Abstract
A prior screen identified dozens of Drosophila melanogaster mutants that possess defective long-term memory (LTM). Using spaced olfactory conditioning, we trained 26 of these mutant lines to associate an odor cue with electric shock and then examined the memory of this conditioning 24 h later. All of the mutants tested revealed a deficit in LTM compared to the robust LTM observed in control flies. We used in vivo functional optical imaging to measure the magnitude of a previously characterized LTM trace, which is manifested as increased calcium influx into the axons of α/β mushroom body neurons in response to the conditioned odor. This memory trace was defective in all 26 of the LTM mutants. These observations elevate the significance of this LTM trace given that 26 independent mutants all exhibit a defect in the trace, and further suggest that the calcium trace is a fundamental mechanism underlying Drosophila LTM.
Introduction
Drosophila melanogaster has been extensively used to study olfactory memory formation at the molecular/cellular and systems neuroscience levels of analysis (Davis, 2005). Flies can be conditioned to avoid an odor that is paired with a negative reinforcer like an electric shock and display this memory for a time period that is dependent on the training regimen. For example, a single cycle of conditioning produces robust short-term memory (STM) that decays over a period of ∼1 d (Tully et al., 1994; Beck et al., 2000; Pascual and Préat, 2001). Long-term memory (LTM) is generated by repeated training sessions separated by a rest period, requires normal protein synthesis and Creb (cAMP response element-binding protein) and calcium-calmodulin-dependent protein kinase II (CaMKII) activity at the time of conditioning, and can endure for several days (Tully et al., 1994; Perazzona et al., 2004; Yu et al., 2006; Akalal et al., 2010).
Combined behavioral and neuroanatomical studies have identified the mushroom bodies (MBs) as crucial for olfactory associative learning and memory in insects. MB axons extend to form five distinct lobes of neuropil (α, β, α′, β′, and γ). The functional diversity of the different classes of MB neurons has long been recognized based on the differential expression of gene and protein markers (Yang et al., 1995; Crittenden et al., 1998) and behavioral experiments that target expression of rescuing transgenes in specific subsets of these neurons (Zars et al., 2000; McGuire et al., 2003; Blum et al., 2009). In vivo functional optical imaging has also allowed the discovery of memory traces (Liu and Davis, 2006), which are often detected as increased calcium influx into the processes of neurons being studied. One cellular memory trace that may underlie LTM was discovered as an increased calcium response in the α collaterals of the α/β MB neurons following spaced olfactory conditioning (Yu et al., 2006). This memory trace requires normal protein synthesis, normal Creb activity, and intact signaling through CaMKII (Yu et al., 2006; Akalal et al., 2010).
Here, we have reevaluated the significance of this memory trace to LTM by assaying the memory trace in 26 independently isolated mutants, initially identified from a screen for mutants that specifically impair LTM (Dubnau et al., 2003). We have confirmed the LTM impairment in all 26 individual mutants and, surprisingly, discovered that all 26 independent mutants abolished the LTM trace in parallel with the impairment of behavioral memory. Given that the gene products of the 26 LTM mutants represent diverse cellular functions, these results suggest that the calcium-based memory trace is fundamental to long-term behavioral memory.
Materials and Methods
Transgenic animals and fly culture.
The Drosophila LTM mutant collection was previously described and obtained from Dr. J. Dubnau (Dubnau et al., 2003). Briefly, these mutants were generated by P-element transposon mutagenesis, yielding mutants possessing a single, homozygous, and adult-viable transposon insertion on either the second or third chromosome. We first mapped the insertion in these mutants to the second or third chromosomes. We then selected mutants with insertions on the third chromosome for these studies since the Uas-G-CaMP and c739-Gal4 elements used for optical imaging are located on the second chromosome. Flies were then constructed to contain all three elements: c739-Gal4; Uas-G-CaMP; LTM mutant insertion/LTM mutant insertion. Flies were cultured on standard medium at room temperature and transferred overnight to a 25°C incubator before training.
Behavioral assays.
Olfactory learning was assayed using an olfactory classical conditioning procedure that allows the performance gains occurring from the pairing of a single odor with electric shock to be measured relative to naive flies (Yu et al., 2006). In this assay, flies were exposed to two odors in succession, one odor [the conditioned stimulus (CS+)] paired with electric shock pulses [unconditioned stimulus (US)] followed by a counter odor (the CS−) without electric shock during the training phase. We used the odor benzaldehyde (Ben) as the CS+ and 3-octanol (Oct) as the CS−. Multiple spaced training trials were performed, which are required to induce a long-lasting, protein-synthesis-dependent memory (Tully et al., 1994; Yu et al., 2006). We used 5× spaced training to generate LTM with an interval of 15 min between each training cycle. After training, the flies were incubated at 25°C for 24 h before testing. Each group of animals was tested only once for distribution between the CS+ and CS− in a T-maze at 24 h after training and their avoidance of the CS+ was quantified. A corresponding naive control for each trained group underwent all of the same manipulations as the trained flies except that they were not administered odor or electric shock at the time that other groups were trained. The performance indices were calculated as a ΔPI by subtracting the naive score from the score of the corresponding trained group. In all cases, only experiments where the naive flies exhibited naive performance scores that were not significantly different from zero (t test) were used. These procedures allowed us to make meaningful comparisons between groups since performance was normalized to naive performance (Yu et al., 2006). A variety of controls, including performance after backwards, CS-only, and US-only conditioning, have been performed here and in prior studies (Yu et al., 2006) to guard against nonassociative effects entering the calculation of the ΔPI. Nevertheless, we cannot eliminate the possibility that nonassociative factors that remain undetected by these controls remain hidden in the performance indices.
Functional cellular imaging.
We have previously described protocols for functional cellular imaging using the calcium reporter transgene Uas-G-CaMP driven by the Gal4 driver, c739-Gal4, an enhancer detector element that drives expression specifically in the α/β MB neurons (Yu et al., 2006). Briefly, mutant and control flies containing both a Gal4 driver and Uas-G-CaMP were separated before behavioral testing from the remainder of the trained flies. The bulk of the trained flies were tested for behavioral memory as described above to obtain behavioral scores. Flies to be imaged were mounted stably under a laser-scanning confocal microscope in pipette tips with their heads exposed and secured to the tip opening with silicon cement. A small region of cuticle was removed from the top of the head capsule and this area subsequently covered with a piece of plastic wrap. Imaging was performed using a 20× objective and an excitation line of 488 nm. The emitted light was collected from 520 ± 15 nm. Under these conditions, the thickness of the optical slice collected was calculated to be 32 μm, which is sufficient to collect the fluorescence throughout the bulbous tip of the α lobe. A small piece of filter paper was placed inside a syringe barrel that was in line with the pressurized air flowing at a rate of 100 ml/min. Concentrated odorants (0.1 μl of Ben or Oct) were diluted with 0.9 μl of mineral oil and spread on the filter paper. For odorant delivery, a three-way Teflon valve under the control of a programmable timer was used, such that fresh air could be delivered to the animals for a predetermined period of time with an instantaneous switch to odor-laced air without altering the overall flow rate. The CS+ and CS− odors were delivered with an interstimulus interval of 5 min. In prior experiments, we have shown that although the α/β neurons respond with calcium influx to odors delivered to the antennae and electric shock pulses delivered to the fly's abdomen, only the α branch of the α/β neurons form an LTM trace (Yu et al., 2006). We collected imaging data across time at a depth in the Drosophila brain at which the tip of the α lobe neuropil exhibits its largest diameter and strongest intensity to ensure that the center of the bulbous tip was found and fluorescence across volume of the tip was collected.
Data analysis.
Images were acquired at ∼5 frames/s at a resolution of 256 × 256 pixels followed by image data analysis as previously described (Yu et al., 2004, 2005, 2006). Regions of interest (the tip of the α branch) were circumscribed and a pseudocolor image of the %ΔF/Fo ratio produced. The value Fo was calculated for each pixel within the region of interest as the fluorescence before odor application as averaged over five successive frames. The value ΔF was calculated for each pixel within the region of interest as the difference between the maximum average intensity during the 3 s odor application for five successive frames and Fo. The maximum average intensity was extracted from the intensities averaged over a five-frame sliding window. The maximum of all these average intensities was used to represent the odor response to calculate the %ΔF/Fo.
Results
LTM mutants exhibited significant reduction in 24 h memory following spaced forward conditioning
Twenty-six lines identified previously to have impaired LTM (Table 1) (Dubnau et al., 2003) were selected for our studies along with control flies that carry c739-Gal4 and Uas-G-CaMP only. These mutants represent genes with diverse cellular functions, including cell adhesion receptors, transcription factors, metabolic enzymes, structural proteins, etc. Flies were trained using a 5× spaced conditioning protocol and tested for 24 h performance in the olfactory conditioning assay. Control flies were tested for 24 h LTM at multiple times across the time period that these studies were performed to verify that the spaced conditioning protocol was effective in producing significant LTM. The mean ΔPI for the control genotype (Fig. 1A) was at ∼0.4. Of the 26 lines we tested, 23 lines exhibited a significant reduction in LTM performance in the initial survey (Fig. 1A).
Table 1.
Long-term memory mutants studied for the long-term memory trace
| Mutant line | Mutant name | Genes associated with P-element insertion positions |
|---|---|---|
| E3945 | Premjera | CG1910 (novel) |
| E2655 | Milord-2 | CG9755 (pumilio; translational regulator) |
| E3548 | Jack | CG2765 (novel) and CG2746 (RPL19; ribosome protein) |
| D0264 | Rafael | CG5208 (novel) and Dad (negative regulator of TGF-β) |
| E0350 | Beluy-2 | CG6310 (novel) and CG7858 (mocs1; molybdenum cofactor biosynthesis) |
| E1023 | Chingis Khan | CG7524 (Src64B; src oncogene family; predicted tyrosine kinase) |
| D0067 | Norka | CG11962 (novel) and CG10901 (oskar; mRNA localization) |
| E0389 | Rijiy I | CG5620 (Zn finger transcription factor) and CG10429 (novel) |
| E0627 | Gryzun | CG17569 (novel) |
| E3803 | Murashka-2 | CG9381 (RING finger protein) |
| E1847 | Box | CG4919 (Gclm; Glutamate-cysteine ligase modifier subunit) |
| E1654 | Chyorny | CG9922 (novel) and CG3143 (foxo; predicted transcription factor) |
| E1715 | Baikal | CG14873 (pxb; Hh/smo signaling and neurogenesis) |
| E1877 | Moladietz | CG4482 (protein binding) |
| E3947 | Premjera | CG1910 (novel) and CG2245 (nero; predicted deoxyhypusine monooxygenase activity) |
| D0283 | Milkah-1 | CG12426 (novel) and CG5017 (nucleosome assembly chaperone) |
| D0417 | Ruslan | CG6669 (klingon; Immunoglobulin superfamily) |
| E4203 | Visgun | CG16707 (endolyn homolog; membrane protein) |
| E3503 | Trezor | CG11711 (Mob2; protein binding) |
| D0981 | Mirta | CG14696 (novel) and CG4674 (novel) |
| D0487 | Milkah-2 | CG12426 (novel) and CG5017 (nucleosome assembly chaperone) |
| E0391 | Diana | CG14696 (novel) and CG4674 (novel) |
| E3145 | Dikar | CG32393 (novel) |
| D0668 | Beluy-1 | CG6310 (novel) and CG7858 (mocs1; molybdenum cofactor biosynthesis) |
| C0167 | Pastrel | CG8588 (novel) |
| C0150 | Krasavietz | CG2922 (exba; translational regulation, ribosome binding, translation initiation factor binding) |
Mutant names, line designations, and the positions of P-element insertion were previously reported (Dubnau et al., 2003, Berger et al., 2008). The table also indicates the genes that are associated with the insertion sites. Gene annotations are from Flybase (http://flybase.bio.indiana.edu).
Figure 1.
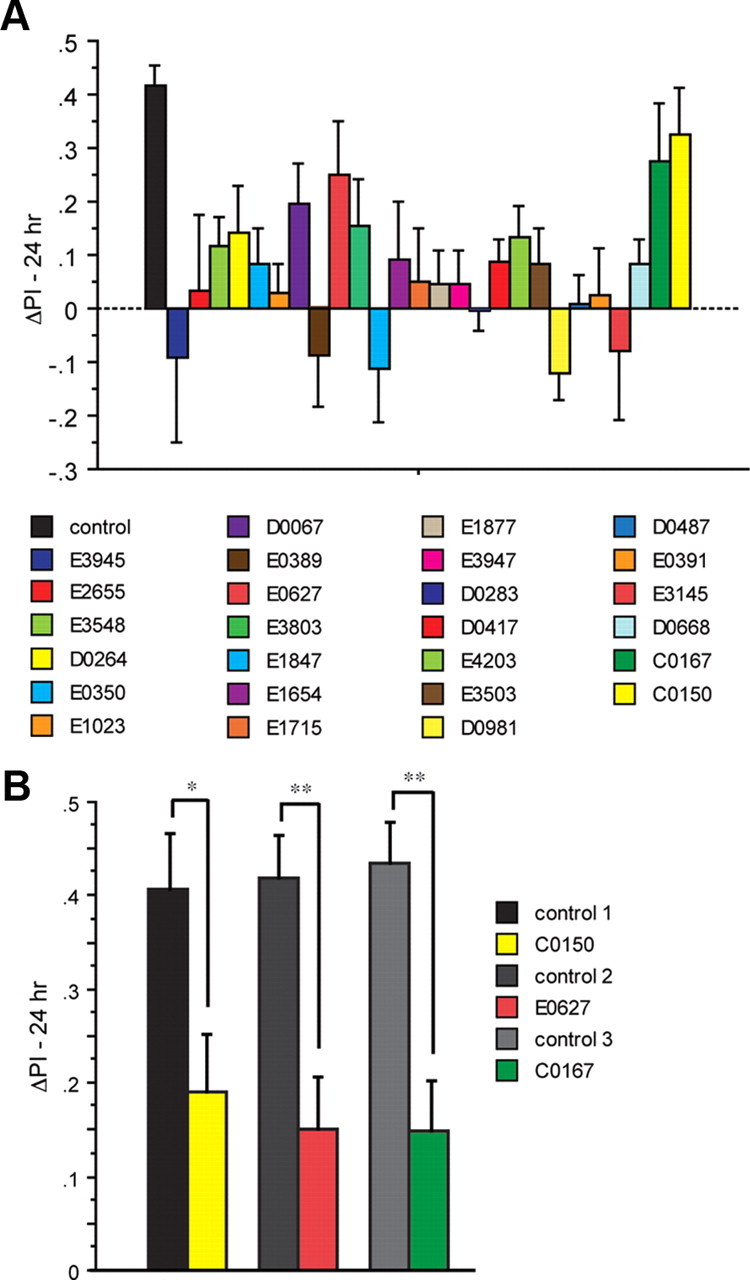
Long-term memory in 26 LTM mutants. A, An initial screen of mutant flies carrying one copy of c739-Gal4 and one copy of Uas-G-CaMP was performed using a 5× spaced conditioning protocol. Flies received forward conditioning with 1 min exposure to the CS+ odor (Ben) with 12 electric shock pulses (90 V), followed by 1 min exposure to the CS− odor (Oct) without electric shock after a 30 s exposure to fresh air. The training protocol was performed a total of five times with a 15 min intertrial interval (ITI). Flies were transferred to a T-maze at 24 h after conditioning and tested for behavioral memory. Naive groups of flies were carried through the same manipulations as the conditioned animals except that they were not exposed to odor and electric shock. Each conditioned group of flies was tested in parallel with a naive group. Some flies were separated before behavioral testing and analyzed for cellular memory by functional imaging (Fig. 2). The performance gains of the trained flies were measured as ΔPI. This was computed by subtracting the score of a naive group of flies from the corresponding conditioned group. In all cases, we chose experiments wherein the average scores of naive animals for each group were not statistically significant (Wilcoxon test) from zero to prevent floor and ceiling effects. For the c739; Uas-G-CaMP control flies, spaced conditioning was performed a total of three times across the several months that this experiment was conducted with an n = 6 for each experiment. The 24 h memory scores for control groups (∼0.4) were within the normal range of LTM scores we have come to expect and comparable to the LTM scores obtained by differential conditioning procedures that average the performance gains using two different CS+ odors (Tully et al., 1994; Perazzona et al., 2004; Yu et al., 2006). The ΔPI values were subjected to nonparametric tests; i.e., a Kruskal–Wallis test for multiple comparisons with genotype as the main factor, Mann–Whitney U test for comparing two independent samples, and Wilcoxon matched pairs test to compare single performance indices against zero. p values were corrected for the multiple comparisons to the control using the Benjamini and Hochberg false discovery rate. Out of the 26 lines tested, 22 yielded p values that were significantly different from the control (Kruskal–Wallis statistic 64.67, p < 0.0001; corrected Mann–Whitney pairwise comparisons, p ≤ 0.023). One mutant (E3803) was marginally nonsignificant (corrected Mann–Whitney pairwise comparison, p ≤ 0.051). This mutant was not selected for retesting given the marginally nonsignificant probability and because this is a replication experiment of Dubnau et al. (2003). The mutants C0150, E0627, and c0167 yielded p values that were not significantly different from the control (corrected Mann–Whitney pairwise comparisons, p ≥ 0.136. n = 12 for all groups). Error bars are the SEM. B, A second test of the three LTM mutants that failed to reach significance. Flies from each mutant genotype (C0150, E0627, and c0167) were trained using the 5× spaced conditioning protocol and compared to the c739; Uas-G-CaMP control. In each case, the mutant group performed significantly differently from its corresponding control group (Mann–Whitney pairwise comparisons, p ≤ 0.0433; n = 12 for all groups). Significance levels are indicated (*p ≤ 0.05, **p ≤ 0.01). Error bars are the SEM.
We retested the three lines that failed to reach significance individually alongside the control c739; GCaMP to determine whether the lines had normal LTM or were LTM mutants but statistical outliers. The retests indicated that each line possessed an LTM deficit compared to the control group (Fig. 1B). These behavioral results confirm the classification of these 26 lines as LTM mutants as originally assigned (Dubnau et al., 2003).
LTM mutants exhibited significant reduction of the memory trace in the α/β MB neurons following spaced forward conditioning
Some flies were removed from each of the trained groups immediately before behavioral testing at 24 h after learning. These flies were mounted for optical imaging and tested for calcium responses to both the CS+ and CS− odors. We have previously shown that although the α and β branches of the α/β neurons respond to odors delivered to the antennae and electric shock pulses delivered to the fly's abdomen, the LTM trace that is revealed as increased calcium influx in response to the CS+ odor is branch specific, occurring only in the α branch of the α/β neurons (Yu et al., 2006). We collected imaging data across time at the tip of the α branch of the α/β neurons. Surprisingly, all of the mutant groups showed significantly reduced calcium influx in response to the CS+ when compared to the response of control flies (Fig. 2). The mutants elicited %ΔF/Fo ratios to the CS+ odor that are typical for naive flies, flies trained using conditioning protocols that fail to produce LTM (backwards conditioning; Yu et al., 2006), and for the response of all genotypes to the CS− odor. This %ΔF/Fo ratio was typically within the 4–7% range, whereas control flies typically present %ΔF/Fo ratios of 12–14% (Fig. 2) (Yu et al., 2006). The CS+ odor-elicited calcium responses for the control group in this experiment were nearly twice as large as any other group (Fig. 2). In contrast, all groups including the control flies exhibited similar %ΔF/Fo ratios for the CS− odor. Thus, these data indicate that spaced forward conditioning induces cellular changes that lead to an increased calcium influx into the α branch of the α/β MB neurons in response to the CS+ odor when measured at 24 h after conditioning and that this LTM trace is significantly decreased in 26 independent LTM mutants.
Figure 2.
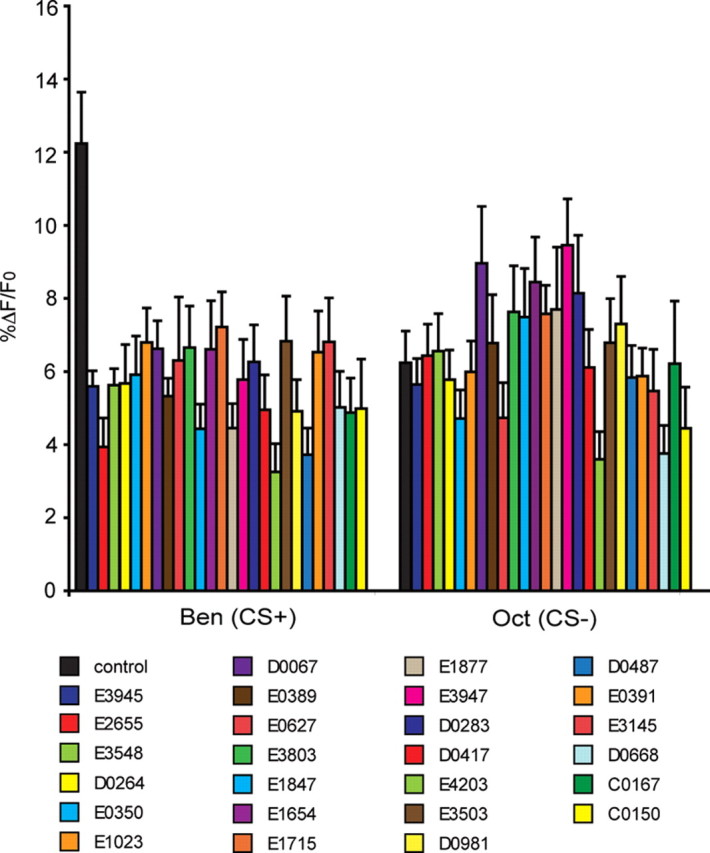
LTM mutants had significantly lower calcium responses than controls in the MB α lobes in response to the CS+ odor (Ben) when tested 24 h after spaced forward conditioning. The calcium response (%ΔF/Fo) of the mutant and control flies (c739; Uas-G-CaMP) to the CS+ and CS− odors 24 h after conditioning. Twenty-four hours after spaced forward conditioning, flies were mounted and tested for calcium responses to the CS+ and CS− odors. As expected, a significant increase in the %ΔF/Fo was detected in the α branch of the α/β MB neurons of control flies to the CS+ (Kruskal–Wallis statistic 73.491, p < 0.0001; Mann–Whitney pairwise comparisons, p < 0.0001). Control flies exhibited odor-elicited calcium responses that were nearly twice as large as any other group. In contrast, all the mutant groups exhibited a blunted calcium response to the CS+ odor, with %ΔF/Fo scores that were similar in magnitude to that for the CS− odor and typical of the responses of naive flies and flies that are trained using conditioning protocols that do not elicit LTM (Yu et al., 2006). The %ΔF/Fo responses to CS− odor in mutants were not significantly different from control (Mann–Whitney pairwise comparisons, p ≥ 0.0974) except for e3947 versus wild type (Mann–Whitney pairwise comparisons, p = 0.0235). Error bars are the SEM. n = 9–28 for all groups.
Discussion
Functional optical imaging has offered invaluable insights about how memories are represented in the Drosophila olfactory nervous system after olfactory classical conditioning. This approach, often using the green fluorescent protein-based sensors synaptopHluorin or G-CaMP coupled with in vivo imaging techniques has highlighted specific neurons that may be involved in distinct stages of memory. For instance, an early memory trace forms immediately after olfactory conditioning in the projection neurons of the antennal lobe and persists for a few minutes (Yu et al., 2004). An intermediate term memory trace potentially related to middle-term memory forms between 30 and 180 min in the dorsal paired medial (DPM) neurons after single cycle training (Yu et al., 2005). Interestingly, the DPM neuron memory trace forms in only one of the two major branches of the DPM neuron process that emanates from the DPM cell body. This process specifically innervates the vertical lobes of the MBs (Yu et al., 2005). As described by Yu et al. (2006) and in this report, an LTM trace forms in the α/β MB neurons after spaced conditioning but not after single cycle or massed conditioning. It is intriguing that this memory trace is also axon branch specific, forming only in the α axon branch of the α/β MB neurons (Yu et al., 2006). The α axon branches are located in the vertical lobes of the MBs, the same neuropil region innervated by the DPM process that exhibits an intermediate-term memory trace. This spatial coincidence suggests that the two memory traces, albeit forming with different time courses, are related to one another functionally.
Prior studies provided evidence that the LTM trace that forms in the α/β MB neurons is much more than a simple correlate of LTM (Yu et al., 2006): (1) The LTM trace forms only with spaced forward conditioning, like long-term behavioral memory. (2) The LTM trace is dependent on the activity of the amnesiac (amn) gene product, like long-term behavioral memory. (3) The LTM trace requires the activity of the transcription factor Creb (disrupted by a Creb dominant negative), like long-term behavioral memory. (4) The LTM trace is disrupted by inhibitors of protein synthesis administered at the time of spaced conditioning like long-term behavioral memory. (5) The LTM trace requires the normal activity of calcium/calmodulin-dependent protein kinase II (CaMKII) (disrupted by an RNA hairpin to CaMKII mRNA), like long-term behavioral memory (Akalal et al., 2010). Thus, the evidence that this LTM trace underlies long-term behavioral memory in a fundamental way was exceptionally strong before this study.
Nevertheless, we wanted to examine the relationship between long-term behavioral memory and the LTM trace in LTM mutants. We entered this study with the prediction that the LTM mutants would fall into two classes: those that disrupted the LTM trace and therefore disrupt gene products that are mechanistically involved in the formation of the LTM trace, and those that exhibit a normal LTM trace despite having impaired long-term behavioral memory. The latter class would presumably represent those gene products that are involved in processes downstream of the LTM trace, such as being involved in the readout of the trace at testing. The gene products disrupted by the LTM mutants were diverse (Table 1). For instance, several of the LTM genes are involved in RNA localization and/or translational regulation. E2655 has an insertion in the Pumilio (pum) gene, a transcript-specific translational suppressor (MacDonald, 1992; Sonoda and Wharton, 1999; Nakahata et al., 2001; Dubnau et al., 2003). The C0150 mutant, also known as krasavietz (kra), contains a molecular lesion within a translation initiation factor and believed to be important for the interaction of eIF-2B, eIF-4γ, and eIF-5 with eIF-2 (Koonin, 1995). Kra has also been shown to be important for axon guidance and is a translational inhibitor (Lee et al., 2007). A third mutant, D0417, is within the Klingon (Klg) gene, which encodes a member of the Ig superfamily of cell adhesion molecules and has also been implicated in photoreceptor development (Butler et al., 1997). Klg has been shown to be acutely required for LTM and regulated by Notch (Matsuno et al., 2009). Oskar is involved in the translocation of mRNA to the posterior pole during oocyte development and assembly of the germ plasm (Ephrussi and Lehmann, 1992; Lehmann and Ephrussi, 1994). One of the mutants examined, E3947 or cheerio (cher), is a Filamin A homolog. Cheerio has been reported to interact with dFMR1 for LTM formation. LTM was reported as defective in dFmr1/cheerio double heterozygotes but normal in the individual heterozygotes for either gene (Bolduc et al., 2010). Thus, actin dynamics may play an important role in LTM.
Remarkably, the LTM mutants assayed here did not fall into the two expected classes. Rather, every one of the mutants impaired the LTM trace in parallel with long-term behavioral memory. Thus, in addition to the four disruptive treatments discussed above (amn, Creb dominant negative, CaMKII RNA hairpin, protein synthesis inhibitors) that block both the LTM trace and long-term behavioral memory, we now add 26 independent LTM mutants that have such parallel effects. The logical implication of the parallel effects of 30 disruptive treatments on the LTM trace and long-term behavioral memory is that this memory trace is fundamental to long-term behavioral memory. Although the large design of the experiment constrained the use of only Ben as the CS+, we believe that the results are likely to extend to other odors as well.
A second major conclusion offered by these results is that the calcium-based LTM trace is well downstream and perhaps near the endpoint in the cellular plasticity of α/β MB neurons in their service of LTM. The LTM mutants alter the function of a diverse set of genes (Table 1), yet they all result in the abolishment of the LTM calcium trace in the α axon branch of the α/β MB neurons. Had increased calcium influx in response to the CS+ represented a cellular event early or midway in the signaling changes that occur in the neurons after training, then a large fraction of the 26 mutants surveyed would have represented functions downstream of the change in calcium dynamics, and exhibited the calcium-based trace found in control flies. The fundamental nature of this calcium-based memory trace to LTM underscores the importance of understanding the mechanics of its formation.
Footnotes
This work was supported by Grants NS19904 and NS52352 from the National Institute of Neurological Disorders and Stroke.
The authors declare no competing financial interests.
References
- Akalal D-BG, Yu D, Davis RL. A late-phase long-term memory trace forms in the γ neurons of Drosophila mushroom bodies after olfactory classical conditioning. J Neurosci. 2010;30:16699–16708. doi: 10.1523/JNEUROSCI.1882-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CDO, Schroeder B, Davis RL. Learning performance of Drosophila after repeated conditioning trials with discrete stimuli. J Neurosci. 2000;20:2944–2953. doi: 10.1523/JNEUROSCI.20-08-02944.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KH, Kong EC, Dubnau J, Tully T, Moore MS, Heberlein U. Ethanol sensitivity and tolerance in long-term memory mutants of Drosophila melanogaster. Alcohol Clin Exp Res. 2008;32:895–908. doi: 10.1111/j.1530-0277.2008.00659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum AL, Li W, Cressy M, Dubnau J. Short- and long-term memory in Drosophila require cAMP signaling in distinct neuron types. Curr Biol. 2009;19:1341–1350. doi: 10.1016/j.cub.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc FV, Bell K, Rosenfelt C, Cox H, Tully T. Fragile x mental retardation 1 and filamin a interact genetically in Drosophila long-term memory. Front Neural Circuits. 2010;3:22. doi: 10.3389/neuro.04.022.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SJ, Ray S, Hiromi Y. Klingon, a novel member of the Drosophila immunoglobulin superfamily, is required for the development of the R7 photoreceptor neuron. Development. 1997;124:781–792. doi: 10.1242/dev.124.4.781. [DOI] [PubMed] [Google Scholar]
- Crittenden JR, Skoulakis EM, Han KA, Kalderon D, Davis RL. Tripartite mushroom body architecture revealed by antigenic markers. Learn Mem. 1998;5:38–51. [PMC free article] [PubMed] [Google Scholar]
- Davis RL. Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Annu Rev Neurosci. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, Broger C, Tully T. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Lehmann R. Induction of germ cell formation by oskar. Nature. 1992;358:387–392. doi: 10.1038/358387a0. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Multidomain organization of eukaryotic guanine nucleotide exchange translation initiation factor eIF-2B subunits revealed by analysis of conserved sequence motifs. Protein Sci. 1995;4:1608–1617. doi: 10.1002/pro.5560040819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Nahm M, Lee M, Kwon M, Kim E, Zadeh AD, Cao H, Kim HJ, Lee ZH, Oh SB, Yim J, Kolodziej PA, Lee S. The F-actin-microtubule crosslinker Shot is a platform for Krasavietz-mediated translational regulation of midline axon repulsion. Development. 2007;134:1767–1777. doi: 10.1242/dev.02842. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Ephrussi A. Germ plasm formation and germ cell determination in Drosophila. Ciba Found Symp. 1994;182:282–296. doi: 10.1002/9780470514573.ch16. [DOI] [PubMed] [Google Scholar]
- Liu X, Davis RL. Insect olfactory memory in time and space. Curr Opin Neurobiol. 2006;16:679–685. doi: 10.1016/j.conb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- MacDonald PM. The Drosophila pumilio gene: an unusually long transcription unit and an unusual protein. Development. 1992;114:221–232. doi: 10.1242/dev.114.1.221. [DOI] [PubMed] [Google Scholar]
- Matsuno M, Horiuchi J, Tully T, Saitoe M. The Drosophila cell adhesion molecule klingon is required for long-term memory formation and is regulated by Notch. Proc Natl Acad Sci U S A. 2009;106:310–315. doi: 10.1073/pnas.0807665106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Nakahata S, Katsu Y, Mita K, Inoue K, Nagahama Y, Yamashita M. Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J Biol Chem. 2001;276:20945–20953. doi: 10.1074/jbc.M010528200. [DOI] [PubMed] [Google Scholar]
- Pascual A, Préat T. Localization of long-term memory within the Drosophila mushroom body. Science. 2001;294:1115–1117. doi: 10.1126/science.1064200. [DOI] [PubMed] [Google Scholar]
- Perazzona B, Isabel G, Preat T, Davis RL. The role of cAMP response element-binding protein in Drosophila long-term memory. J Neurosci. 2004;24:8823–8828. doi: 10.1523/JNEUROSCI.4542-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T, Preat T, Boynton SC, Del Vecchio M. Genetic dissection of consolidated memory in Drosophila. Cell. 1994;79:35–47. doi: 10.1016/0092-8674(94)90398-0. [DOI] [PubMed] [Google Scholar]
- Yang MY, Armstrong JD, Vilinsky I, Strausfeld NJ, Kaiser K. Subdivision of the Drosophila mushroom bodies by enhancer-trap expression patterns. Neuron. 1995;15:45–54. doi: 10.1016/0896-6273(95)90063-2. [DOI] [PubMed] [Google Scholar]
- Yu D, Ponomarev A, Davis RL. Altered representation of the spatial code for odors after olfactory classical conditioning. Neuron. 2004;42:437–449. doi: 10.1016/s0896-6273(04)00217-x. [DOI] [PubMed] [Google Scholar]
- Yu D, Keene AC, Srivatsan A, Waddell S, Davis RL. Drosophila DPM neurons form a delayed and branch-specific memory trace after olfactory classical conditioning. Cell. 2005;123:945–957. doi: 10.1016/j.cell.2005.09.037. [DOI] [PubMed] [Google Scholar]
- Yu D, Akalal DB, Davis RL. Drosophila α/β mushroom body neurons form a branch-specific, long-term cellular memory trace after spaced olfactory conditioning. Neuron. 2006;52:845–855. doi: 10.1016/j.neuron.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zars T, Fischer M, Schulz R, Heisenberg M. Localization of a short-term memory in Drosophila. Science. 2000;288:672–675. doi: 10.1126/science.288.5466.672. [DOI] [PubMed] [Google Scholar]


