Abstract
In the testicular cancer cell line, NT2, we previously demonstrated that differentially methylated regions were located in introns or intergenic regions, and postulated these might regulate non-coding RNAs. Three micro-RNAs and three small nucleolar RNAs were differentially methylated; one, miR-199a, was associated with the progression and prognosis of gastric and ovarian cancers. In this report we document, by epigenomic profiling of testicular tissue, that miR-199a is transcribed as antisense of dynamin 3 (chromosome 1q24.3), and hypermethylation of this region is correlated with miR-199a-5p/3p repression and tumor malignancy. Re-expression of miR-199a in testicular cancer cells led to suppression of cell growth, cancer migration, invasion and metastasis. The miR-199a-5p, one of two mature miRNA species derived from miR-199a, is associated with tumor malignancy. We further identified the embryonal carcinoma antigen podocalyxin-like protein 1 (PODXL), an anti-adhesive protein expressed in aggressive tumors, as a target of miR-199a-5p. We demonstrated PODXL is overexpressed in malignant testicular tumor, and cellular depletion of PODXL resulted in suppression of cancer invasion. The inverse relationship between PODXL and miR-199a-5p expression suggests PODXL is a downstream effector mediating the action of miR199a-5p. This report identifies DNA methylation, miR-199a dysregulation and PODXL as critical factors in tumor malignancy.
Keywords: intronic methylation, miR-199a, testicular tumor, PODXL, malignancy
Introduction
Altered gene and/or non-coding RNA expression are key features of cancer. Genetic and epigenetic modulation is an essential phenomenon of carcinogenesis. DNA methylation, a fundamental epigenetic modification, allows cells of different tissues to stably maintain diverse characteristics despite the same genetic makeup (Jones and Takai, 2001). In cancer cells, hypermethylation of tumor suppressor genes, and/or hypomethylation of oncogenes or heterochromatin results in aberrant expression of genes leading to tumorigenesis, genomic instability or the promotion of cell proliferation (Cheung et al., 2009). Recent reports suggested methylation may have a role in the regulation of tumor malignancy (Li et al., 2001; Aleman et al., 2008; Watts et al., 2008).
Testicular cancer is a malignant, highly aggressive neoplasm in young males. The molecular mechanisms operative in this malignancy have not been fully understood. We hypothesize that aberrant DNA methylation is a factor for development of testicular malignancy. In our previous study, we profiled differential methylation of a testicular cancer cell line NTera-2 (NT2), a cell line originally isolated from pulmonary metastasis in a patient with primary embryonal carcinoma of the testis (Andrews et al., 1984). The majority of the identified differentially methylated regions are located in introns or intergenic regions (Cheung et al., 2010). We postulated these differentially methylated regions might link to regulation of non-coding RNAs. When these differentially methylated regions were mapped to a non-coding RNA database, we identified three microRNAs (miRNA) and three small nucleolar RNAs that were differentially methylated. One miRNA, miR-199a, was previously implicated in the progression and prognosis of gastric and ovarian cancers (Nam et al., 2008; Ueda et al., 2009; Yin et al., 2010).
In this study we document that miR-199a was generally hypermethylated in malignant testicular tumor, which correlated with its downregulation. Expression of miR-199a resulted in suppression of its invasive phenotype. We identified podocalyxin-like protein 1 (PODXL) as a target of miR-199a-5p. PODXL expression was aberrantly upregulated in malignant testicular tumor and inversely correlated with miR-199a-5p expression. Depletion of PODXL suppressed cancer invasion. Our data imply that an epigenetic change in a previously unrecognized intronic region contributes to the aggressive behavior of testicular tumors via dysregulation of miR-199a and its corresponding target, PODXL.
Results
Identification of a hypermethylated intronic region and its association with testicular tumor malignancy
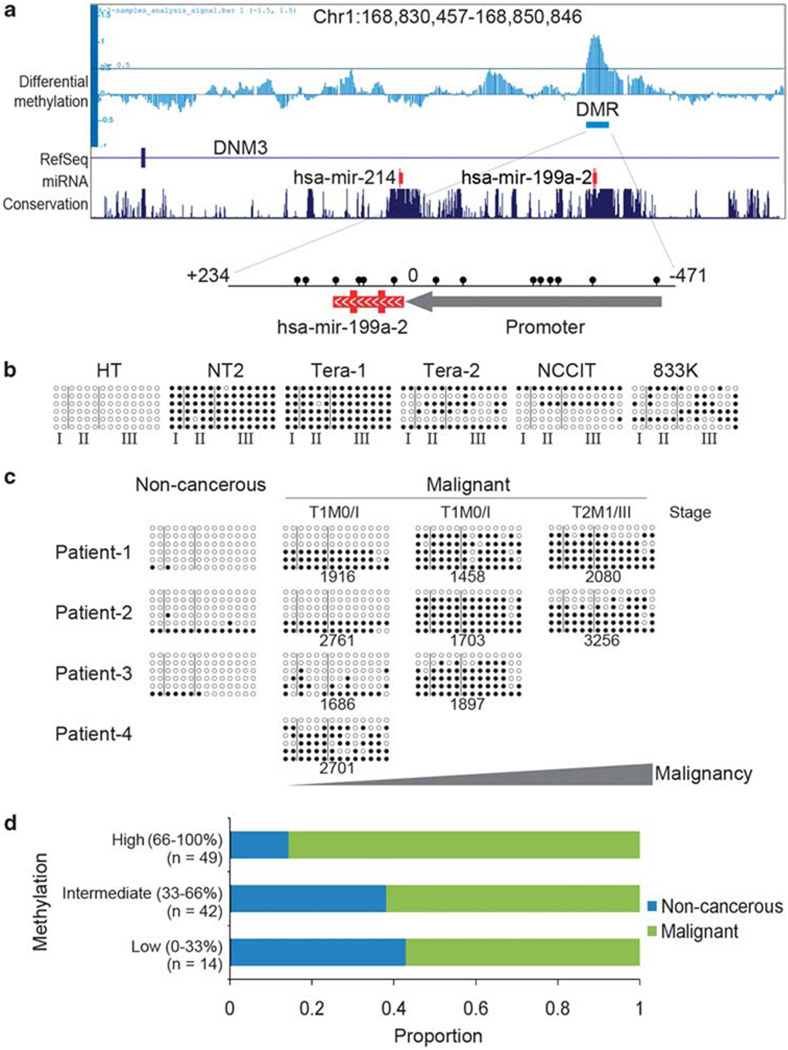
Genomic analysis revealed that our previously identified differentially methylated region, a conserved hypermethylated region of ~700 bp spanning miR-199a and its upstream region, is embedded in intron-14 of dynamin 3 (DNM3) at 1q24.3 (Figure 1a). The miR-199a is transcribed as antisense of the host gene DNM3. Luciferase assay indicated that the upstream region of miR-199a (+9:−471) has promoter activity (Supplementary Figure 1a). We performed bisulfite sequencing on several testicular cancer cell lines (NT2, Tera-1, Tera-2, NCCIT and 833 K) and a non-cancerous fetal testicular cell line (HT). The miRNA-199a locus in all the cancer lines was highly or partially methylated, whereas in the non-cancerous testis cell lines it was unmethylated (Figure 1b). To investigate whether aberrant methylation of miR-199a is related to tumor malignancy, we obtained tumor DNA from patients with testicular cancer with different degrees of metastasis and with three normal specimens as controls. Bisulfite sequencing analysis revealed an acquired methylation pattern as the tumor becomes more malignant and metastatic (Figure 1c). Hypermethylation of miR-199a was confirmed by high-throughput methylation assay (Methylight) for determination of relative DNA methylation in samples extracted from tissue arrays of a testicular cancer panel (n = 105) (Figure 1d). The results indicated that methylation was increased in malignant testicular tumors (P = 0.0005 for non-cancerous vs malignant).
Figure 1.
Methylation of miR-199a is associated with testicular tumor malignancy. (a) Genomic representation of differential methylation from chr1:168 830 456–168 850 846 (hg17). A hypermethylated differentially methylated region (DMR) embedded in intron-14 of DNM3 was identified by MeDIP-chip and mapped to miR-199a and its upstreaming promoter (+234:−471). Relative location of all CpG sites (lollipops) in the DMR are indicated. (b) Genomic bisulfite sequencing covering upstream (region III), intermediate (region II) and downstream (region I) regions of miR-199a in different testicular cancer cell lines (NT2, Tera-1, Tera-2, NCCIT and 833K) and a normal fetal testicular cell line (HT). (c) Genomic bisulfite sequencing of miR-199a in testicular cancer patients at different stages of malignancy (stage and case# were indicated). Normal testicular tissues were included as non-cancerous control. Samples were obtained from different patients. T1: the tumor has not spread beyond the testicle and epididymis; T2: the tumor has spread to blood or lymph vessels near the tumor or tunica vaginalis; M0: no distant metastasis; M1: distant metastasis is present. (d) Analysis of miR-199a methylation in patients from tissue microarray (n = 105). Samples were divided into three groups according to their methylation: low (0–33%), intermediate (33–66%) and high (66–100%). Proportion of non-cancerous and malignant cases was represented in each group. Methylation was significantly lower in malignant group compared with non-cancerous group (P = 0.0005, Mann–Whitney Test).
Expression of miR-199a-5p is associated with testicular tumor malignancy and negatively correlated with methylation
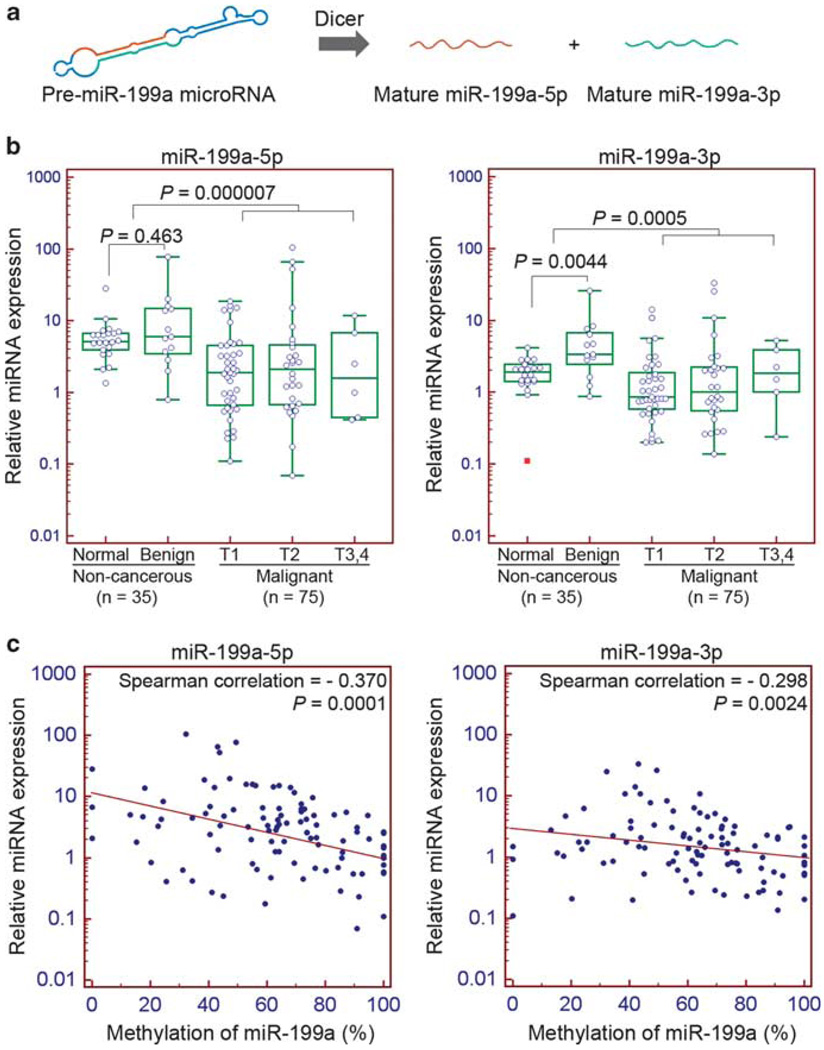
Two mature miRNA species are derived from the miR-199a precursor, namely miR-199a-5p and miR-199a-3p (Figure 2a). However, they have different seed sequences that regulate different targets. To determine whether the expression of these miRNAs is related to testicular tumor malignancy, we measured their expression by quantitative reverse transcription PCR. Comparison of the non-cancerous and malignant groups indicated miR-199a-5p was significantly downregulated in malignant tumors (P = 0.000007). The difference between normal and non-invasive benign tumors, however, was insignificant (P = 0.463). The miR-199a-3p, although processed from the same precursor RNA, was not significantly changed as contrasted to miR-199a-5p in cancer (P = 0.0005). We observed a significant upregulation of miR-199a-3p in benign tumors (P = 0.0044). These results indicate the expression of miR-199a-5p, but not miR-199a-3p, is changed during neoplastic development (Figure 2b).
Figure 2.
Expression of miR-199a is associated with testicular tumor malignancy and negatively correlated with methylation. (a) Biogenesis of miR-199a. (b) Expression of miR-199a-5p and miR-199a-3p in non-cancerous and malignant testicular tumors. The difference between non-cancerous and malignant groups is significant (miR-199a-5p: P = 0.000007; miR-199a-3p: P = 0.0005; Mann–Whitney test). T3: the tumor invades the spermatic cord; T4: the tumor invades the scrotum. (c) Scatter plots of miR-199a-5p and miR-199a-3p expression against methylation. Expression of both miR-199a-5p and miR-199a-3p correlates negatively with methylation (miR-199a-5p: Spearman correlation = −0.370, P = 0.0001; miR-199a-3p: Spearman correlation = −0.298, P = 0.0024).
Increased methylation in promoters is one mechanism for transcriptional silencing. The relationship between methylation and expression was demonstrated by correlation analysis of the genomic DNA and RNA isolated from the same individuals. Spearman’s rank correlation analysis of methylation and expression indicated inverse correlations for both miR-199a-5p (correlation = −0.370, P = 0.0001) and miR-199a-3p (correlation = −0.298, P = 0.0024), suggesting that methylation is a negative regulator of miR-199a (Figure 2c). The role of methylation as a transcriptional inhibitor was supported by treatment of cultured NT2 cells with the demethylation agent 5-aza-2′-deoxycytidine (5-aza). The 5-aza inhibits de novo methyltransferase to reverse the acquired methylation lesion. As anticipated, 5-aza treatment restored miR-199a expression by more than 40-fold (Figure 5b). In addition, in vitro methylation of the cloned miR-199a promoter ligated to a luciferase gene suppressed the luciferase activity by 80%, as compared with the unmethylated promoter control (Supplementary Figure 1b).
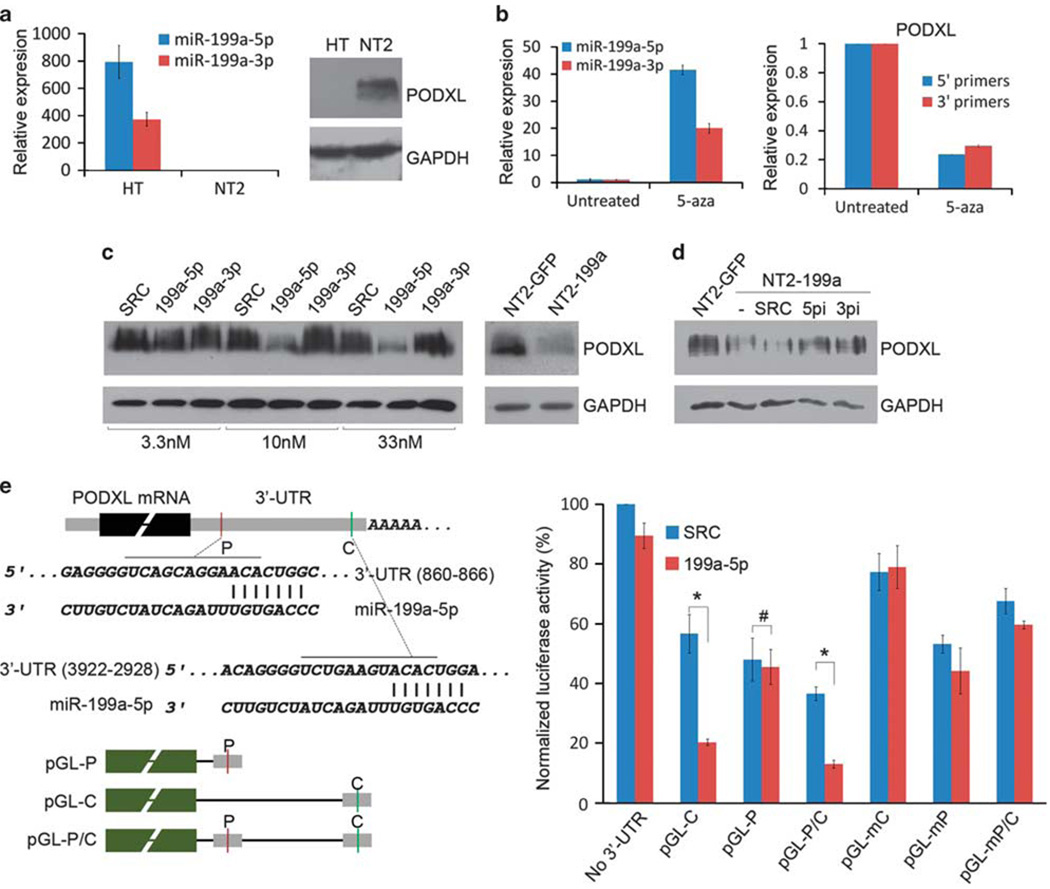
Figure 5.
Identification of PODXL as the target of miR-199a-5p. (a) Reciprocal expression of miR-199a-5p and PODXL in normal (HT) and cancerous (NT2) cell lines. (b) Expression of miR-199a-5p, miR-199a-3p and PODXL in NT2 cells treated with 5-aza. Both 5′ and 3′ end primers for PODXL mRNA were shown. (c) PODXL level in NT2 cells transiently transfected with miR-199a-5p (199a-5p), miR-199a-3p mimics (199a-3p) or stably express miR-199a (NT2-199a). SRC: scramble miRNA. (d) PODXL level in NT2-199a cells transiently transfected with inhibitors of miR-199a-5p (5pi) or miR-199a-3p (3pi). (e) Luciferase reporter assay of PODXL 3′-UTR. Two miR-199a-5p targeting sites (P: poorly conserved site; C: conserved site) were cloned to the 3′-end of Firefly luciferase (pGL-P and pGL-C). The mutant constructs (pGL-mC and pGL-mP) were generated by changing the binding sites to complementary sequences. The plasmids were co-transfected with miR-199a-5p mimics (199a-5p) or scramble miRNA control (SRC). Luciferase activity was measured 48 h after transfection (*P < 0.001; #P = 0.806, two-tailed Student’s t-test). Error bar: s.e.m. of triplicates.
Expression of miR-199a suppresses cancer migration, invasion and cell growth
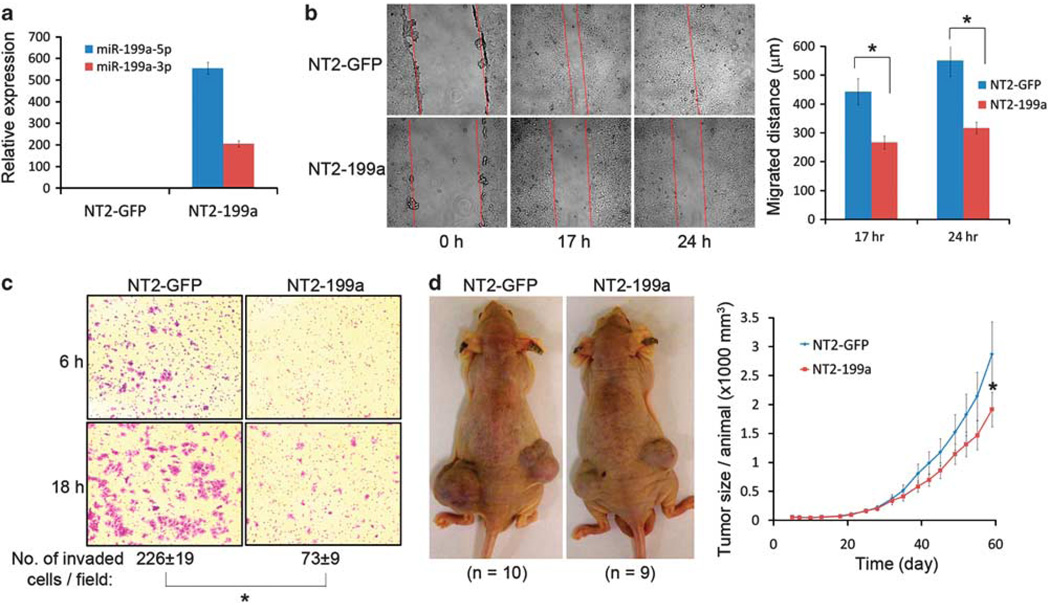
Previous reports showed that miR-199a is changed in several aggressive tumor types in addition to testicular tumor (Iorio et al., 2007; Nam et al., 2008; Worley et al., 2008; Ichimi et al., 2009; Ueda et al., 2009). To study the function of miR-199a, we induced constitutive expression of miR-199a in cancer cells with lentivirus. Cells positively expressing miR-199a were sorted by flow cytometry. These cells (NT2-199a) demonstrated greater than 500-fold increase in miR-199a-5p and 200-fold of miR-199a-3p expression when compared with vector infected control cells (NT2-GFP) (Figure 3a). A change of cell motility is one characteristic of metastasis (Sahai, 2005). Using the wound-healing assay, we found that NT2-199a cells migrated more slowly than NT2-GFP cells (P < 0.005) (Figure 3b). Another feature of metastasis is its ability to invade extracellular matrix (Sahai, 2005). Matrigel invasion assay indicated that expression of miR-199a significantly suppressed the ability of NT2 cells to invade the matrigel basement (P < 0.005) (Figure 3c). We also examined the effect of miR-199a on tumor growth. Two months after subcutaneous implantation of transfected cells in athymic nude mice, the average size of the tumors in the NT2-199a group was ~33% smaller than that in the control group (P = 0.145) (Figure 3d). In addition, reduced cell growth was confirmed by direct counting of cultured cells grown on fibronectin-coated plates (Supplementary Figure 2).
Figure 3.
The miR-199a suppresses cancer migration, invasion and decreases cancer growth. (a) Ectopic expression of miR-199a in a metastatic testicular cancer cell line NT2. NT2 cells were infected with lentivirus carrying GFP (NT2-GFP), or GFP/miR-199a (NT2-199a). (b) Wound healing assay for assessment of cancer cell migration. Difference of migration between the two groups is significant (*P < 0.005, two-tailed Student’s t-test). Error bar: s.e.m. of triplicates. (c) Matrigel invasion assay for assessment of cancer invasion. Invaded cells were stained with crystal violet and counted. The difference is significant (*P < 0.005, two-tailed Student’s t-test). (d) Tumor size of NT2-GFP and NT2-199a cells in athymic nude mice. Cancer cells were injected subcutaneously into two groups of nude mice (n = 10 for NT2-GFP; n = 9 for NT2-199a) and the tumor size was monitored at different time points. Mean size of the tumors per animal was plotted (*P = 0.145 at day 60, 2-tailed Student’s t-test). Error bar: s.e.m.
miR-199a suppresses cancer metastasis in mouse xenograft model
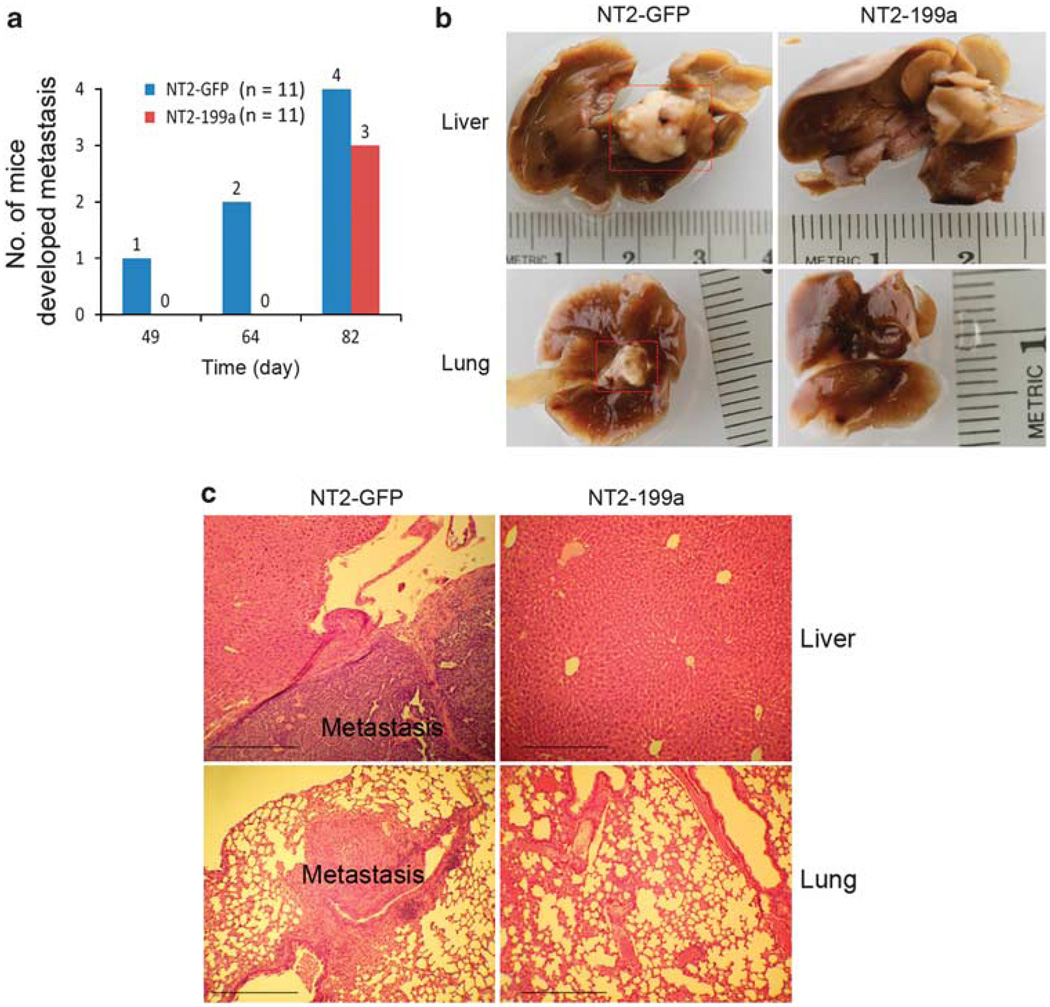
To confirm the anti-metastastic property of miR-199a, we used a xenograft animal model to study its function in vivo. Equal numbers of NT2-GFP and NT2-199a cells were injected intravenously in athymic nude mice via tail vein (n = 11 for each group). Mice were killed at day 49, 64 and 82 after injection. At day 49 and 64, three mice out of six from the control group (NT2-GFP) developed pulmonary and liver metastasis. No metastases were found in the NT2-199a group. At day 82, all the remaining mice (5 from each group) were killed. Four mice from the control group developed metastasis, compared with four mice from the NT2-199a group (Figure 4a). Metastasis developed in organs such as lung and liver, which are common metastatic sites for human secondary testicular cancer (Figure 4b). Histologic analysis indicated invasion of xenografted cancer cells (NT2-GFP) in liver and lung, and no invasion of cells expressing miR-199a (NT2-199a) at day 64 (Figure 4c). At later stage (day 82), miR-199a appeared to be less effective in suppressing metastasis. The lung and liver metastases from NT2-199a group at day 82 expressed miR-199a-5p/3p at a comparable level to those of cultured NT2-199a cells (Supplementary Figure 5).
Figure 4.
The miR-199a suppresses cancer metastasis in mouse xenograft model. (a) Number of mice developed metastasis at different time points. NT2-GFP and NT2-199a cells were injected intravenously in athymic nude mice via tail vein (n = 11 for each group). Animals were killed and necropsied for metastasis at days 49, 64 and 82 after injection. (b) Gross view of cancer metastasis in lungs and livers. No metastasis was observed in NT2-199a group at days 49 and 64. (c) Hematoxylin and eosin stained sections of lungs and livers in NT2-GFP and NT2-199a groups at day 64. Metastasis from human cell line (NT2-GFP) was indicated. Scale bar: 100 µm.
Identification of PODXL as the target of miR-199a-5p
As only miR-199a-5p was related to tumor malignancy, we sought to identify targets of miR-199a-5p compatible with its function (Figure 2b). We presumed the targets would be significantly upregulated in malignant NT2 cells. Analysis of our previous microarray expression data with multiple miRNA target prediction algorithms (TargetScan and PicTar) generated a list of upregulated predicted target genes (Supplementary Table 1). A search of the target genes revealed PODXL as a gene important in various malignant tumors including testicular cancer (Schopperle et al., 2003; Casey et al., 2006; Hayatsu et al., 2008; Koch et al., 2008). Notably, PODXL was one of the significantly upregulated target genes. It is an anti-adhesive transmembrane sialoglyco-protein implicated in the development of aggressive forms of cancer (He and Hannon, 2004; Sahai, 2005; Iorio et al., 2007; Worley et al., 2008). Western blot analysis confirmed overexpression of this protein in NT2 cells, as well as a reciprocal relationship with miR-199a-5p levels (Figure 5a). Furthermore, demethylation of NT2 cells by 5-aza restored the miR-199a-5p level and suppressed PODXL expression, suggesting a link between methylation, miR-199a-5p expression and PODXL level (Figure 5b). To demonstrate the effect of the miRNA on the PODXL level, we transfected NT2 cells with different concentrations of miR-199a-5p or miR-199a-3p mimics. Seventy-two hours after transfection, the PODXL protein was significantly decreased following miR-199a-5p, but not miR-199a-3p treatment. The same effect was observed when NT2 cells stably expressed miR-199a (NT2-199a) (Figure 5c). When NT2-199a cells were transfected with miR-199a-5p inhibitor (5pi), the PODXL level was restored. Surprisingly, miR-199a-3p inhibitors (3pi) also restored PODXL, probably because both inhibitors target the same primary miRNA precursor molecules (Figure 5d). Regulation of PODXL by miR-199a-5p probably occurs through binding of miRNA at its 3′-UTR. To validate this speculation, we cloned the two predicted binding sites in PODXL 3′-UTR and linked them to firefly luciferase vectors. When these luciferase vectors were co-transfected with miR-199a-5p mimics in NT2 cells, luciferase activity of the vector carrying the conserved binding site was significantly suppressed. However, miR-199a-5p did not suppress the vector carrying a poorly conserved binding site. To show that the suppression of luciferase activity is due to binding of the miRNA to the seed sequence, we generated the mutant constructs by mutating the seed sequence. As expected, miR-199a-5p had little effect on the mutant constructs (Figure 5e). These data show that miR-199a-5p regulates PODXL through a conserved binding site in its 3′-UTR.
PODXL is highly expressed in malignant testicular tumor and negatively correlated with miR-199a-5p
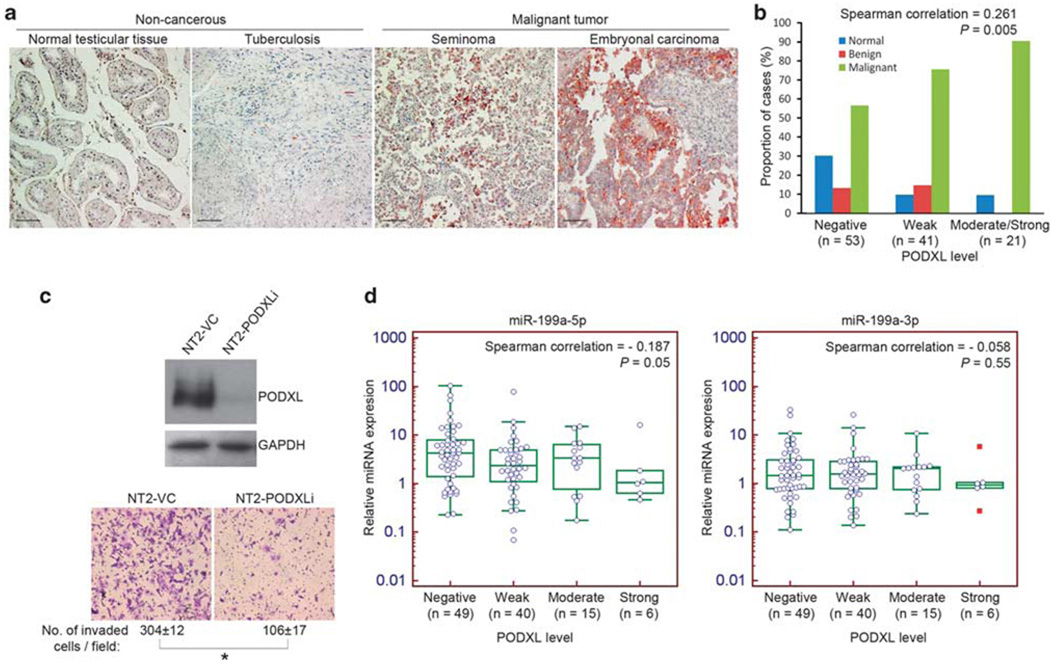
Given that PODXL is a target of miR-199a-5p, the expression and its correlation with miR-199a-5p in primary tissue remains unclear. Using the same tissue arrays, we applied immunohistochemistry to analyze the level of PODXL protein in testicular tumors. We found high levels of PODXL in malignant tumors including seminoma, non-seminomatous embryonal carcinoma and yolk sac tumor, but not in non-invasive normal or benign tissue (Figure 6a). Although PODXL is not expressed in all cases of malignant tumors, when samples were grouped according to PODXL intensity, we observed an increased number of malignant tumors with high levels of PODXL (Figure 6b). Spearman’s rank correlation test showed a positive correlation between PODXL level and the occurrence of malignancy (correlation = 0.286, r = 0.286, P = 0.002). An inverse relationship between miR-199a-5p and PODXL was observed in cultured cells (Figures 5a–d). We further confirmed this relationship in tissues (n = 110) by correlation analysis. Based on the immunohistochemistry staining intensity, PODXL level was divided into four groups (negative, weak, moderate and strong). A scatter plot of miR-199a-5p or miR-199a-3p against PODXL level was created. The mean value of both miRNA species decreased with increasing levels of PODXL. Spearman’s rank correlation test indicated a negative correlation for miR-199a-5p only (correlation = −0.187, P = 0.05). The correlation of miR-199a-3p with PODXL was not significant (correlation = −0.058, P = 0.55) (Figure 6d). The difference of the correlation coefficient agrees with the finding that PODXL is a target of miR-199a-5p, but not miR-199a-3p.
Figure 6.
PODXL protein is highly expressed in malignant testicular tumor and negatively correlated with miR-199a-5p level. (a) Immunohistochemistry of PODXL in representative non-cancerous and malignant testicular tumor sections. Scale bar: 100 µm. (b) Expression of PODXL protein in non-cancerous and malignant testicular specimens. Samples were divided into three groups (negative, weak, moderate or strong) based on the PODXL level and the proportion of different grades of tumors was shown. (c) Depletion of PODXL protein by RNAi suppresses cell invasion in vitro as revealed by matrigel invasion assay. Number of invaded cells per field was counted and represented as mean ± s.d. (*P < 0.001, two-tailed Student’s t-test). NT2-PODXLi: PODXL knockdown; NT2-VC: vector control. (d) Scatter plots of miR-199a-5p and miR-199a-3p expression against PODXL level. Expression of miR-199a-5p, but not 3p, correlates negatively with PODXL level (miR-199a-5p: Spearman correlation = −0.187, P = 0.05; miR-199a-3p: Spearman correlation = −0.058, P = 0.55).
PODXL knockdown suppresses cancer invasion in vitro
As a target of miR-199a-5p, PODXL might participate in the anti-metastatic function of this miRNA. To validate this hypothesis, we stably knocked down PODXL in NT2 cells with RNAi. The stable PODXL knockdown cells (NT2-PODXLi) displayed slower migration as revealed in the wound-healing assay (Supplementary Figure 3a). Moreover, the matrigel invasion assay showed that NT2-PODXLi was less invasive than the vector control cells (NT2-VC) (Figure 6c). The invasiveness of NT2-PODXLi cells was similar to that of NT2-199a cells. However, in NT2-PODXLi cells the level of miR-199a was invariable relative to the parent NT2 cells (Supplementary Figure 3b). Thus, we demonstrated that knockdown of PODXL alone without changing the level of its riboregulator miR-199a-5p would suppress cancer invasion in a manner similar to the effect of overexpression of miR-199a, implying that PODXL is a downstream target of miR-199a-5p.
Methylation and expression of miR-199a and PODXL level in seminomas and non-seminomas
We also analyzed the samples according to their histologic subtypes (seminomas vs non-seminomas). These subtypes show similar proportion of different invasiveness (Supplementary Figure 4e). We found that miR-199a is more highly methylated in seminomas than in non-seminomas (P = 0.04) (Supplementary Figure 4a). A comparison of the miR-199a-5p and miR-199-3p levels between seminomas and non-seminomas showed that seminomas express in general a lower level of both miRNAs than non-seminomas (P < 0.05) (Supplementary Figures 4b–c). A comparison of PODXL between seminomas and non-seminomas showed that the proportion of positive staining in malignant non-seminomas is slightly higher than seminomas (Supplementary Figure 4d); however, the difference is not statistically significant (P = 0.127, χ2-test).
Discussion
Epigenetic alteration is a mechanism for carcinogenesis. Genome-wide profiling reveals global changes across cancer genomes, however, the function and consequence of such changes have not been intensively studied. In particular, epigenetic changes occurring in non-regulatory regions such as introns and intergenic regions, have largely been ignored. Here we describe DNA methylation linked dysregulation of a conserved miR-199a caused by aberrant methylation in an intronic region of DNM3 at 1q24.3. We found that hypermethylation in the DNM3 intron leads to miR-199a depression. Both miR-199a methylation and expression are associated with testicular tumor malignancy. We demonstrated the relationship of miR-199a to anti-invasive and anti-metastatic properties. Subsequently we identified an embryonal carcinoma tumor antigen, PODXL, as a target of miR-199a-5p. PODXL is an anti-adhesive protein upregulated in many aggressive tumors (Somasiri et al., 2004; Casey et al., 2006; Hayatsu et al., 2008), but the mechanism for this phenomenon is unknown. We showed that miR-199a-5p is a negative regulator of PODXL. Based on our data we propose that epigenetic alteration in an intron of DNM3 leads to dysregulation of miR-199a and PODXL, and that this is one mechanism for development of testicular cancer.
The miRNAs have an important role in tumorigenesis. They can be oncogenic or tumor suppressive (Esquela-Kerscher and Slack, 2006). Specifically, some miRNA such as miR-122, miR-148a, miR-34b/c, miR-21, miR-373 and miR-520 (Huang et al., 2008; Lujambio et al., 2008; Zhu et al., 2008; Tsai et al., 2009) have been shown to be important in cancer metastasis. However, few miRNAs for testicular cancer metastasis/invasion are known. The miR-199a was initially identified to be an evolutionarily conserved small RNA essential for development (Chakrabarty et al., 2007; Friedman et al., 2009; Lee et al., 2009; Lin et al., 2009). Recently it was reported to be linked to other aggressive tumor types, such as gastric cancer (Ueda et al., 2009), bladder cancer (Ichimi et al., 2009), uveal melanoma (Worley et al., 2008) and ovarian cancer (Iorio et al., 2007; Chen et al., 2008; Nam et al., 2008). The anti-invasion/metastasis property of miR-199a demonstrated in this study further supports the tumor suppressive role of this miRNA. Although both miR-199a-5p and miR-199a-3p are derived from the same precursor RNA, only miR-199a- 5p was identified to be downregulated in testicular tumor malignancy. The reason why only a miRNA is correlated with a phenotype whereas the other remains uncorrelated is not clear, possibly due to different stability of the mature miRNA molecules. Few reports from literature demonstrate co-dysregulation of both miRNA specimens. The miR-199a-3p is known to target mTOR and proto-oncogene c-MET in cancers and Smad1 during chondrogenesis (Kim et al., 2008; Lin et al., 2009; Fornari et al., 2010). It is differentially expressed during renal ischemia reperfusion injury and viral infection (Godwin et al., 2010; Santhakumar et al., 2010). The miR-199a-5p targets IKKβ in ovarian cancers, and Hif-1-α and Sirt1 in cardiac myocytes (Chen et al., 2008; Rane et al., 2009). It is upregulated in cervical carcinomas, and human embryonic stem cells treated with activin A (Lee et al., 2008; Tsai et al., 2010). The role of miR-199a-5p/3p in a variety of cellular events suggests that it is an important disease-related miRNA.
PODXL is another frequently cancer-upregulated protein (Somasiri et al., 2004; Casey et al., 2006; Hayatsu et al., 2008). It is an anti-adhesion transmembrane protein that inhibits cell–cell interaction through the charge-repulsive effects (Takeda et al., 2000). Disruption of cell–cell interaction at primary sites is a crucial step in developing an invasive phenotype. A previous study indicated that forced expression of PODXL in MCF-7 breast carcinoma cells perturbed cell-cell interaction (Somasiri et al., 2004). In this report, we demonstrate the link between PODXL and miR-199a-5p. By using luciferase assay, we showed miR-199a-5p could suppress PODXL through binding to a conserved site (Figure 5e). For the scramble miRNA control groups, the constructs carrying a conserved (pGL-C) or a poorly conserved (pGL-P) miR-199a-5p-binding site showed a lower luciferase activity than the no 3′-UTR control, probably due to the presence of other background miRNA-binding sites that interact with endogenous miRNAs (for example miR-145 and miR-181 are found in the flanking miR-199a-5p conserved site based on TargetScan and PicTar). Although we have shown that PODXL is a target of miR-199a-5p, we cannot rule out other targets that might modulate tumor invasion. In the other report, miR-199a-5p targets IKKβ in ovarian cancer (Chen et al., 2008). Our previous microarray data and others indicated that IKKβ was not changed in testicular cancers (data not shown). Therefore, we believe the same miRNA specimen may regulate different targets in different types of cancer.
In our xenografted animal model, expression of miR-199a in NT2 cells suppressed metastasis at day 49 and 64. However at a later stage (day 82), miR-199a was less effective. Lung and liver metastases expressing miR-199a-5p and miR-199a-3p in the NT2-199a group at day 82 were comparable to those of cultured NT2-199a cells (Supplementary Figure 5). The reason why miR-199a failed to suppress metastasis at this stage is elusive. It may be because the neoplastic cells have an alternative strategy to circumvent tumor suppression by miR-199a.
Comparison of methylation of miR-199a between seminomas and non-seminomas indicated that seminomas were generally more methylated than non-seminomas (Supplementary Figure 4a). However, the P-value of ‘non-cancerous vs cancerous’ comparision (P = 0.0005) is smaller than that of ‘seminomas vs nonseminomas’ comparison (P = 0.04). Although previous studies demonstrated that seminomas are relatively more hypomethylated than non-seminomas (Smiraglia et al., 2002; Netto et al., 2008), these studies reported methylation on a genome-wide scale. However, in our study we specifically focused on a specific locus of miR-199a. Similar to the observation for methylation, the P-values of ‘cancerous vs non-cancerous’ comparisons for miR-199a-5p/3p expressions are smaller (P < 0.00001) than those of ‘seminoma vs non-seminoma’ (P < 0.05). Our statistical analyses imply that methylation and expression of miR-199a are relevant to both histologic subtypes, however, seminomas seem to show a stronger statistical difference.
Collectively, we report that aberrant DNA methylation in an intron of a host gene DNM3 as a critical factor for testicular tumor malignancy by modulating the level of anti-metastatic miR-199a and its corresponding target PODXL.
Materials and methods
Cell lines and cell culture
NT2, Tera-1, Tera-2, NCCIT and HT cell lines were purchased from ATCC (Manassas, VA, USA). The 833K was provided by Dr YF Lau. HT, NT2 and its sublines (NT2-GFP, NT2-199a, NT2-VC and NT2-PODXLi) were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum. NCCIT and 833K were cultured in RPMI-1640 medium (Invitrogen) supplemented with 10% fetal bovine serum. Tera-1 and Tera-2 cells were cultured in McCoy’s 5a medium (modified) (ATCC) supplemented with 15% fetal bovine serum. All cells were maintained in a humidified incubator at 37°C with 5% CO2.
Normal and tumor tissues
DNA of testicular normal and metachronous tumor tissues were purchased from Oncomatrix (San Marcos, CA, USA). Testicular tumor tissue microarrays (T231 and TE2081) were purchased from US Biomax (Rockville, MD, USA). This panel includes 22 cases of normal, 13 cases of benign and 81 cases of malignant tumors (50 seminomas and 31 non-seminomas). Clinical stage of all tumor tissues was TNM graded and the pathology of patients were summarized in Supplementary Table 2.
Isolation of RNA and DNA from archived tissues and cultured cells
RNA was isolated from formalin-fixed paraffin-embedded tissues using the RecoverAll Total Nucleic Acid Isolation Kit (Ambion, Austin, TX, USA). RNA was isolated from cultured cells using mirVana miRNA Isolation Kit (Ambion) for miRNA expression analysis or using Trizol Reagent (Invitrogen) for mRNA expression analysis. Genomic DNA was isolated from formalin-fixed paraffin-embedded tissues using the EZ DNA Methylation-Direct Kit (Zymo Research, Orange, CA, USA), followed directly by bisulfite treatment. All procedures were performed according to manufacturers’ instructions.
Genomic bisulfite sequencing and Methylight quantitative PCR
Genomic bisulfite sequencing was performed as previously described (Cheung et al., 2010). For Methylight quantitative PCR, bisulfite-converted DNA was used for real-time PCR using a pair of custom-made TaqMan probes (Applied Biosystems, Foster City, CA, USA) specific for either methylated (M) or unmethylated (U) region of the promoter of miR-199a. Sequences of the probes are as follows: M: 6FAM-5′-TGCGTTGTGTCGTTGGAGAGATCG-3′-MGBNFQ; U: VIC-5′-TGTGTTGTGTTGTTGGAGAGAT TGTTA-3′-G-MGBNFQ. Methylation of miR-199a was calculated by: Cmeth = 100/(1 + 2(CtCG−CtTG))%, where CtCG and CtTG are the threshold cycles of M (FAM channel) and U (VIC channel) detectors, respectively (Eads et al., 2000).
Reverse transcription and real-time PCR of miRNA
Reverse transcription and real-time PCR of miRNA and mRNA Reverse transcription and real-time PCR of miRNA and mRNA were performed as previously described (Cheung et al., 2010). Relative miRNA expression was normalized by miR-191 (Peltier and Latham, 2008).
miRNA transfection and establishment of stable cell lines
The miR-199a-5p mimics, miRNA scramble control and miR-199a inhibitors were purchased from Ambion. Cells were transfected with indicated amount of miRNA molecules using Lipofectamine 2000 (Invitrogen). Cells were harvested for RNA or protein extraction 72 h after transfection. For establishment of stable cell lines NT2-GFP and NT2-199a, NT2 parent cells were infected with lentivirus carrying a vector of GFP (NT2-GFP) or GFP/miR-199a (NT2-199a) (System Biosciences, Mountain View, CA, USA). Seventy-two hours after infection, cells were sorted by GFP marker. For establishment of stable PODXL knockdown cell lines NT2-VC and NT2-PODXLi, NT2 parent cells were transfected with vectors expressing short-hairpin RNAs against PODXL or GFP (vector control) (Origene, Rockville, MD, USA). Stable RNAi sublines were selected by Puromycin. Four different short-hairpin RNA sequences were tested and the vector with highest RNAi efficiency was employed in subsequent experiments.
Wound healing assay
Monolayer cells on 12-well plate were scratched to generate the ‘wounds’ using a P10 pipette tip. The wells were gently washed and incubated at 37°C for 17–24 h. Images were captured with a microscope at × 10 (Carl Zeiss, Thornwood, NY, USA). Distance between the edges was measured by software AxioVison (Carl Zeiss). Three independent experiments, each with six replicates, were performed.
Cell invasion assay
In vitro cell invasion assay was performed using Growth Factor Reduced Matrigel Invasion Chambers with 8 µm pore size (BD Biosciences, San Diego, CA, USA). Cells (5 × 104) resuspended in serum-free Dulbecco’s modified Eagle’s medium were added to matrigel-coated inserts and placed in wells containing 0.6 ml of complete medium supplemented with 10% fetal bovine serum as chemoattractant. After 6 and 18 h of incubation at 37°C, cells that had not invaded the matrigel were removed from the interior sides of the inserts by cotton swabs. Invaded cells on the exterior sides were stained with crystal violet and counted with a microscope (Carl Zeiss). Three independent experiments, each with three replicates, were performed.
Cloning of 3′-UTR and luciferase reporter assay
The flanking sequences containing the predicted miRNA binding sites were cloned to the Firefly luciferase vector pGL4.13 (Promega, Madison, WI, USA). The seed sequence in the mutant constructs was changed to its complementary base using the Phusion Site-directed Mutagenesis Kit (Finnzymes, Woburn, MA, USA). A volume of 100 ng of pGL4.13-UTR was co-transfected with 33 nm of miRNA mimics or scramble control (12-well format in triplicate) in NT2 cells. Forty-eight hours after transfection, cells were lysed and luciferase activity was measured by Luminometer (Promega, Sunnyvale, CA, USA) using the Dual-Luciferase Reporter Assay System Kit (Promega). Renilla luciferase was used as normalization control.
Cloning of miR-199a promoter and in vitro methylation
The promoter region of miR-199a (+9:−471) was amplified by PCR and ligated to Firefly luciferase reporter vector pGL3-Basic (Promega). A volume of 100 ng of pGL3 and 2 ng of pGL4.73 plasmids were co-transfected to NIH-3T3 cells. Primers for cloning of promoter are as follows: forward 5′-AAG AGCTCACTTTCATTTGGCCACA-3′; reverse 5′-A TCTCCAGAAGCTTCCTTCTATGT-3′. In vitro methylation was performed as previously described (Singal et al., 1997). Briefly, promoter region of miR-199a was restriction-cut from pGL3 construct, gel-purified and in vitro methylated by CpG methyltransferase M.SssI (New England Biolabs, Ipswich, MA, USA). Methylation of promoter was verified by methylation sensitive restriction enzyme HhaI. Either methylated or unmethylated promoter DNA was ligated to the luciferase vector in a molar ratio of 1:1. The ligated products (3 µg) were used directly for transfection to NIH-3T3 cells.
Western blot analysis
Western blot analysis was performed as previously described (Pang et al., 2009). Primary antibodies used were as follows: PODXL (clone 3D3, 3 µg/ml, Santa Cruz Biotechnology, Santa Cruz, CA, USA); GAPDH (1:8000, Genway, San Diego, CA, USA). Secondary antibodies (1:10000) were purchased from Bio-Rad (Hercules, CA, USA).
Immunohistochemistry
Immunohistochemistry was performed as previously described (Li et al., 2007). Briefly, formalin-fixed paraffin-embedded tissue arrays were deparafinized in xylenes and hydrated in a gradual series of ethanol. Antigen retrieval was done by heating the slides in citrate buffer at 100°C. The slides were probed with anti-PODXL antibody (1:200, Atlas Antibodies, Stockholm, Sweden) overnight at room temperature. Signal was developed using DAB Histochemistry Kit (Invitrogen). Cells were counter stained with hematoxylin. Expression of PODXL was scored as 0 (negative: no observable positive), 1 (weak: < 25% positive), 2 (moderate: 25–75% positive) and 3 (strong: > 75% positive) by two experienced scientists separately. Triplicate experiments were performed.
Animal studies of tumor growth and metastasis
For in vivo tumor growth study, 5-week-old male athymic nude mice (Charles River, Wilmington, MA, USA) were injected subcutaneously with 1 × 107 NT2-GFP or NT2-199a (n = 10 for each group) in each flank of each mouse. Mouse weight and tumor size were measured every Monday and Thursday. Tumor volume was calculated as follows: length × width2 × 1/2. All mice were killed 60 days after implantation. The mean tumor volume ± s.e.m. of each group was calculated. For metastasis study, 5-week-old male athymic nude mice were injected intravenously with 1 × 106 of NT2-GFP or NT2-199a cells (n = 11 for each group) via tail vein. Three mice from each group were killed 49 and 64 days after implantation. The remaining mice were killed 82 days after implantation. Metastasis was examined in major organs including brain, liver, kidney, lung and testis by necropsy and confirmed by histochemistry and expression of GFP.
Statistical analysis
Mann–Whitney Test was used to compare the difference of methylation, miR-199a-5p and miR-199a-3p expressions between non-cancerous and malignant groups. The differences for wound healing assay, invasion assay, tumor growth and luciferase assay were analyzed by two-tailed Student’s t-test, assuming equal variance. The correlations between miR-199a expression and methylation, miR-199a expression and PODXL level, tumor invasiveness and PODXL level, were analyzed by Spearman’s rank correlation. The difference in PODXL expression in seminomas and non-seminomas was analyzed by χ2-test. P < 0.05 is considered statistically significant.
Acknowledgements
We thank Dr Yun-Fai Chris Lau for providing the 833K cell line. This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), Eunice Kennedy Shriver National Institute of Child Health and Human Development and the Chinese University of Hong Kong.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Aleman A, Cebrian V, Alvarez M, Lopez V, Orenes E, Lopez-Serra L, et al. Identification of PMF1 methylation in association with bladder cancer progression. Clin Cancer Res. 2008;14:8236–8243. doi: 10.1158/1078-0432.CCR-08-0778. [DOI] [PubMed] [Google Scholar]
- Andrews PW, Damjanov I, Simon D, Banting GS, Carlin C, Dracopoli NC, et al. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab Invest. 1984;50:147–162. [PubMed] [Google Scholar]
- Casey G, Neville PJ, Liu X, Plummer SJ, Cicek MS, Krumroy LM, et al. Podocalyxin variants and risk of prostate cancer and tumor aggressiveness. Hum Mol Genet. 2006;15:735–741. doi: 10.1093/hmg/ddi487. [DOI] [PubMed] [Google Scholar]
- Chakrabarty A, Tranguch S, Daikoku T, Jensen K, Furneaux H, Dey SK. MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc Natl Acad Sci USA. 2007;104:15144–15149. doi: 10.1073/pnas.0705917104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Alvero AB, Silasi DA, Kelly MG, Fest S, Visintin I, et al. Regulation of IKKbeta by miR-199a affects NF-kappaB activity in ovarian cancer cells. Oncogene. 2008;27:4712–4723. doi: 10.1038/onc.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HH, Lee TL, Davis AJ, Taft DH, Rennert OM, Chan WY. Genome-wide DNA methylation profiling reveals novel epigenetically regulated genes and non-coding RNAs in human testicular cancer. Br J Cancer. 2010;102:419–427. doi: 10.1038/sj.bjc.6605505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HH, Lee TL, Rennert OM, Chan WY. DNA methylation of cancer genome. Birth Defects Res C Embryo Today. 2009;87:335–350. doi: 10.1002/bdrc.20163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Fornari F, Milazzo M, Chieco P, Negrini M, Calin GA, Grazi GL, et al. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70:5184–5193. doi: 10.1158/0008-5472.CAN-10-0145. [DOI] [PubMed] [Google Scholar]
- Friedman LM, Dror AA, Mor E, Tenne T, Toren G, Satoh T, et al. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc Natl Acad Sci USA. 2009;106:7915–7920. doi: 10.1073/pnas.0812446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin JG, Ge X, Stephan K, Jurisch A, Tullius SG, Iacomini J. Identification of a microRNA signature of renal ischemia reperfusion injury. Proc Natl Acad Sci USA. 2010;107:14339–14344. doi: 10.1073/pnas.0912701107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayatsu N, Kaneko MK, Mishima K, Nishikawa R, Matsutani M, Price JE, et al. Podocalyxin expression in malignant astrocytic tumors. Biochem Biophys Res Commun. 2008;374:394–398. doi: 10.1016/j.bbrc.2008.07.049. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. at Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345–352. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- Kim S, Lee UJ, Kim MN, Lee EJ, Kim JY, Lee MY, et al. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) J Biol Chem. 2008;283:18158–18166. doi: 10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- Koch LK, Zhou H, Ellinger J, Biermann K, Holler T, von Rucker A, et al. Stem cell marker expression in small cell lung carcinoma and developing lung tissue. Hum Pathol. 2008;39:1597–1605. doi: 10.1016/j.humpath.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Lee DY, Shatseva T, Jeyapalan Z, Du WW, Deng Z, Yang BB. A 3′-untranslated region (3′UTR) induces organ adhesion by regulating miR-199a* functions. PLoS One. 2009;4:e4527. doi: 10.1371/journal.pone.0004527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Choi CH, Choi JJ, Park YA, Kim SJ, Hwang SY, et al. Altered MicroRNA expression in cervical carcinomas. Clin Cancer Res. 2008;14:2535–2542. doi: 10.1158/1078-0432.CCR-07-1231. [DOI] [PubMed] [Google Scholar]
- Li LC, Zhao H, Nakajima K, Oh BR, Ribeiro Filho LA, Carroll P, et al. Methylation of the E-cadherin gene promoter correlates with progression of prostate cancer. J Urol. 2001;166:705–709. [PubMed] [Google Scholar]
- Li Y, Tabatabai ZL, Lee TL, Hatakeyama S, Ohyama C, Chan WY, et al. The Y-encoded TSPY protein: a significant marker potentially plays a role in the pathogenesis of testicular germ cell tumors. Hum Pathol. 2007;38:1470–1481. doi: 10.1016/j.humpath.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin EA, Kong L, Bai XH, Luan Y, Liu CJ. miR-199a, a bone morphogenic protein 2-responsive MicroRNA, regulates chondrogenesis via direct targeting to Smad1. J Biol Chem. 2009;284:11326–11335. doi: 10.1074/jbc.M807709200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- Netto GJ, Nakai Y, Nakayama M, Jadallah S, Toubaji A, Nonomura N, et al. Global DNA hypomethylation in intratubular germ cell neoplasia and seminoma, but not in nonseminomatous male germ cell tumors. Mod Pathol. 2008;21:1337–1344. doi: 10.1038/modpathol.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang AL, Peacock S, Johnson W, Bear DH, Rennert OM, Chan WY. Cloning, characterization, and expression analysis of the novel acetyltransferase retrogene Ard1b in the mouse. Biol Reprod. 2009;81:302–309. doi: 10.1095/biolreprod.108.073221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, et al. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104:879–886. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E. Mechanisms of cancer cell invasion. Curr Opin Genet Dev. 2005;15:87–96. doi: 10.1016/j.gde.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Santhakumar D, Forster T, Laqtom NN, Fragkoudis R, Dickinson P, Abreu-Goodger C, et al. Combined agonist-antagonist genome-wide functional screening identifies broadly active antiviral microRNAs. Proc Natl Acad Sci USA. 2010;107:13830–13835. doi: 10.1073/pnas.1008861107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopperle WM, Kershaw DB, DeWolf WC. Human embryonal carcinoma tumor antigen, Gp200/GCTM-2, is podocalyxin. Biochem Biophys Res Commun. 2003;300:285–290. doi: 10.1016/s0006-291x(02)02844-9. [DOI] [PubMed] [Google Scholar]
- Singal Rc, Ferris R, Little JA, Wang SZ, Ginder GD. Methylation of the minimal promoter of an embryonic globin gene silences transcription in primary erythroid cells. Proc Natl Acad Sci USA. 1997;94:13724–13729. doi: 10.1073/pnas.94.25.13724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiraglia DJ, Szymanska J, Kraggerud SM, Lothe RA, Peltomaki P, Plass C. Distinct epigenetic phenotypes in seminomatous and nonseminomatous testicular germ cell tumors. Oncogene. 2002;21:3909–3916. doi: 10.1038/sj.onc.1205488. [DOI] [PubMed] [Google Scholar]
- Somasiri A, Nielsen JS, Makretsov N, McCoy ML, Prentice L, Gilks CB, et al. Overexpression of the anti-adhesin podocalyxin is an independent predictor of breast cancer progression. Cancer Res. 2004;64:5068–5073. doi: 10.1158/0008-5472.CAN-04-0240. [DOI] [PubMed] [Google Scholar]
- Takeda T, Go WY, Orlando RA, Farquhar MG. Expression of podocalyxin inhibits cell-cell adhesion and modifies junctional properties in Madin-Darby canine kidney cells. Mol Biol Cell. 2000;11:3219–3232. doi: 10.1091/mbc.11.9.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- Tsai ZY, Singh S, Yu SL, Kao LP, Chen BZ, Ho BC, et al. Identification of microRNAs regulated by activin A in human embryonic stem cells. J Cell Biochem. 2010;109:93–102. doi: 10.1002/jcb.22385. [DOI] [PubMed] [Google Scholar]
- Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2009;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts GS, Futscher BW, Holtan N, Degeest K, Domann FE, Rose SL. DNA methylation changes in ovarian cancer are cumulative with disease progression and identify tumor stage. BMC Med Genomics. 2008;1:47. doi: 10.1186/1755-8794-1-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley LA, Long MD, Onken MD, Harbour JW. Micro-RNAs associated with metastasis in uveal melanoma identified by multiplexed microarray profiling. Melanoma Res. 2008;18:184–190. doi: 10.1097/CMR.0b013e3282feeac6. [DOI] [PubMed] [Google Scholar]
- Yin G, Chen R, Alvero AB, Fu HH, Holmberg J, Glackin C, et al. TWISTing stemness, inflammation and proliferation of epithelial ovarian cancer cells through MIR199A2/214. Oncogene. 2010;29:3545–3553. doi: 10.1038/onc.2010.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]