Abstract
Purpose
Bone is the most common metastatic site for prostate cancer, and osseous metastases are the leading cause of morbidity from this disease. Recent autopsy studies prove that 100% of men who die of prostate cancer have bone involvement. Understanding the biology of prostate cancer and its evolution to an incurable androgen independent phenotype requires an understanding of the genetic and cellular alterations that lead to the seeding and proliferation of tumor foci in bone, as well as the microenvironment in which these metastases arise. No intensive studies, however, have been conducted on osseous metastatic tissues from patients with metastatic prostate cancer due to lack of access to such tissues for profiling and other research.
Experimental Design
We demonstrate, for the first time, a reproducible methodology to obtain high quality clinical tumor tissues metastatic to the bone. This technique allowed the procurement of viable metastatic tumor tissue from involved bones in 13 recent autopsies conducted at the University of Michigan, and analyzed the gene expression of these tissues using real time PCR and microarrays.
Results
We present here the discovery of non-ossified bone metastases from multiple patients with advanced prostate cancer and their subsequent characterization and comparison to non-osseous metastases from the same patients
Conclusion
This represents a versatile and practical approach that may be employed to characterize the steps in metastasis and the phenotypic characteristics of osseous metastasis of prostate cancer and to profile RNA, DNA and cDNA from tumor samples metastatic to the bone.
Keywords: Bone marrow, tumor, metastatic prostate cancer
Introduction
Bone metastases are ultimately responsible for the death of patients with androgen independent metastatic prostate cancer (1). Investigators have been unable to study the molecular events underlying bone metastasis in these patients because of the lack of quality tissue available for study. For example, profiling studies have been hindered by the need for microgram quantities of RNA, which are difficult to obtain, as tumor tissues are lodged in the calcified, dense bone infrastructure.
Investigation of metastatic prostate cancer at the phenotypic, molecular and immunohistochemical level has led to discovery of several gene candidates implicated in the etiology, pathogenesis, progression, and evolution of prostate cancer, and has provided unique diagnostic and therapeutic targets. Using tissues harvested at the time of autopsy as a part of our Rapid Autopsy series at the University of Michigan, our group has identified several such crucial markers which offer an insight into the biology of prostate cancer (2–7). Such tissue samples have also successfully produced xenografts (8). These studies thus highlight the value of efficient procurement of metastatic prostate cancer tissues; however, all of these studies utilized tumor primarily from non-osseous metastatic sites (for example, metastatic cancer in liver, lymph nodes, brain, lungs, etc.).
Profiling and research analysis of metastatic prostate cancer tissue in bone (henceforth called osseous metastatic prostate cancer) is bound to give us further understanding into the biology of this disease, as skeletal manifestations of prostate cancer are responsible for the majority of complications, and also mark progression of patient to a ‘pre- lethal’ stage. Though previous studies have aimed at identifying metastatic tumor to bone at the time of autopsies, recovery of tumor tissues in a fashion that allow procurement of RNA in a substantial quantity or high quality from bone metastases has proven difficult (7–10). We present here the recovery of non-ossified bone metastases from multiple patients with advanced prostate cancer and their subsequent characterization and comparison to non-osseous metastatic prostate cancer from the same patients.
Materials and Methods
Rapid Autopsy Bone Tissue Procurement Protocol
The rapid autopsy program at the University of Michigan has been previously described (7). These autopsies are referred to as “rapid” because of the short average interval of <3h between death of the patient and onset of autopsy. The detailed clinical features of the patients included in this study are provided in Table 1.
Table 1.
Clinical characteristics of Rapid Autopsy patients in this cohort.
| Sites of Metastasis at Autopsy | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case No. | Age at Diagnoses | Total Gleason | Treatment | Survival from Hormone Treatment | Survival from 1st Chemotherapy | Survival from Diagnoses (mo) | Bone | Ly Node | Liver | Lungs | Adrenal | Dura | Brain |
| 37 | 60 | 9 | H, C | 39 | 40 | 41 | Yes | No | Yes | No | Yes | Yes | No |
| 38 | 63 | 9 | P, X, H, C | 78 | 32 | 131 | Yes | No | Yes | Yes | Yes | Yes | No |
| 39 | 56 | Unknown | P, H, C | 87 | 123 | 168 | Yes | Yes | Yes | Yes | No | No | No |
| 40 | 72 | 10 | H, C | 45 | 44 | 47 | Yes | No | Yes | Yes | No | No | Yes |
| 41 | 61 | 7 | P, H, C | 78 | 35 | 108 | Yes | No | No | No | No | Yes | No |
| 42 | 56 | 7 | H, C | 42 | 30 | 60 | Yes | Yes | Yes | Yes | Yes | Yes | No |
| 43 | 47 | 9 | P, R | 60 | 40 | 70 | Yes | Yes | No | Yes | Yes | Yes | No |
| 46 | 66 | 9 | P, X, H, C | 69 | 30 | 69 | Yes | Yes | Yes | No | No | No | No |
| 47 | 61 | 7 | H, C | 45 | 43 | 46 | Yes | No | No | Yes | Yes | No | Yes |
| 48 | 69 | 8 | P, R, H, C | 15 | 120 | 158 | Yes | No | No | No | No | No | No |
| 49 | 60 | 7 | P, H, C | 81 | 73 | 98 | Yes | Yes | No | Yes | No | No | No |
| 50 | 62 | Unknown | P, R, H, C | 110 | 113 | 192 | Yes | Yes | No | Yes | Yes | No | No |
| 51 | 54 | 7 | P, X, H, C | 122 | 120 | 133 | Yes | No | Yes | No | No | No | No |
Treatment types: H = androgen ablation, C = Chemotherapy, P= prostatectomy, R = radiation therapy to prostate, X = palliative radiation
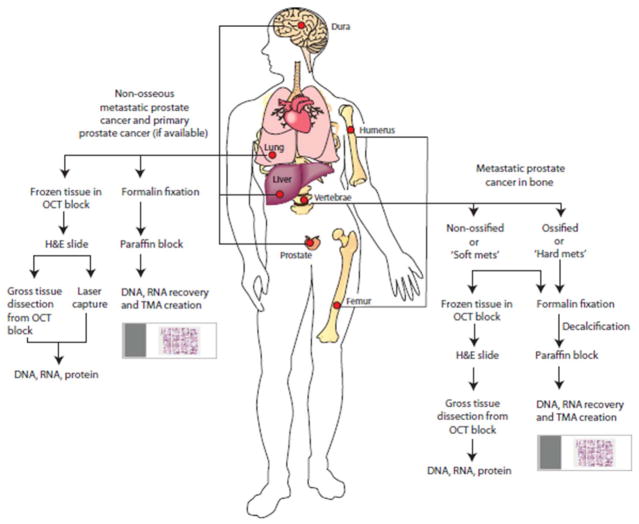
As a part of a new tissue procurement protocol during these autopsies, multiple osseous sites including the vertebrae, femurs, and humeri of all patients were longitudinally sectioned using motorized saws and examined for presence of grossly visible metastases. Examination for bone metastases was also guided by the patient’s bone scan. Bone marrow tissues harboring grossly evident tumor and foci suspicious for tumor, as indicated by presence of marrow with firm texture and areas of hemorrhage/necrosis, were grossly dissected or scooped out using surgical blades. Further, in addition to collecting tumor in bone marrow from grossly evident foci, we also performed a systematic procurement of bone marrow from bilateral humeri, femur and vertebrae (Figure 1). All tissues from the marrow cavities of these bones including the grossly evident tumor foci, as well as matched adjacent normal bone marrow (when present) were procured for RNA extraction and phenotypic assessment.
Figure 1.
Schematic overview of sampling and procurement of osseous and non-osseous metastatic prostate cancer by the University of Michigan ‘Rapid Autopsy’ program.
These fresh tissue samples, once scooped/dissected out of the bone marrow (henceforth referred to as ‘soft bone metastatic tissues’), were sliced into 1.0 × 1.0 × 0.2 cm sections for freezing in OCT media for RNA recovery or overnight fixation in 10% buffered-formalin. For OCT embedding, the tissues were placed into moulds filled with OCT which are then immersed into isopentane cooled over dry ice; the OCT blocks embedded as such are stored for future use at −80°C. Histological sections stained with H&E were later prepared from the OCT embedded frozen blocks containing bone marrow tissues, and formalin-fixed, paraffin-embedded bone tissues to confirm histology and identify tumor areas of interest for further gross dissection and RNA recovery. Slices of dense bone with or without evidence of gross involvement by tumor from bilateral humeri, femur and vertebrae were also procured, these bone samples (henceforth referred to as ‘hard bone metastatic tissues’) were fixed and decalcified as per standard protocol (10). These hard bone metastatic tissues were paraffin embedded and serve as a source of tissues for possible DNA and RNA recovery and tissue micro array (TMA) construction (Figure 1).
Pathology evaluation and analysis
Detailed histopathologic evaluation of metastatic tissues from bone marrow (soft bone marrow metastatic tissues) and hard bone (hard bone marrow metastatic tissues) was performed independently by three pathologists (B.H., R.B.S. and R.M.). The tissues were histologically evaluated for presence of viable tumor, areas of necrosis, hemorrhage, stroma, hard bone, as well as normal bone marrow. Based on areas of interest from this histopathology assessment, metastatic foci with high tumor density were grossly dissected out from within the bone marrow (the tissues being embedded in OCT blocks) and preserved within RNA later.
RNA extraction and Real Time PCR validation from metastatic prostate cancer tissue within bones
RNA was extracted from the tissues using Qiazol using Qiagen’s miRNeasy Minikit following the manufacturer’s instructions (Qiagen). Quantitative PCR (QPCR) was performed using Applied Biosystems’ inventoried Taqman assays (20x Primer Probe mix) corresponding to AMACR (Assay ID Hs02786742_s1), ERG (Assay ID Hs01554634_m1), KLK3 (Assay ID Hs02576345_m1), PCA3 (Assay ID Hs01371939_g1), TMPRSS2:ERG (Assay ID Hs03063375_ft) and GAPDH (Assay ID Hs00266705_g1). Briefly, 1 μg of total RNA was reverse transcribed into cDNA using SuperScript III (Invitrogen) in the presence of oligo dT and random primers. All reactions were performed with 2x Taqman Universal PCR Master Mix (Applied Biosystems, Foster City, CA) on an Applied Biosystems Step One Plus Real Time PCR System according to standard protocols. The amount of each target gene relative to the housekeeping gene GAPDH was determined using the comparative threshold cycle (Ct) method (Applied Biosystems User Bullet in #2, http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf). For each sample, the relative amount of each target gene was calibrated against prostate epithelial cell line (PREC). All assays were performed in duplicate. To quantitate the differentially expressed genes between the soft bone metastatic samples and matched non-osseous metastatic samples, primers corresponding to the different genes were used, the sequences of which are detailed in Supplementary Table 2.
Array CGH Analysis
Array CGH data (Agilent Human Genome CGH 244K microarray) was analyzed for three matched pairs of soft bone metastatic samples and non-osseous metastatic samples for cases: WAF43, WAF49, and WAF51. Log2 ratios for each aCGH probe were computed as log2(rProcessedSignal/gProcessedSignal), and after filtering out control probes, log-ratios for replicate probes in the same sample were summarized by the median. The resulting 235,834 log-ratios for each sample were segmented into regions of constant copy number using the Circular Binary Segmentation algorithm of Olshen et al(11), as implemented in the R package DNAcopy(12) using a p-value cutoff of p<0.001 for each binary split. Segments whose mean log-ratios were greater than log2(2.75/2) ≈ 0.46 or less than log2(1.25/2) ≈ −0.68 were classified as gains or losses, respectively; these cutoffs were chosen to be smaller in absolute value than the predicted cutoffs of log2(3/2) ≈ 0.58 and log2(1/2) = −1 corresponding to 1-copy gain and 1-copy loss, respectively. By choosing liberal cutoffs, we decreased the likelihood of missing potentially interesting regions of gain or loss at the cost of introducing false positives; subsequent filtering of regions by recurrence across samples was performed to reduce the incidence of such false positives. By intersecting regions of gain or loss across samples, continuous segments of gain or loss that were recurrent in at least 2 samples were identified.
Gene Expression Analysis
Gene expression data (Agilent Human Gene Expression 44K microarray) was analyzed for five matched pairs of soft bone metastatic samples and non-osseous metastatic samples for cases: WAF40, WAF42, WAF43, WAF49, and WAF51. The metastatic samples were labeled with Cy5 fluorescent dye, while the reference sample (pool of benign prostate, Clonetech) was labeled with Cy3 fluorescent dye. Log2 ratios for each probe were computed as log2(rProcessedSignal/gProcessedSignal), and after filtering of control probes, log-ratios for replicate probes in the same sample were summarized by the median. The resulting 41,000 log-ratios per sample were tested for differential expression between the soft bone marrow metastatic tissues and non-osseous metastases using paired Student’s t-tests. The false discovery rate was controlled at 10% using Storey’s Q-value method(13) as implemented in the R package qvalue(14).
Results
Clinical information regarding the rapid autopsy patients of this cohort is presented in Table 1. In general, the autopsies revealed widely disseminated osseous and non-osseous metastases. Bone marrow was variably present at the site of osseous metastases; the tumor was most commonly found to involve the marrow cavity, but was also found in periosteal location, as well as the skull table extending into the dura (Figure 2). Tumor necrosis was identified in some of these metastatic foci; areas of hemorrhage were also evident within these tissues, as well as adjacent bone marrow presumably involved with the same cancerous process (Figure 3).
Figure 2.
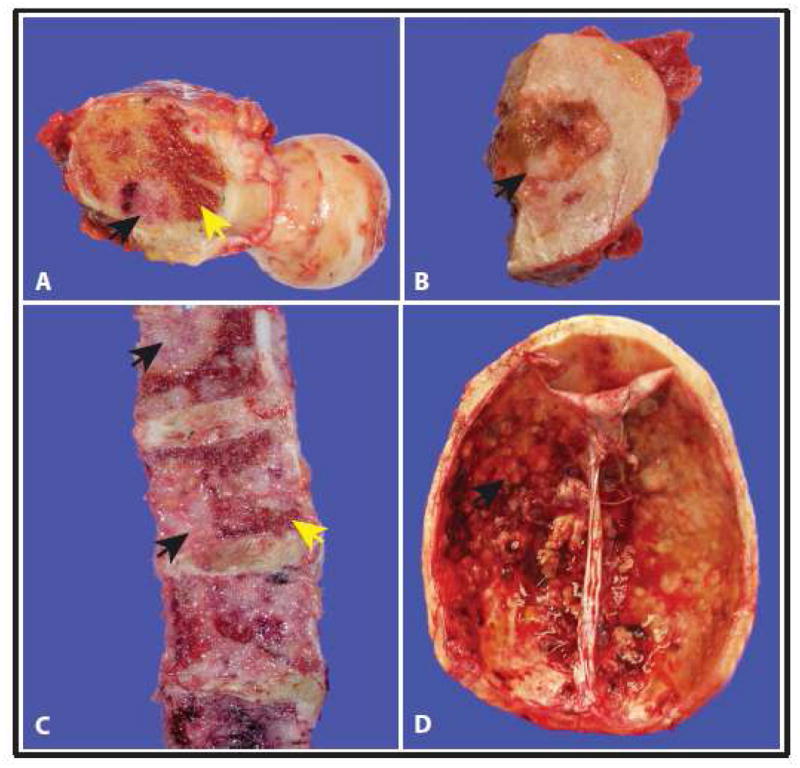
Gross representation of bone metastasis at different sites from our warm autopsy series of androgen independent prostate cancer patients. (A) and (B) Femoral head (C) Vertebrae (D) Skull and dura. For each image, black arrows indicate metastatic tumor foci and yellow arrows indicate residual bone marrow.
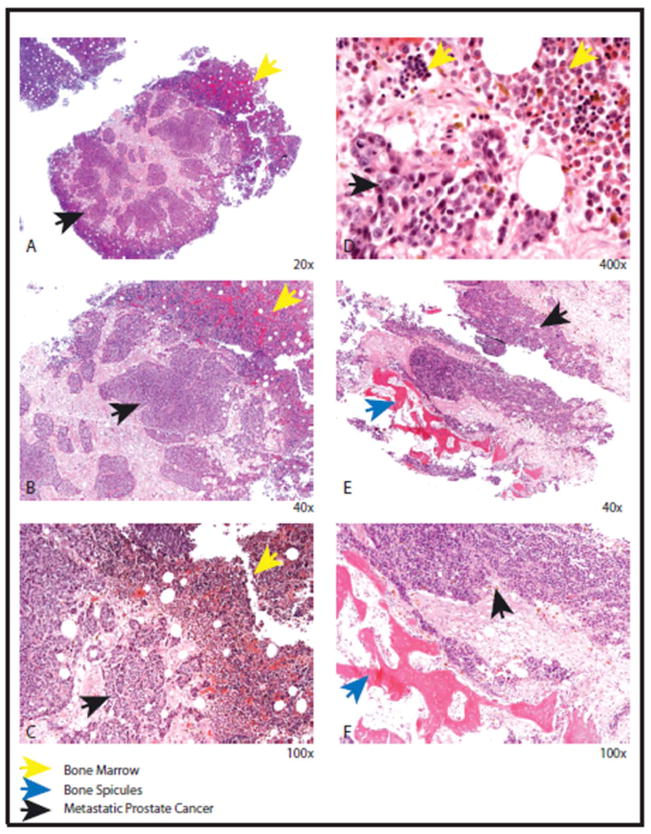
Figure 3.
Histopathology of androgen independent metastatic osseous metastases. Solid sheets and nests of uniform tumor cells indicated by black arrow (A, B, C and D) adjacent to residual normal bone (yellow arrow). The tumor cells have round nuclei with prominent nucleoli compared to the residual bone with normal hematopoietic maturation (D). The tumor cells (black arrow) can be seen adjacent to the residual bone spicules (blue arrow) ( E and F).
These metastatic tumor tissues exhibited a variable histological pattern as expected from our previous study (7). While the tumor tissues retrieved by surgical dissection from the marrow cavity showed areas of high tumor density with scarce bony areas, the hard bone metastatic tissues showed a preponderance of calcified trabeculae intermingled with islands of entrapped tumor. The soft bone marrow metastatic prostate cancer tissues showed presence of occasional bone spicules (Figure 3) which did not hinder overall RNA extraction using standard protocols. Such high density and high volume tumor areas were used as an index for grossly dissecting out tissues to be utilized for RNA recovery. RNA extracted from these samples were run through the Agilent Bioanalyzer to assess the quality and integrity of the samples. A representative image of RNA extracted by this procedure from soft bone prostate metastatic samples is shown in Supplementary Figure 1.
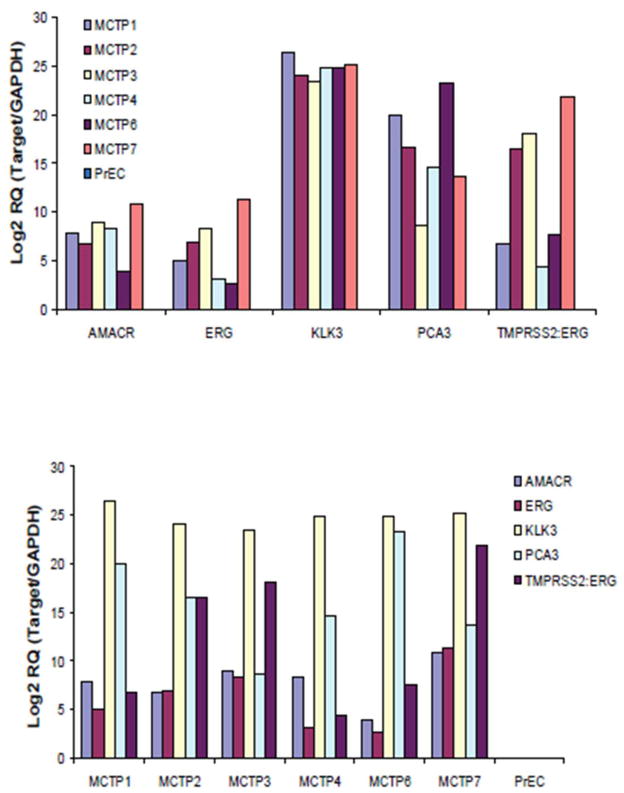
Total RNA was isolated from the soft bone marrow metastatic prostate cancer tissues excavated from bones by qiagen mRNAeasy protocol and the expression of a panel of prostate cancer biomarkers was assessed by Taqman based real time RT-PCR. This panel of genes included common prostate cancer specific genes such as prostate specific antigen, PSA (kallikrein-related peptidase 3, KLK3) and alpha-methylacyl-CoA racemase, AMACR; the prostate specific non-protein coding transcript, prostate cancer antigen, PCA3; v-ets erythroblastosis virus E26 oncogene homolog, ERG and prostate cancer gene fusion, transmembrane serine protease 2, TMPRSS2-ERG. As expected, PSA and PCA3 show very high levels of expression in all of the samples examined, while the expression of AMACR, ERG, and TMPRSS2-ERG showed variable expression across the sample set, when compared against their expression in normal prostate epithelial cell line PrEC (Figure 4).
Figure 4.
Expression profile of prostate cancer biomarker genes ( AMACR, ERG, KLK3, PCA3, and TMPRSS2:ERG fusion) in the bone metastatic tissues as assessed by quantitative RT-PCR. Log 2 fold change in Target gene/GAPDH is plotted with all samples normalized to the PREC control sample.
aCGH analysis
312 recurrent copy number aberrations were identified, consisting of 194 gains and 118 losses. These aberrations were characterized by type of recurrence to determine if soft bone metastases and non-osseous metastases from the same patient exhibited similar patterns. A majority of recurrent segments (200 of 312) were specific to an individual patient and occurred in both sample types for that patient. Additionally, segments specific to a sample type and recurrent across patients were found but were small in number (6 of 312 segments; 3 in soft bone metastases and 3 in soft tissue metastases). The remaining 106 segments were uncategorized, as the recurrences were across different pairs and across different sample types. Visual inspection of the copy number profiles revealed that all six recurrent segments specific to sample type were likely to be artifacts rather than real copy number changes. Specifically, differences in inferred copy number profiles for the same patient appeared as a result of small differences in log-ratios that happened to lie on opposite sides of the thresholds used to define gain and loss. In summary, then, this aCGH analysis failed to identify any regions of interest.
Gene Expression Analysis
Gene expression profiling data from five soft bone-metastatic cancer prostate tissues compared with patient-matched non- osseous metastatic cancer tissues revealed 672 probes, corresponding to 664 unique genes, differentially expressed in the lethal bone-metastatic tissues (Supplementary Table 1). Intriguingly, global metastasis signature genes including EZH2, MMPs, CXCRs, and stromal derived factor 1 (SDF1) etc., are not found to be differentially expressed between non-osseous metastatic tissues and bone metastatic tissues, albeit many cadherin family genes such as PCDHGA5, CDH17, PCDH11X, and PCDH15 are. Other noteworthy genes differentially overexpressed in soft bone-metastatic tissues include Cysteine rich transmembrane BMP regulator 1 (CRIM1), Insulin like Growth Factor Binding protein 2 (IGFBP2) etc. Bone morphogenetic proteins (BMPs) belong to the TGF-Beta superfamily and are vital bone inductive factors (15). BMPs also play important roles during embryonic development and the postnatal homeostasis of various organs and tissues, by controlling cellular differentiation, proliferation and apoptosis. One of the most highly expressed genes in soft bone-metastatic tissues as compared to non-osseous metastatic tissues is Wolf-Hirschhorn syndrome candidate 2 (WHSC2), that shows over 50-fold upregulation. Wolf-Hirschhorn syndrome (WHS) is a malformation syndrome associated with a hemizygous deletion of a 165 kb WHS critical region in the distal short arm of chromosome 4 that harbors genes WHSC1 and WHSC2. WHSC2 is highly expressed in fetal tissues, and is a member of the NELF (negative elongation factor) protein complex that regulates RNA polymerase II transcription elongation and may function as a transcriptional repressor (16). Incidentally, WHSC1 gene (more commonly known as MMSET) is also expressed in early development, and is a transcriptional repressor; it is overexpressed in a wide variety of aggressive cancers (17), and is involved in the chromosomal translocation t(4;14)(p16.3;q32.3) in multiple myelomas (18). Among the genes downregulated in soft bone-metastatic tissues, as compared to non-osseous metastatic tissues, a most interesting observation appears to be the downregulation of Dicer1. Dicer1 encodes a protein that functions as a ribonuclease, and is required by the RNA interference to produce the active small RNA component that represses gene expression (http://www.ncbi.nlm.nih.gov/gene/23405). Interestingly, Dicer1 has been shown to be downregulated in several cancer types (19–21), although one report observed Dicer up-regulation in prostate adenocarcinoma (22). Our observation of Dicer downregulation in soft-bone metastatic tissues suggests it will be important to follow-up as miRNA is being considered for therapeutic strategies (23). Another noteworthy gene seen to be downregulated in soft-bone metastatic tissues is Ubiquitin specific peptidase 10 (USP10) that was recently shown to be downregulated in clear cell carcinomas (24). USP10 was shown to suppress tumor cell growth in cells with wild type p53, and thus, its downregulation represents a loss of p53 phenotype. A summary list of genes significantly up and downregulated in soft-bone metastatic tissues is provided in the Supplementary Table 1.
Discussion
By cutting the long bones of prostate cancer patients in a sagittal instead of cross -sectional manner, non-ossified metastases have been identified in a majority of patients at the time of death. This method of bone tissue sampling offers several opportunities over other previously described methods for the collection of suitable research samples. It allows the collection of sample set which has eluded characterization secondary to the technical hurdles in retrieving viable tumor tissues from dense, calcified bones. The use of a motorized saw to collect samples yields a larger section of tissue as compared to drilled sections with enhanced visibility for discerning the presence of metastatic foci. The larger surface area of frozen soft bone marrow tumor thin slices (4–5 micron average thickness) that are collected and the ease of sectioning them at the cryotome renders less compression artifact and preservation of tissue structure, allowing the samples to be more readily amenable to conventional histological tissue preparation with enhanced morphological assessment. In comparison, hard bone metastatic tissues are practically very resistant to sectioning at the cryotome; RNA can be collected from such fresh or frozen bone tissues using a drilling technique which allows contamination of tumor tissue with normal bone, or from paraffin embedded decalcified bone tissues where RNA is virtually completely degraded because of the acid treatment.
Not only is the quantity of tissue available for study enhanced as a result of the present protocol, but the RNA extracted from this tissue is of high quality. This may be due to fewer intermediary processing steps, such as pulverization or bone decalcification procedures, that may adversely affect RNA yield and quality, as well as reduced contamination. Given the possible small sample size of a micro-metastasis, it is particularly crucial to maintain stringent purity of the RNA sample for subsequent analysis.
The sampling method described here was validated by the expression of a panel of prostate cancer associated genes, which included PSA PCA3, E26, ERG, TMPRSS2-ERG, and AMACR. These observations further confirmed the prostatic origin of the tissues and strongly suggested that the tissue extraction protocol described here was effective in providing experimentally useful material. This would enable integrative studies of osseous metastatic tissues in parallel with metastatic tissues from other non-osseous sites, and parental localized prostate cancer. Such studies could also be coupled with analysis of bone turnover, as was examined during hormonal treatment for metastatic prostate cancer (25), as well as bone morphogenic proteins and Wnts (26) and effects of prostate cancer cells on the control of the cell cycle (27). Indeed, analysis of the micrometastasis to the bone in prostate cancer, as revealed during autopsy, may reveal parallels to other cancers, such as recent identification of disseminated tumor cells (DTC) in breast cancer, assisting clinicians in determining prognosis and therapeutic monitoring (28). The availability of high quality bone marrow tissue samples post-mortem may also answer important questions as to the relationships between the bone marrow and DTC (29).
Comparative genomic hybridization data revealed no major differences between the paired soft and hard bone metastases from the same patients suggesting that these metastases belong on the same tumorigenesis pathway. Gene expression analysis revealed reproducible differences between non-osseous and osseous metastases across patients. Many intriguing gene expression changes are observed, including upregulation of WHSC2, CRIM1 and cadherin genes as well downregulation of Dicer1 and USP10 among others, which will merit follow up studies. The genes upregulated in the soft bone metastatic tumor samples by the microarray analysis were further validated using quantitative PCR, shown in Supplementary Fig. 2. Further, analysis of these genes in 2 of the matched non-osseous metastatic tissues did not show upregulation, indicating the specific control of these transcripts in metastasis to the bone (data not shown). It remains to be determined if the soft, non-osseous metastases are pre-ossified bone lesions, or if they represent bone metastases that do not have the capacity to form calcified lesions.
In summary, the method described herein provides access to high quality tissue that is an invaluable contribution from patients to the study of bone metastasis. The validation of this method, as evidenced by the presence of prostate cancer biomarkers, opens important new possibilities for comparisons between clinical monitoring through biomarker evaluation during treatment, with the presence of bone metastasis at the time of death.
Supplementary Material
672 differentially expressed genes in the bone metastatic samples compared to the matched soft tissues samples. A two fold increase in the level of the transcript across the 5 matched samples analyzed was considered as upregulated and a ratio of 0.5 was considered as down regulated.
Primer sequences used for quantitative PCR analysis.
Electropherogram obtained from the Agilent Bioanalyzer showing the integrity of a representative RNA sample extracted from soft bone metastatic sample. A RIN number here indicates the RNA integrity with a value of 10 being the most intact and a value of 4 being acceptable.
Validation of the microarray analysis using quantitative PCR analysis for the genes upregulated in the soft bone metastatic samples. Each transcript was normalized to the level of GAPDH in the sample. All the samples were compared to the level of the respective transcript in the prostate pool obtained from Clonetech (CPP).
Acknowledgments
KJP, ACM supported by NIH SPORE in Prostate Cancer 2 P50 CA69568 and Prostate Cancer Foundation, KJP supported by American Cancer Society as a Clinical Research Professor, and by NIH 1 PO1 CA093900. The authors also thank Dr. Lakshmi P. Kunju for her participation in the performance of rapid autopsy, Dr. Jason C. Carvalho, M.D. for edition of images, Robin Kunkel for assistance in preparing figures and Rhonda L. Hotchkin for editorial assistance.
The authors wish to thank the patients and their families and are indebted to them for supporting prostate cancer research through the SPORE initiative.
References
- 1.Taichman RS, Loberg RD, Mehra R, Pienta KJ. The evolving biology and treatment of prostate cancer. J Clin Invest. 2007;117(9):2351–61. doi: 10.1172/JCI31791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 3.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 4.Rubin MA, Zhou M, Dhanasekaran SM, et al. alpha-Methylacyl coenzyme A racemase as a tissue biomarker for prostate cancer. JAMA. 2002;287(13):1662–70. doi: 10.1001/jama.287.13.1662. [DOI] [PubMed] [Google Scholar]
- 5.Mehra R, Tomlins SA, Yu J, et al. Characterization of TMPRSS2-ETS gene aberrations in androgen-independent metastatic prostate cancer. Cancer Res. 2008;68(10):3584–90. doi: 10.1158/0008-5472.CAN-07-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loberg RD, Day LL, Harwood J, et al. CCL2 is a potent regulator of prostate cancer cell migration and proliferation. Neoplasia. 2006;8(7):578–86. doi: 10.1593/neo.06280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah RB, Mehra R, Chinnaiyan AM, et al. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Res. 2004;64(24):9209–16. doi: 10.1158/0008-5472.CAN-04-2442. [DOI] [PubMed] [Google Scholar]
- 8.Rubin MA, Putzi M, Mucci N, et al. Rapid (“warm”) autopsy study for procurement of metastatic prostate cancer. Clin Cancer Res. 2000;6(3):1038–45. [PubMed] [Google Scholar]
- 9.Suprun H, Rywlin AM. Metastatic carcinoma in histologic sections of aspirated bone marrow: a comparative autopsy study. South Med J. 1976;69(4):438–9. doi: 10.1097/00007611-197604000-00018. [DOI] [PubMed] [Google Scholar]
- 10.Roudier MP, True LD, Higano CS, et al. Phenotypic heterogeneity of end-stage prostate carcinoma metastatic to bone. Hum Pathol. 2003;34(7):646–53. doi: 10.1016/s0046-8177(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 11.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics (Oxford, England) 2004;5(4):557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 12.Seshan VE, Olshen A. DNAcopy: DNA copy number data analysis R package version 1.22.1. 2009 http://www.bioconductor.org/packages/2.3/bioc/html/DNAcopy.html.
- 13.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(16):9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dabney A, Storey J, Warnes G. qvalue: Q-value estimation for false discovery rate control R package version 1.24.0. 2010 http://CRAN.R-project.org/package=qvalue.
- 15.Ye L, Lewis-Russell JM, Kyanaston HG, Jiang WG. Bone morphogenetic proteins and their receptor signaling in prostate cancer. Histol Histopathol. 2007;22(10):1129–47. doi: 10.14670/HH-22.1129. [DOI] [PubMed] [Google Scholar]
- 16.Mariotti M, Manganini M, Maier JA. Modulation of WHSC2 expression in human endothelial cells. FEBS Lett. 2000;487(2):166–70. doi: 10.1016/s0014-5793(00)02335-8. [DOI] [PubMed] [Google Scholar]
- 17.Kassambara A, Klein B, Moreaux J. MMSET is overexpressed in cancers: link with tumor aggressiveness. Biochem Biophys Res Commun. 2009;379(4):840–5. doi: 10.1016/j.bbrc.2008.12.093. [DOI] [PubMed] [Google Scholar]
- 18.Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92(9):3025–34. [PubMed] [Google Scholar]
- 19.Pampalakis G, Diamandis EP, Katsaros D, Sotiropoulou G. Down-regulation of dicer expression in ovarian cancer tissues. Clinical biochemistry. 43(3):324–7. doi: 10.1016/j.clinbiochem.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 20.Sekine S, Ogawa R, Ito R, et al. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology. 2009;136(7):2304–15. e1–4. doi: 10.1053/j.gastro.2009.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng ZH, Sun XJ, Fu WN, et al. Decreased expression of DICER1 in gastric cancer. Chin Med J (Engl) 2007;120(23):2099–104. [PubMed] [Google Scholar]
- 22.Chiosea S, Jelezcova E, Chandran U, et al. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169(5):1812–20. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy SD, Gajula RP, Pakala SB, Kumar R. MicroRNAs and cancer therapy: The next wave or here to stay? Cancer biology & therapy. 9(7) doi: 10.4161/cbt.9.7.11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 140(3):384–96. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansen JS, Brasso K, Iversen P, et al. Changes of biochemical markers of bone turnover and YKL-40 following hormonal treatment for metastatic prostate cancer are related to survival. Clin Cancer Res. 2007;13(11):3244–9. doi: 10.1158/1078-0432.CCR-06-2616. [DOI] [PubMed] [Google Scholar]
- 26.Dai J, Hall CL, Escara-Wilke J, Mizokami A, Keller JM, Keller ET. Prostate cancer induces bone metastasis through Wnt-induced bone morphogenetic protein-dependent and independent mechanisms. Cancer Res. 2008;68(14):5785–94. doi: 10.1158/0008-5472.CAN-07-6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinski J, Parikh A, Bova GS, Isaacs JT. Therapeutic implications of enhanced G(0)/G(1) checkpoint control induced by coculture of prostate cancer cells with osteoblasts. Cancer Res. 2001;61(17):6372–6. [PubMed] [Google Scholar]
- 28.Vincent-Salomon A, Bidard FC, Pierga JY. Bone marrow micrometastasis in breast cancer: review of detection methods, prognostic impact and biological issues. J Clin Pathol. 2008;61(5):570–6. doi: 10.1136/jcp.2007.046649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alix-Panabieres C, Riethdorf S, Pantel K. Circulating tumor cells and bone marrow micrometastasis. Clin Cancer Res. 2008;14(16):5013–21. doi: 10.1158/1078-0432.CCR-07-5125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
672 differentially expressed genes in the bone metastatic samples compared to the matched soft tissues samples. A two fold increase in the level of the transcript across the 5 matched samples analyzed was considered as upregulated and a ratio of 0.5 was considered as down regulated.
Primer sequences used for quantitative PCR analysis.
Electropherogram obtained from the Agilent Bioanalyzer showing the integrity of a representative RNA sample extracted from soft bone metastatic sample. A RIN number here indicates the RNA integrity with a value of 10 being the most intact and a value of 4 being acceptable.
Validation of the microarray analysis using quantitative PCR analysis for the genes upregulated in the soft bone metastatic samples. Each transcript was normalized to the level of GAPDH in the sample. All the samples were compared to the level of the respective transcript in the prostate pool obtained from Clonetech (CPP).