of bacteria survived after 15 minutes. The data suggest that cell killing by Homopolymer-11 is very slow.
Membrane Disruption as a Function of Lipid Content
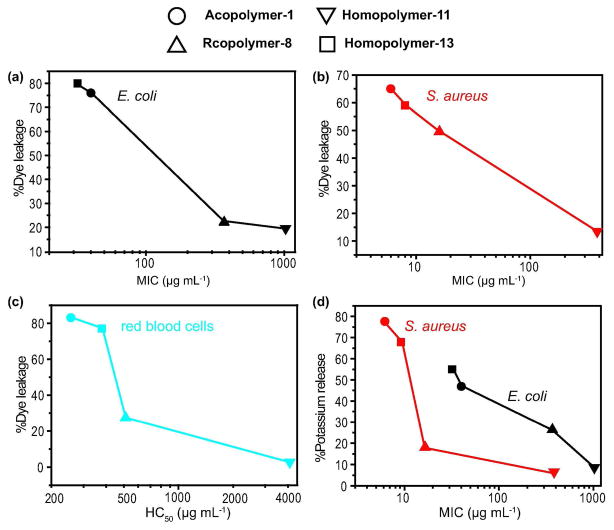
Next, we examined whether membrane disruption by the polymers is specific for the types of lipids present in E. coli and S. aureus versus those in red blood cell membranes. We prepared lipid vesicles composed of a) POPE/POPG (3/1), b) cardiolipin (CL), and c) DOPC that encapsulated calcein dye to mimic E. coli, S. aureus, and red blood cells, respectively. Percent dye leakage was measured relative to total vesicle lysis that was induced with 20% Triton X-100 (Figure 5A–C, Table S7). A comparison of the level of lysis reveals that Acopolymer-1, Rcopolymer-8, and Homopolymer-13 are not specific for the negatively charged POPE/POPG and cardiolipin vesicles over the neutral DOPC vesicles. Therefore, the selectivity of these polymers for bacteria is due to more than a simple electrostatic interaction with the anionic bacterial plasma membrane. The low levels of vesicle lysis observed with Homopolymer-11 support the idea that at least an 8–10 Å backbone spacing between cation presenting groups is critical for plasma membrane interaction.
Figure 5.
a) Percent dye leakage from E. coli model membrane vesicles: POPE/POPG = 3/1, [lipid] = 4.5 μM, [polymer] = 4 μg mL−1 vs E. coli MIC. b) Percent dye leakage from S. aureus model membrane vesicles: [CL] = 4.5 μM, [polymer] = 4 μg mL−1 vs S. auerus MIC. c) Percent dye leakage from red blood cell model membrane vesicles: [DOPC] = 4.5 μM, [polymer] = 1 μg mL−1 vs HC50 data. d) Percent potassium release vs MIC. The data shown are the average of triplicate measurements for two independent replicates. Standard error of the mean (SEM) are reported in Table S7.
Release of potassium from the bacterial cell occurs upon disruption of the membrane potential (39). We investigated the levels of potassium release incurred by our polymers using potassium ion sensitive fluorophore, potassium-binding benzofuran isophthalate-AM (PBFI-AM). Percent potassium release was measured 16 minutes after the addition of polymer and reported relative to valinomycin (100%) (Table S8). Acopolymer-1 and Homopolymer-13 were the most effective at causing a high level of potassium release (> 50%) consistent with their lower MICs (Figure 5D). Thus, the ability of the polymers to disrupt membrane potential correlates with potassium release.
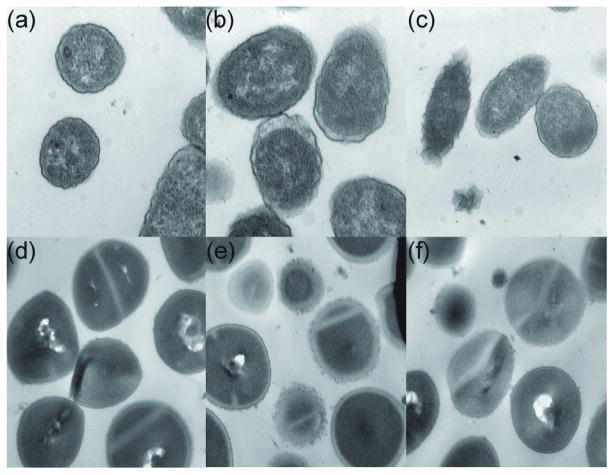
To determine to what extent Acopolymer-1 disrupts bacterial lipid membranes, thin-section TEM was employed (Figure 6). The cell surface of E. coli and S. aureus bacteria treated with Acopolymer-1 (1× MIC or 10× MIC) exhibited severely damaged cell surfaces containing membrane blebs (Figure 6B, C, E and F) as compared to untreated bacteria (Figure 6A and D). Similar levels of membrane disruption are observed with antibacterial peptide-treated bacteria (40–42).
Figure 6.
Cross-section TEM images of bacteria. Red circles label bacteria with disrupted membranes. a) E. coli without treating with Acopolymer-1; b) E. coli treated with 1×MIC of Acopolymer-1; c) E. coli treated with 10×MIC of Acopolymer-1; d) S. aureus without treating with Acopolymer-1; e) S. aureus treated with 1×MIC of Acopolymer-1; f) S. aureus treated with 10×MIC of Acopolymer-1.
CONCLUSION
Like antimicrobial peptides and their reported mimics (43), the AROMP and ROMP polymers described in this work caused rapid membrane depolarization and potassium release, followed by cell death. We conclude that an ordered microstructure is required for optimal antibacterial activity even in the context of a polymer in which the backbone may have an irregular conformation. Moreover, the hydrophobic spacer distance along the backbone between neighboring ammonium-presenting groups must be greater than 4 Å and at least 8–10 Å. In our study, the relative importance of ester versus amide attachments, distribution of alkenes within the backbone, and longer spacing between side-chains could not be addressed with the polymer chemistry available. In addition to the balance of spacer distance, hydrophobicity, and charge studied here, further adjustment of these additional parameters may improve the attachment of the polymers to the bacterial membrane or to an unidentified cellular receptor.
Many types of polymers, dendrimers, and rigid, dicationic small molecules have been employed as antimicrobial peptide mimics (24, 29, 34, 44–55). Each polymer has a different backbone and consequently different conformational preferences and different partitioning properties. Given the multitude of transforming applications envisioned for antimicrobial polymers, we expect that materials based on different scaffolds may be optimized for different functions. The polymers prepared by ruthenium-catalyzed AROMP provide the necessary spacing of cations for antibacterials and offer the advantage of facile incorporation of additional functionality at regular intervals. They are a new and powerful addition to the pharmacopeia of scaffolds.
METHODS
Detailed Experimental Procedures are available in the Supporting Information
General Procedure for NMR AROMP Reactions
An NMR tube was evacuated under high vacuum for 15 min, and then was purged with Ar for another 15 min. Under an Ar atmosphere, a solution of monomer A (1-cyclobutenecarboxylate ester) in CD2Cl2 (300 μL) was added to the NMR tube. Then a solution of 15 in CD2Cl2 (300 μL) was added to the NMR tube. After complete mixing of the solution, the NMR tube was spun for 4–30 min at 25 °C in the NMR spectrometer (400, 500 or 600 MHz) until the precatalyst had reacted. Then monomer B (cyclohexene derivatives) in CD2Cl2 (300 μL) was added to the NMR tube. After all of monomer A was converted, the reaction was quenched with ethylvinyl ether (50 μL) and was stirred for 1 h. The monomer feed ratio is 25:1, and the final degree of polymerization (DP) is approximately 4.
General Procedure for NMR Tube Random Copolymer ROMP Reactions
An NMR tube was evacuated under high vacuum for 15 min, and then was purged with Ar for another 15 min. Under an Ar atmosphere, a solution of monomer A (1-cyclobutenecarboxamide) and monomer B (cyclooctene) in CD2Cl2 (300 μL) was added to the NMR tube. Then a solution of 15 in CD2Cl2 (300 μL) was added to the NMR tube, and the NMR tube was placed into the 400 MHz, 500 MHz or 600 MHz NMR spectrometer, and the reaction was monitored for several hours at 25 ºC. After all the monomers had been consumed, the reaction was quenched with ethylvinyl ether (50 μL) and was stirred for 1 h.
General Procedure for NMR Tube Homopolymer ROMP Reactions
An NMR tube was evacuated for 15 min, and then was purged with Ar for another 15 min. Under an Ar atmosphere, a solution of monomer in CD2Cl2 (300 μL) was added to the NMR tube. A solution of 15 in CD2Cl2 (300 μL) was added to the NMR tube. After complete mixing of the solution, the NMR tube was placed into the 400 MHz, 500 MHz or 600 MHz NMR spectrometer, and the reaction was monitored for several hours at 25 °C until almost all of the monomer had been consumed. Then the reaction was quenched with ethylvinyl ether (50 μL), and was monitored for an additional 1 h.
Polydispersity Index (PDI) determination
Polymers (before flash column chromatography purification) were analyzed by gel permeation chromatography.
ClogP calculation
CLogP’s of quaternary ammonium polymers were calculated according to Crippen’s fragmentation (28) using ChemDraw Ultra 12.0 for the ring-opened AB dyads (copolymers) or A monomers (homopolymer).
MIC and Hemolysis Assays
The minimal inhibitory concentration (MIC) for the polymers towards six bacterial species were determined using the broth microdilution method as described by protocol M7-A7 of the Clinical and Laboratory Standards Institute (56).
Dye Leakage Experiments of Lipid Vesicles
Dye leakage experiments were performed according to the literature (46, 57).
Potassium Release Assay
Potassium ion release assays were performed by following the method of Silverman et al. (58)
Thin-section TEM of Bacteria
Sections were then counterstained with uranyl acetate and lead citrate and viewed with a FEI Tecnai12 BioTwinG2 electron microscope. Digital images were acquired with an AMT XR-60 CCD Digital Camera system.
Supplementary Material
Acknowledgments
This research was supported by NIH grants R01HD38519 (N.S.), R01GM74776 (K.A.P), S10RR021008 (N.S.), NIH/NCRR S10 RR025072 (Orbitrap), NYSTAR Award (FDPC040076, NS), and NSF grant CHE0131146 (NMR). We are grateful to J. P. Schneider (National Cancer Institute at Frederick) for stimulating discussions; we thank J. Marecek (Stony Brook University) for his assistance with NMR spectroscopy and S. Van Horn (Stony Brook University) for her help with the thin-section TEM imaging.
Footnotes
Supporting Information Available: Spectra, additional data, and detailed experimental procedures. This material is free of charge via the Internet at http://pubs.acs.org.
References
- 1.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 2.Opar A. Bad bugs need more drugs. Nat Rev Drug Discov. 2007;6:943–944. [Google Scholar]
- 3.Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010;362:1804–1813. doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burman WJ. Rip Van Winkle wakes up: development of tuberculosis treatment in the 21st century. Clin Infect Dis. 2010;50(Suppl 3):S165–172. doi: 10.1086/651487. [DOI] [PubMed] [Google Scholar]
- 6.Haydel SE. Extensively drug - resistant tuberculosis : a sign of the times and an impetus for antimicrobial discovery. Pharmaceuticals. 2010;3:2268–2290. doi: 10.3390/ph3072268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, Bartlett JG, Edwards J., Jr The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis. 2008;46:155–164. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- 8.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 9.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 10.Easton DM, Nijnik A, Mayer ML, Hancock RE. Potential of immunomodulatory host defense peptides as novel anti-infectives. Trends Biotechnol. 2009;27:582–590. doi: 10.1016/j.tibtech.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yount NY, Bayer AS, Xiong YQ, Yeaman MR. Advances in antimicrobial peptide immunobiology. Pep Sci. 2006;84:435–458. doi: 10.1002/bip.20543. [DOI] [PubMed] [Google Scholar]
- 12.Marchand C, Krajewski K, Lee HF, Antony S, Johnson AA, Amin R, Roller P, Kvaratskhelia M, Pommier Y. Covalent binding of the natural antimicrobial peptide indolicidin to DNA abasic sites. Nucleic Acids Res. 2006;34:5157–5165. doi: 10.1093/nar/gkl667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hale JD, Hancock RE. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev Anti Infect Ther. 2007;5:951–959. doi: 10.1586/14787210.5.6.951. [DOI] [PubMed] [Google Scholar]
- 14.Schneider T, Kruse T, Wimmer R, Wiedemann I, Sass V, Pag U, Jansen A, Nielsen AK, Mygind PH, Raventos DS, Neve S, Ravn B, Bonvin AM, De Maria L, Andersen AS, Gammelgaard LK, Sahl HG, Kristensen HH. Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science. 2010;328:1168–1172. doi: 10.1126/science.1185723. [DOI] [PubMed] [Google Scholar]
- 15.Buckling A, Brockhurst M. Microbiology: RAMP resistance. Nature. 2005;438:170–171. doi: 10.1038/438170a. [DOI] [PubMed] [Google Scholar]
- 16.Perron GG, Zasloff M, Bell G. Experimental evolution of resistance to an antimicrobial peptide. Proc Biol Sci. 2006;273:251–256. doi: 10.1098/rspb.2005.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenawy e-R, Worley SD, Broughton R. The chemistry and applications of antimicrobial polymers: a state-of-the-art review. Biomacromolecules. 2007;8:1359–1384. doi: 10.1021/bm061150q. [DOI] [PubMed] [Google Scholar]
- 18.Marr AK, Gooderham WJ, Hancock RE. Antibacterial peptides for therapeutic use: obstacles and realistic outlook. Curr Opin Pharmacol. 2006;6:468–472. doi: 10.1016/j.coph.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Smith D, Pentzer EB, Nguyen ST. Bioactive and therapeutic ROMP polymers. Polym Rev. 2007;47:419–459. [Google Scholar]
- 20.Hennig A, Gabriel GJ, Tew GN, Matile S. Stimuli-responsive polyguanidino-oxanorbornene membrane transporters as multicomponent sensors in complex matrices. J Am Chem Soc. 2008;130:10338–10344. doi: 10.1021/ja802587j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dohm MT, Mowery BP, Czyzewski AM, Stahl SS, Gellman SH, Barron AE. Biophysical mimicry of lung surfactant protein B by random nylon-3 copolymers. J Am Chem Soc. 2010;132:7957–7967. doi: 10.1021/ja909734n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song A, Parker KA, Sampson NS. Synthesis of copolymers by alternating ROMP (AROMP) J Am Chem Soc. 2009;131:3444–3445. doi: 10.1021/ja809661k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dathe M, Nikolenko H, Meyer J, Beyermann M, Bienert M. Optimization of the antimicrobial activity of magainin peptides by modification of charge. FEBS Lett. 2001;501:146–150. doi: 10.1016/s0014-5793(01)02648-5. [DOI] [PubMed] [Google Scholar]
- 24.Arnt L, Nusslein K, Tew GN. Nonhemolytic abiogenic polymers as antimicrobial peptide mimics. J Polym Sci Pol Chem. 2004;42:3860–3864. [Google Scholar]
- 25.Song A, Parker KA, Sampson NS. Cyclic Alternating ROMP (CAROMP). Rapid Access to Functionalized Cyclic Polymers. Org Lett. 2010;12:3729–3731. doi: 10.1021/ol101432m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song A, Lee JC, Parker KA, Sampson NS. Scope of the ring opening metathesis polymerization (ROMP) reaction of 1-substituted cyclobutenes. J Am Chem Soc. 2010;132:10513–10520. doi: 10.1021/ja1037098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JC, Parker KA, Sampson NS. Amino acid-bearing ROMP polymers with a stereoregular backbone. J Am Chem Soc. 2006;128:4578–4579. doi: 10.1021/ja058801v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghose AK, Crippen GM. Atomic physicochemical parameters for three-dimensional-structure-directed quantitative structure-activity relationships. 2 Modeling dispersive and hydrophobic interactions. J Chem Inf Comput Sci. 1987;27:21–35. doi: 10.1021/ci00053a005. [DOI] [PubMed] [Google Scholar]
- 29.Ilker MF, Nusslein K, Tew GN, Coughlin EB. Tuning the hemolytic and antibacterial activities of amphiphilic polynorbornene derivatives. J Am Chem Soc. 2004;126:15870–15875. doi: 10.1021/ja045664d. [DOI] [PubMed] [Google Scholar]
- 30.Zasloff M, Martin B, Chen HC. Antimicrobial activity of synthetic magainin peptides and several analogues. Proc Natl Acad Sci U S A. 1988;85:910–913. doi: 10.1073/pnas.85.3.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hancock REW. Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative gram-negative bacteria. Clin Infect Dis. 1998;27:S93–S99. doi: 10.1086/514909. [DOI] [PubMed] [Google Scholar]
- 32.Choi S, Isaacs A, Clements D, Liu DH, Kim H, Scott RW, Winkler JD, DeGrado WF. De novo design and in vivo activity of conformationally restrained antimicrobial arylamide foldamers. Proc Natl Acad Sci U S A. 2009;106:6968–6973. doi: 10.1073/pnas.0811818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radzishevsky IS, Rotem S, Bourdetsky D, Navon-Venezia S, Carmeli Y, Mor A. Improved antimicrobial peptides based on acyl-lysine oligomers. Nat Biotechnol. 2007;25:657–659. doi: 10.1038/nbt1309. [DOI] [PubMed] [Google Scholar]
- 34.Mowery BP, Lindner AH, Weisblum B, Stahl SS, Gellman SH. Structure-activity relationships among random Nylon-3 copolymers that mimic antibacterial host-defense peptides. J Am Chem Soc. 2009;131:9735–9745. doi: 10.1021/ja901613g. [DOI] [PubMed] [Google Scholar]
- 35.Zhang LJ, Scott MG, Yan H, Mayer LD, Hancock REW. Interaction of polyphemusin I and structural analogs with bacterial membranes, lipopolysaccharide, and lipid monolayers. Biochemistry. 2000;39:14504–14514. doi: 10.1021/bi0011173. [DOI] [PubMed] [Google Scholar]
- 36.Anderson RC, Hancock REW, Yu PL. Antimicrobial activity and bacterial-membrane interaction of ovine-derived cathelicidins. Antimicrob Agents Chemother. 2004;48:673–676. doi: 10.1128/AAC.48.2.673-676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silvestro L, Weiser JN, Axelsen PH. Antibacterial and antimembrane activities of cecropin A in Escherichia coli. Antimicrob Agents Chemother. 2000;44:602–607. doi: 10.1128/aac.44.3.602-607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang LJ, Dhillon P, Yan H, Farmer S, Hancock REW. Interactions of bacterial cationic peptide antibiotics with outer and cytoplasmic membranes of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2000;44:3317–3321. doi: 10.1128/aac.44.12.3317-3321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herranz C, Cintas LM, Hernandez PE, Moll GN, Driessen AJM. Enterocin P causes potassium ion efflux from Enterococcus faecium T136 cells. Antimicrob Agents Chemother. 2001;45:901–904. doi: 10.1128/AAC.45.3.901-904.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fjell CD, Jenssen H, Hilpert K, Cheung WA, Pante N, Hancock RE, Cherkasov A. Identification of novel antibacterial peptides by chemoinformatics and machine learning. J Med Chem. 2009;52:2006–2015. doi: 10.1021/jm8015365. [DOI] [PubMed] [Google Scholar]
- 41.Malina A, Shai Y. Conjugation of fatty acids with different lengths modulates the antibacterial and antifungal activity of a cationic biologically inactive peptide. Biochem J. 2005;390:695–702. doi: 10.1042/BJ20050520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sawyer JG, Martin NL, Hancock RE. Interaction of macrophage cationic proteins with the outer membrane of Pseudomonas aeruginosa. Infect Immun. 1988;56:693–698. doi: 10.1128/iai.56.3.693-698.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wimley WC. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem Biol. 2010;5:905–917. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamuro Y, Schneider JP, DeGrado WF. De novo design of antibacterial beta-peptides. J Am Chem Soc. 1999;121:12200–12201. [Google Scholar]
- 45.Lienkamp K, Madkour AE, Musante A, Nelson CF, Nusslein K, Tew GN. Antimicrobial polymers prepared by ROMP with unprecedented selectivity: a molecular construction kit approach. J Am Chem Soc. 2008;130:9836–9843. doi: 10.1021/ja801662y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Al-Badri ZM, Som A, Lyon S, Nelson CF, Nusslein K, Tew GN. Investigating the effect of increasing charge density on the hemolytic activity of synthetic antimicrobial polymers. Biomacromolecules. 2008;9:2805–2810. doi: 10.1021/bm800569x. [DOI] [PubMed] [Google Scholar]
- 47.Kuroda K, Caputo GA, DeGrado WF. The role of hydrophobicity in the antimicrobial and hemolytic activities of polymethacrylate derivatives. Chem-Eur J. 2009;15:1123–1133. doi: 10.1002/chem.200801523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuroda K, DeGrado WF. Amphiphilic polymethacrylate derivatives as antimicrobial agents. J Am Chem Soc. 2005;127:4128–4129. doi: 10.1021/ja044205+. [DOI] [PubMed] [Google Scholar]
- 49.Gelman MA, Weisblum B, Lynn DM, Gellman SH. Biocidal activity of polystyrenes that are cationic by virtue of protonation. Org Lett. 2004;6:557–560. doi: 10.1021/ol036341+. [DOI] [PubMed] [Google Scholar]
- 50.Endo Y, Tani T, Kodama M. Antimicrobial activity of tertiary amine covalently bonded to a polystyrene fiber. Appl Environ Microb. 1987;53:2050–2055. doi: 10.1128/aem.53.9.2050-2055.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chongsiriwatana NP, Patch JA, Czyzewski AM, Dohm MT, Ivankin A, Gidalevitz D, Zuckermann RN, Barron AE. Peptoids that mimic the structure, function, and mechanism of helical antimicrobial peptides. Proc Natl Acad Sci U S A. 2008;105:2794–2799. doi: 10.1073/pnas.0708254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li GJ, Shen JR, Zhu YL. Study of pyridinium-type functional polymers. II. Antibacterial activity of soluble pyridinium-type polymers. J Appl Polym Sci. 1998;67:1761–1768. [Google Scholar]
- 53.Som A, Vemparala S, Ivanov I, Tew GN. Synthetic mimics of antimicrobial peptides. Biopolymers. 2008;90:83–93. doi: 10.1002/bip.20970. [DOI] [PubMed] [Google Scholar]
- 54.Chen CZ, Beck-Tan NC, Dhurjati P, van Dyk TK, LaRossa RA, Cooper SL. Quaternary ammonium functionalized poly(propylene imine) dendrimers as effective antimicrobials: structure-activity studies. Biomacromolecules. 2000;1:473–480. doi: 10.1021/bm0055495. [DOI] [PubMed] [Google Scholar]
- 55.Arnt L, Tew GN. New poly(phenyleneethynylene)s with cationic, facially amphiphilic structures. J Am Chem Soc. 2002;124:7664–7665. doi: 10.1021/ja026607s. [DOI] [PubMed] [Google Scholar]
- 56.CLSI. Approved Standard M7-A6. 6. Clinical and Laboratory Standards Institute; Wayne, PA: 2006. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. [Google Scholar]
- 57.Mason AJ, Moussaoui W, Abdelrahman T, Boukhari A, Bertani P, Marquette A, Shooshtarizaheh P, Moulay G, Boehm N, Guerold B, Sawers RJ, Kichler A, Metz-Boutigue MH, Candolfi E, Prevost G, Bechinger B. Structural determinants of antimicrobial and antiplasmodial activity and selectivity in histidine-rich amphipathic cationic peptides. J Biol Chem. 2009;284:119–133. doi: 10.1074/jbc.M806201200. [DOI] [PubMed] [Google Scholar]
- 58.Silverman JA, Perlmutter NG, Shapiro HM. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob Agents Chemother. 2003;47:2538–2544. doi: 10.1128/AAC.47.8.2538-2544.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.