Abstract
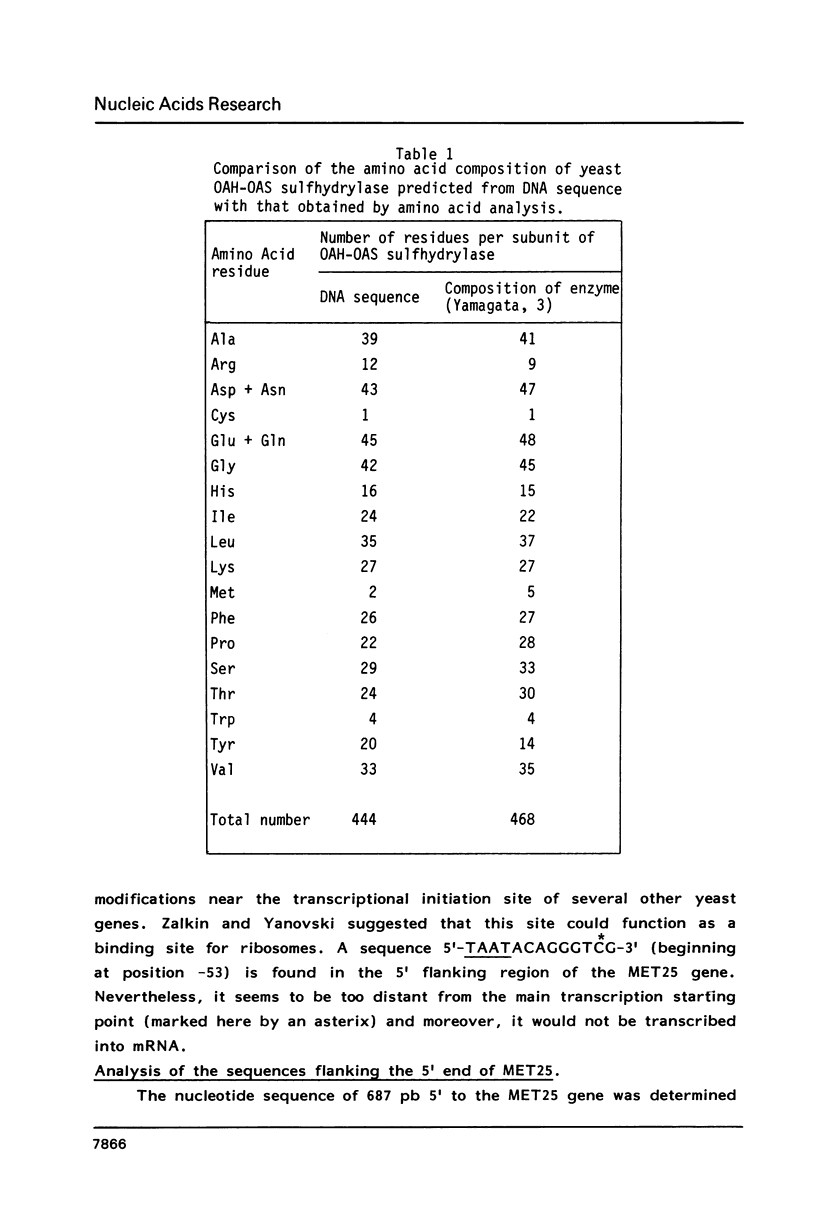
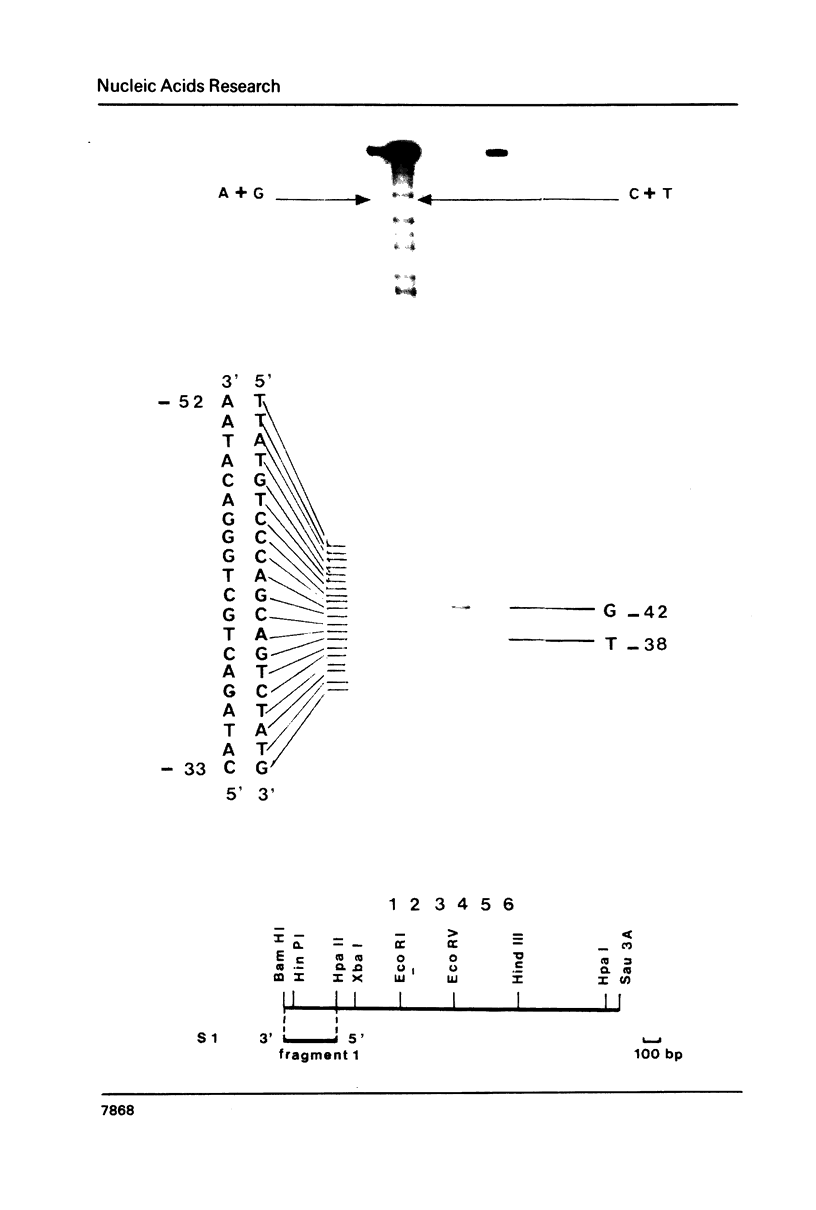
To elucidate further the molecular basis of the specific regulatory mechanism modulating the expression of the genes implicated in methionine metabolism, we have cloned and characterized two genes, MET3 and MET25, and shown that the regulation of their expression is transcriptional. The sequence of the cloned yeast MET25 gene which encodes the O-acetyl homoserine - O-acetyl serine (OAH-OAS) sulfhydrylase is reported here along with its 5' and 3' flanking regions. The amino acid composition predicted from the DNA sequence is in good agreement with that determined by hydrolysis of the purified enzyme. In the 5' flanking region the signal for general amino acid control was not found, corroborating our previous finding that the synthesis of OAH-OAS sulfhydrylase is not submitted to general control. The transcription start points have been determined. The 5' and 3' flanking regions of the MET25 gene suggest initiation and termination signals similar to those associated with other yeast genes.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Cherest H., Eichler F., Robichon-Szulmajster H. Genetic and regulatory aspects of methionine biosynthesis in Saccharomyces cerevisiae. J Bacteriol. 1969 Jan;97(1):328–336. doi: 10.1128/jb.97.1.328-336.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherest H., Eichler F., Robichon-Szulmajster H. Genetic and regulatory aspects of methionine biosynthesis in Saccharomyces cerevisiae. J Bacteriol. 1969 Jan;97(1):328–336. doi: 10.1128/jb.97.1.328-336.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherest H., Nguyen N. T., Surdin-Kerjan Y. Transcriptional regulation of the MET3 gene of Saccharomyces cerevisiae. Gene. 1985;34(2-3):269–281. doi: 10.1016/0378-1119(85)90136-2. [DOI] [PubMed] [Google Scholar]
- Cherest H., Surdin-Kerjan Y., Antoniewski J., de Robichon-Szulmajster H. Effects of regulatory mutations upon methionine biosynthesis in Saccharomyces cerevisiae: loci eth2-eth3-eth10. J Bacteriol. 1973 Sep;115(3):1084–1093. doi: 10.1128/jb.115.3.1084-1093.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delforge J., Messenguy F., Wiame J. M. The regulation of arginine biosynthesis in Saccharomyces cerevisiae. The specificity of argR- mutations and the general control of amino-acid biosynthesis. Eur J Biochem. 1975 Sep 1;57(1):231–239. doi: 10.1111/j.1432-1033.1975.tb02295.x. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nicolet C. M., Chenevert J. M., Friedberg E. C. The RAD2 gene of Saccharomyces cerevisiae: nucleotide sequence and transcript mapping. Gene. 1985;36(3):225–234. doi: 10.1016/0378-1119(85)90177-5. [DOI] [PubMed] [Google Scholar]
- Oberto J., Davison J. Expression of chicken egg white lysozyme by Saccharomyces cerevisiae. Gene. 1985;40(1):57–65. doi: 10.1016/0378-1119(85)90024-1. [DOI] [PubMed] [Google Scholar]
- Sangsoda S., Cherest H., Surdin-Kerjan Y. The expression of the MET25 gene of Saccharomyces cerevisiae is regulated transcriptionally. Mol Gen Genet. 1985;200(3):407–414. doi: 10.1007/BF00425724. [DOI] [PubMed] [Google Scholar]
- Schürch A., Miozzari J., Hütter R. Regulation of tryptophan biosynthesis in Saccharomyces cerevisiae: mode of action of 5-methyl-tryptophan and 5-methyl-tryptophan-sensitive mutants. J Bacteriol. 1974 Mar;117(3):1131–1140. doi: 10.1128/jb.117.3.1131-1140.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Wolfner M., Yep D., Messenguy F., Fink G. R. Integration of amino acid biosynthesis into the cell cycle of Saccharomyces cerevisiae. J Mol Biol. 1975 Aug 5;96(2):273–290. doi: 10.1016/0022-2836(75)90348-4. [DOI] [PubMed] [Google Scholar]
- Yamagata S. O-Acetylserine and O-acetylhomoserine sulfhydrylase of yeast. Subunit structure. J Biochem. 1976 Oct;80(4):787–797. doi: 10.1093/oxfordjournals.jbchem.a131339. [DOI] [PubMed] [Google Scholar]
- Yamagata S., Takeshima K., Naiki N. Evidence for the identity of O-acetylserine sulfhydrylase with O-acetylhomoserine sulfhydrylase in yeast. J Biochem. 1974 Jun;75(6):1221–1229. doi: 10.1093/oxfordjournals.jbchem.a130505. [DOI] [PubMed] [Google Scholar]
- Zalkin H., Yanofsky C. Yeast gene TRP5: structure, function, regulation. J Biol Chem. 1982 Feb 10;257(3):1491–1500. [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]



