Abstract
Regulation of histone H3 lysine 4 and 79 methylation by histone H2B lysine 123 monoubiquitination is an evolutionarily conserved trans-histone crosstalk mechanism, which demonstrates a functional role for histone ubiquitination within the cell. The regulatory enzymes, factors and processes involved in the establishment and dynamic modulation of these modifications and their genome-wide distribution patterns have been determined in many model systems. Rapid progress in understanding this trans-histone crosstalk has been made using the standard experimental tools of chromatin biology in budding yeast (Saccharomyces cerevisiae), a highly tractable model organism. Here, we provide a set of modified and refined experimental procedures that can be used to gain further insights into the underlying mechanisms that govern this crosstalk in budding yeast. Importantly, the improved procedures and their underlying principles can also be applied to other model organisms. Methods presented here provide a rapid and efficient means to prepare enriched protein extracts to better preserve and assess the steady state levels of histones, non-histone proteins and their modifications. Improved chromatin immunoprecipitation and double immunoprecipitation protocols are provided to measure the occupancy and distribution of proteins and their modified forms at specific chromatin regions or loci. A quick and easy method to measure overall protein abundance and changes in protein-protein and protein-DNA interactions on native chromatin is also described.
Introduction
In a eukaryotic nucleus, DNA is wrapped around an octamer of basic histone proteins (H2A, H2B, H3 and H4) to form nucleosomes, the building blocks of chromatin. An important regulatory mechanism that governs overall chromatin structure for factor access is the covalent posttranslational modifications of histones. Histones are extensively adorned with a wide variety of modifications, including acetylation, phosphorylation, methylation, ubiquitination and sumoylation (1–2). Methylation of lysine residues (K) in histones can be present as a mono- (me1), di- (me2) or trimethylated (me3) form, further diversifying the function and complexity of this modification. While histone modifications serve as “marks” for cellular processes (2–3) and are recognized by factors with specific interaction modules (1, 4), their regulation is highly dynamic with the presence of different types and families of enzymes that can either add or remove them (5). Additionally, these enzymes, which might exist as components of multi-protein complexes, are themselves regulated by both endogenous and exogenous cues. Another interesting facet to the regulation of histone modifications is that one modification can regulate another within the same histone (in cis) or on different histones (in trans), a phenomenon termed the “histone crosstalk” (6–8). Regulation of H3K4 and -K79 methylation by H2BK123 monoubiquitination (H2Bub1) during transcription is a well-studied example of a trans-histone modification crosstalk.
This trans-histone regulatory circuit is evolutionarily conserved, as H2Bub1 is important for H3K4 and -K79 methylation in most eukaryotes (9). However, several valuable characteristics, such as the short life cycle, powerful genetics, ease of genome manipulation and being easily amenable to biochemical and high-throughput genome-wide studies, have made budding yeast (Saccharomyces cerevisiae) as the model system of choice to identify and extensively investigate the regulatory pathways, factors and enzymes involved in mediating this trans-histone crosstalk. Rad6, the E2 ubiquitin-conjugating enzyme and Bre1, the E3 ubiquitin ligase are required to conjugate ubiquitin to K123 present in the H2B C-terminal helix (10–11). Ubp8 and Ubp10 are the deubiquitinases involved in the removal of this conjugated ubiquitin to maintain the total H2Bub1 levels in the cell (9). Set1-COMPASS, a multi-protein complex consisting of the methyltransferase (Set1) and seven subunits (Swd1, Swd2, Swd3, Bre2, Sdc1, Spp1 and Shg1), catalyzes all forms of H3K4 methylation (H3K4me). Dot1, a non-SET domain methyltransferase, catalyzes all forms of H3K79 methylation (H3K79me) (1, 11). A role for H2Bub1 in modulating the enzymatic functions of these methyltransferases by affecting their ability to catalyze the different forms of H3K4 and K79 methylation is now well established (9, 11): H2Bub1 is required for H3K4me2, H3K4me3 and H3K79me3. Additionally, H3K4me1 and H3K79me2 are severely reduced in the absence of H2Bub1.
To explain the mechanism of trans-histone crosstalk, it was proposed that the ubiquitin conjugated onto H2B might act as a “bridge” to directly recruit the methyltransferases (12). Since Set1 and Dot1 associate with chromatin even in the absence of H2Bub1 (13–14), their recruitment does not appear to be the basic mechanism by which H2Bub1 participates in the crosstalk. Two studies have alluded to Swd2, a Set1-COMPASS subunit, as a key link in this crosstalk (15–16), but their conflicting findings and conclusions have left the regulation of methyltransferase functions by H2Bub1 as an open question. H2Bub1 has also been proposed to act as a “wedge” to open-up the chromatin and allow access to the enzymes (12, 17). However, contrary to its supposed role in opening up chromatin, using chromatin immunoprecipitation assays (ChIP) and salt-dependent nucleosome disruption assays, we recently showed that H2Bub1 stabilizes the nucleosome by preventing the constant H2A-H2B eviction (18). This finding addresses a longstanding question in chromatin biology as to whether conjugation of bulky ubiquitin moiety onto histones affects nucleosome structure. Further, it has provided a new working model for the trans-histone crosstalk: addition of ubiquitin onto H2B acts as a “glue” to hold the nucleosome together and provides a stable platform for the prolonged chromatin association of Set1-COMPASS and Dot1 to promote their processive methylation. However, understanding this trans-histone crosstalk is an ongoing saga that is far from completion.
Several questions pertaining to the mechanism of nucleosome stabilization by H2Bub1 and the chromatin association of Set1-COMPASS and Dot1 remain to be explored. The basic patch in H4 N-terminal plays a role in a novel trans-histone pathway by controlling the chromatin binding and functions of Dot1 (19). However, the question as to how H2Bub1 controls Dot1 function remains unanswered. While a “docking site” for Dot1 on chromatin via the H4 tail region is now known, how Set1-COMPASS associates with chromatin and how this multi-subunit protein complex is assembled on chromatin remain unknown. We recently found that residues R119 and T122 in the H2B C-terminal helix interact with Spp1, a Set1-COMPASS subunit, and they modulate the chromatin association, integrity and overall stability of Set1-COMPASS independent of H2Bub1 (20). Importantly, we have uncovered a “docking” surface for only Set1-COMPASS, since mutations in R119 and T122 do not affect the functions of Dot1; thereby, revealing an uncoupling of the H2Bub1-mediated co-regulation of H3K4 and -K79 methylation. Therefore, a simple model that can be proposed for the trans-histone crosstalk between H2Bub1 and H3K4 methylation is as follows: H2Bub1 stabilizes the nucleosome by preventing H2A-H2B eviction. In turn, this leads to the retention of a “docking site” for Set1-COMPASS present in H2B on chromatin, culminating in increased complex integrity and stability of Set1-COMPASS needed for high levels of processive H3K4 methylation. While considerable effort has been invested in understanding how the methyltransferases associate with chromatin, the binding of Rad6/Bre1 and Ubp8/Ubp10 to chromatin remains poorly understood and needs further investigation.
In this report, we provide detailed procedures used in our previous studies to address some of the questions mentioned above. An improved method is provided to assess the steady state levels of histones and any other proteins in total cell extracts or nuclear extracts isolated under native or denaturing condition (sections 1.1 and 1.2). A quick and easy method to isolate and detect H2Bub1, a highly labile modification, employing a simple boiling procedure is described in section 1.3. To measure changes in the chromatin association of histones, histone modifying enzymes and their regulatory factors, two different assays are described in section 2. The chromatin association assay measures global changes in protein levels on chromatin obtained from isolated nuclei (section 2.1). On the other hand, local changes in the occupancy and distribution of histones, histone modifications and factors on genes or in different regions of a gene can be assessed employing the ChIP assay (section 2.2.1). A procedure to evaluate changes in the distribution and occupancy of H2Bub1 using chromatin-double immunoprecipitation is provided in section 2.3.
Methods
1. Assessment of global H3K4 and -K79 methylation and H2Bub1 by Western blotting
1.1. Preparation of yeast whole cell extracts by bead-beat procedure
-
1.
Cultures grown overnight are used to re-inoculate fresh 50 ml medium at a starting OD600 0.2 and grown to a final OD600 0.8–1.2. Cells (30–40 × 107) are harvested by centrifugation at 3,000 rpm for 2 min and washed once with 1 ml water while transferring to a 2.0 ml screw-cap tube.
-
2.
Cells are collected by centrifugation at 3,000 rpm for 2 min and the water is completely discarded. The pellets are stored at −80°C until further use.
-
3.
The cells are thawed by vigorous vortexing in the presence of 400 µl SWBNG buffer [10mM Tris.Cl, pH 7.4, 300 mM Sorbitol, 100 mM NaCl, 5mM MgCl2, 5 mM EDTA, 10% glycerol, 0.1% Igepal-30 (NP40), protease inhibitors (1 mM PMSF, 1 µg/ml aprotinin, 1 µg/ml leupeptin, 1 µg/ml pepstatin A)] or SUME buffer (10 mM MOPS, pH 6.8, 0.5% SDS, 8 M Urea, 10 mM EDTA, protease inhibitors) (21).
Technical Notes:- SWBNG buffer is used to isolate proteins under non-denaturing or native condition and SUME buffer is used to isolate proteins under denaturing conditions.
- A variety of buffers can be used to effectively isolate proteins under native conditions as reported previously (22). In SWBNG buffer, the inclusion of a detergent (NP-40) helps to better solubilize proteins and addition of glycerol prevents protein aggregation.
- A low initial NaCl concentration (100 mM) is maintained to prevent excessive sample heating during the bead-beating process.
-
4.
Acid-washed glass beads (Sigma) are added to the top of the cell suspension. Cells are lysed by agitation four times for 30 sec each in a Mini-beadbeater (Biospec) set at “homogenize” mode. The lysates are cooled on ice for 30 sec between successive agitations.
Technical Note: Cooling on ice should be avoided for samples prepared in SUME buffer to prevent precipitation of SDS.
-
5.
A hole is punched at the bottom of the 2 ml screw-cap tube using an 18-guage needle. The lysate is collected by brief centrifugation into a 15 ml collection tube fitted with an adaptor, which is made by cutting the top part of a 5 ml syringe.
-
6.
The lysate is resuspended by brief vortexing and an aliquot (300 µl) is transferred to a labeled 1.5 ml microfuge tube. 26 µl 5M NaCl is added to the tube to obtain a final NaCl concentration of 500 mM. The lysate is vortexed vigorously for 30–40 sec.
Technical Note: Increased NaCl concentration to 500 mM aids in disrupting the nucleosome and solubilization of histones (18, 23).
-
7.
Following two successive centrifugations in a table top centrifuge (13,200 rpm for 20 min at 4°C), the clarified lysate is collected. Care is taken to avoid transfer of any lipid layer that floats at the top of the supernatant.
-
8.Protein concentration
- Protein concentration is measured using a BioRad protein assay kit for samples prepared using SWBNG buffer as per the manufacturer’s instructions.
- For samples prepared using SUME buffer, protein concentration is measured using BioRad DC protein assay kit. Dilute the denatured sample 5-fold by adding 5 µl to 20 µl water. Mix well by pipetting and transfer 5 µl of the diluted sample to 20 µl water. Add kit reagents to perform protein estimation as per the protocol provided by the manufacturer.
- Use known concentrations of BSA as the reference standard for both protein concentration estimation assays.
-
8.
Prepare aliquots of desired protein concentration (50–80 µg) in a total volume of 15–20 µl.
Technical Note: Adjust the final volume using SWBNG or SUME buffer without adding any extra NaCl. It is preferable to dilute the NaCl concentration to less than 500 mM to prevent sample aggregation following the heat denaturation step prior to loading onto SDS-PAGE.
1.2. Quick and easy preparation of yeast nuclear extracts
1.2.1 Cell growth and harvesting
Yeast cultures (50 ml) initiated from overnight cultures are grown to exponential phase (OD600 0.8). For slow-growing yeast strains or strains sensitive to spheroplasting, two 50 ml cultures per yeast strain can be initiated.
Cell growth is stopped by the addition of sodium azide (0.1%) and 10–20 × 107 cells are harvested in a 50 ml tube per culture.
1.2.2. Spheroplasting
Cells are washed one-two times with 25 ml sterile water and incubated at room temperature in 2 ml pre-spheroplasting buffer (100 mM PIPES.KOH, pH 9.4, 10 mM DTT, 0.1% sodium azide).
-
Following centrifugation at 3,000 rpm for 2 min, cells are resuspended in 1.5 ml spheroplasting buffer (50 mM potassium phosphate buffer, 600 mM sorbitol, 10 mM DTT) and incubated in a 30°C water-bath with agitation for 1–1.5 h after the addition of 60–80 units of the yeast lytic enzyme, Quantazyme™ (MP Biomedicals) or recombinant Lyticase (Sigma). If these pure enzymes are unavailable, 100 units of partially purified lyticase from Arthrobacter luteus (Sigma) can be used.
Technical Note: Efficient spheroplasting is monitored by measuring the OD600 of a 1:100 dilution of spheroplasts with 1% SDS. Even a 50% drop in OD600 value yields optimal signal intensity for various histone or histone modification-specific antibodies. Prolonged spheroplasting might affect protein stability due to protease-mediated degradation.
-
Spheroplasts are harvested by centrifugation at 3,000 rpm for 2 min and washed with 500 µl extraction buffer [50 mM HEPES.KOH, pH 7.6, 100 mM KCl, 2.5 mM MgCl2, protease inhibitors (1 mM PMSF, 1 µg/ml aprotinin, 1 µg/ml leupeptin, 1 µg/ml pepstatin A)] by repeated pipetting using a wide-bore tip.
Technical Note: Spheroplasts obtained using partially purified lyticase are washed extensively with 10 ml wash buffer to remove proteases.
Spheroplasts are resuspended gently in 500 µl extraction buffer and lysed by adding 6.5 µl 20% Triton X-100 and incubation on ice for 5 min with periodic gentle mixing.
1.2.3. Nuclear extract preparation
500 µl 30% Sucrose prepared in extraction buffer containing 0.25% Triton X-100 is then added slowly below the lysate and crude nuclear pellet is obtained by centrifugation at 12,000 rpm, 4°C for 10 min.
The crude nuclear pellet is washed once with 1 ml extraction buffer containing 0.25% Triton X-100 and centrifuged at 10,000 rpm, 4°C for 5 min.
-
The supernatant is completely discarded and the pellet is resuspended by vigorous vortexing and repeated pipetting in 400 µl SUME buffer. Samples are briefly sonicated in a Branson Sonifier 450 (30% output, 50% duty cycle, 6 pulses each, two times) to reduce viscosity and centrifuged at maximum speed in a table top centrifuge. Clarified nuclear lysate is transferred to a fresh tube.
Technical Note: For some antibodies, such as α-H3K56ac, denaturing the extract using SUME buffer yields high background signal with cross-reaction and non-specific binding to several proteins. In these cases, extracts are prepared using SWBNG buffer (section 1.1 step 3).
-
Protein concentration of the nuclear extract is measured using BioRad DC Protein Reagent as described in section 1.1 step 8. 5–25 µg of extract is used for Western blotting to detect histones, histone modifications and chromatin-associated proteins.
Note: Nuclei can also be obtained by resuspending the spheroplasts in a buffer containing 0.34 M sucrose followed by lysis using Triton X-100 and centrifugation (for details, see section 2.1 step 3).
1.3. Preparation of yeast whole cell extracts by boiling procedure
-
Cultures grown overnight are used to re-inoculate fresh 25 ml medium at a starting OD600 0.2 and grown to a final OD600 0.6–0.8. Cells (8 × 107) are harvested by centrifugation at 3,000 rpm for 2 min and washed once with 1 ml water while transferring to a 1.5 ml screw-cap tube.
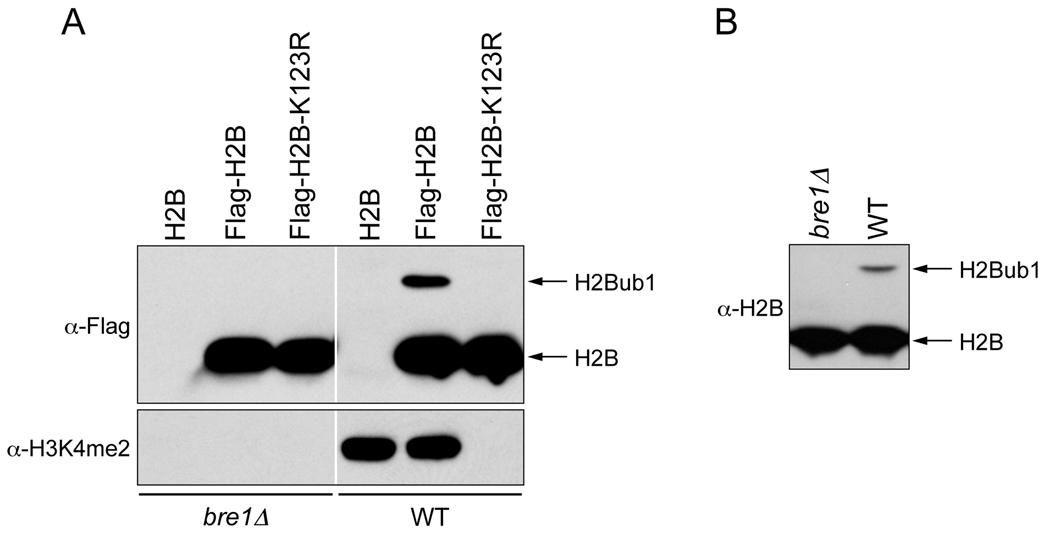
Technical Note: To detect high levels of H2Bub1 as shown in Fig.1, excessive sterilization of growth medium leading to caramelization of carbohydrates and prolonged growth for extended periods should be avoided. Moreover, the steady state level of H2Bub1 is affected by the carbon source (24) and cell cycle phase (21).
While harvesting the cells, start heating water (200 ml) to boil in a 500 ml beaker over a hotplate.
Centrifuge the 1.5 ml screw-cap tube at 3,000 rpm for 2 min to pellet cells. Discard water completely. Add 250 µl lysis buffer (50 mM Tris.Cl, pH 7.5 and 2% SDS) and resuspend cells thoroughly by vortexing.
Place the tubes in a holder (800 ml Beaker Rack, USA Scientific) and tighten the lid. Lyse cells by boiling for 8 minutes.
Centrifuge the boiled samples at 13,200 rpm for 5 min and transfer 200 µl cleared lysate to a fresh, labeled 1.5 ml microfuge tube containing 50 µl of 50% glycerol. Vortex to mix and boil for an additional 5 min.
-
Measure protein concentration using BioRad DC Protein Reagent (section 1.1 step 8). For checking H2Bub1 in Flag-epitope tagged H2B containing strains, prepare 50 µg aliquots and adjust the final volume to 15 µl using the lysis buffer. To more accurately assess any changes in the steady state levels of H2Bub1 between various strains or in different conditions, a range of aliquots with increasing protein concentration (12.5 µg, 25 µg and 50 µg) can also be prepared. Add 2.5 µl 6× loading dye (300 mM Tris.Cl, pH 7.5, 12% SDS, 60% glycerol, 0.12% bromophenol blue, 6% β-mercaptoethanol). Resolve the proteins in a 15% SDS-PAGE, run at 120 V. Following transfer, the PVDF membrane is blocked overnight at 4°C in 5% non-fat dry milk in 1× TBS (50 mM Tris.Cl, pH 7.6, 150 mM NaCl). Blots are probed with α-Flag (1:5000 dilution) in 5% milk in 1× TBS containing 0.05% Tween-20 (Fig. 1A). Alternatively, membranes are blocked overnight at 4°C with 5% non-fat dry milk in 1× TBS containing 0.1% Tween-20 (TBST) and probed with α-H2B (1:1000–1:2500 dilution; Active Motif) added to 2.5% non-fat dry milk in TBST (Fig. 1B).
Technical Notes: H2Bub1 can also be detected following isolation by immunoprecipitation of Flag-H2B from neutralized extracts prepared under denaturing condition using tricholoroacetic acid (TCA) as described previously (25). Additionally, this IP protocol allows enrichment of any histone ubiquitination or sumoylation that might occur in low abundance. However, the boil procedure described above is rapid and is suitable for any abundant ubiquitinated or sumoylated protein. Moreover, even in the presence of SDS in the extracts, protein concentrations can be easily measured using BioRad DC Protein Reagent unlike in the protocols described previously (17, 25).
Fig.1.
Detection of H2Bub1 in lysates prepared using the modified boiling method. (A) Yeast whole cell lysates were prepared using the modified boiling procedure from the wild type or mutant (bre1Δ) strains harboring H2B (no tag control), Flag-H2B or Flag-H2B-K123R. Equal amounts of lysates (50 µg) were resolved in a 15% acrylamide-SDS polyacrylamide gel (15% SDS-PAGE), transferred to a PVDF membrane and probed with an antibody that detects the Flag epitope (α-Flag, 1:5000 dilution, Sigma, catalog no. F3165) or with an antibody that recognizes H3 dimethyl lysine 4 (α-H3K4me2, 1:5000 dilution, Millipore, catalog no. 07–030). (B) Yeast whole cell lysates were prepared from wild type (BY4742) or mutant (bre1Δ) strains and subjected to Western blotting as described for panel A, but the blots were probed with an antibody that detects yeast H2B (α-H2B, 1:2500 dilution, Active Motif, catalog no. 39237).
2. Measuring chromatin association, occupancy and occurrence of histone modifications and their regulatory factors
Many histone-modifying enzymes (HME) and chromatin-remodeling factors (CRF) exist as multi-subunit protein complexes. Their intermolecular interactions and chromatin association are likely subjected to highly dynamic regulation in vivo with high turnover or on/off rates. Therefore, it is conceivable that the integrity and overall stability of a given complex and its biochemical activity purified from total cell populations might not necessarily reflect the events that occur on chromatin in vivo. The fractionation procedure described below in section 2.1, which is derived from previous studies (26–28), allows direct assessment of the global chromatin-bound levels of the components of a specific protein complex containing HME/CRF under a given experimental condition. Additionally, a refined protocol for ChIP assay to examine local, gene and region-specific changes in the chromatin binding of components present in HME/CRF-containing complexes is provided in section 2.2.
2.1. Chromatin association assay for measuring global chromatin-bound levels of histones or factors
Two 50 ml cultures per yeast strain are initiated from overnight cultures and grown to exponential phase (OD600 0.8). For slow-growing yeast strains or strains sensitive to spheroplasting, four 50 ml cultures per yeast strain can be initiated. Cell growth is stopped by the addition of sodium azide (0.1%) and 4 × 108 cells are harvested in a 50 ml tube per culture.
Cells are washed twice with 25 ml sterile water and spheroplasting is performed as described in section 1.2.2. Spheroplasts are harvested and washed with 1 ml wash buffer [50 mM HEPES.KOH, pH 7.6, 100 mM KCl, 2.5 mM MgCl2, 400 mM Sorbitol, protease inhibitors (1 mM PMSF, 1 µg/ml aprotinin, 1 µg/ml leupeptin, 1 µg/ml pepstatin A), 5 mM N-ethylmaleimide (NEM)]. Spheroplasts obtained using partially purified lyticase are washed extensively, two times with 10 ml wash buffer each, to remove proteases.
Spheroplasts are resuspended gently in 1.3 ml Buffer A (10 mM HEPES.KOH (pH 7.6), 10 mM KCl, 1.5 mM MgCl2, 10% glycerol, 0.34 M Sucrose, 1 mM DTT, protease inhibitors, 5 mM NEM). Spheroplasts are lysed by the addition of 6.5 µl 20% Triton X-100 and incubation on ice for 5 min with gentle mixing. Crude nuclear pellet is obtained from the lysate by centrifugation at 3,000 rpm, 4°C for 5 min and resuspended in 1 ml purification buffer (PB) (10 mM Tris.Cl, pH 7.5, 100 mM KCl, 0.1 mM EDTA, 1 mM DTT, 10% glycerol, protease inhibitors, 5 mM NEM) with gently tapping the tubes or with a wide-bore tip. The two 1 ml nuclear suspensions per yeast strain are combined and diluted with PB to a total volume of 7.5 ml.
Nuclei are then mixed with 7.5 ml 66% Percoll (GE Healthcare) prepared in PB, mixed well, transferred to a Beckman polycarbonate centrifuge bottle and centrifuged at 16,500 rpm, 4°C using a Type 50.2 Ti rotor in a Beckman Ultracentrifuge for 40 min. A white band of nuclei seen at the top is carefully collected using a plastic disposable dropper (~4 ml) and transferred to a 30 ml Sorvall centrifuge tube. The nuclei are diluted to 12 ml with recovery buffer (PB without glycerol) and centrifuged at 10,000 rpm using a SS34 rotor 4°C for 15 min in a Sorvall centrifuge. The pellet is resuspended in 1 ml recovery buffer using a wide-bore tip, transferred to a microfuge tube and centrifuged at 13,200 rpm, 4°C for 15 min. The nuclei-containing pellet is resuspended in 1 ml recovery buffer, split into two equal aliquots prior to centrifugation at 13,200 rpm, 4°C for 15 min. One aliquot is then resuspended in 400 µl nuclei storage buffer (recovery buffer + 40% glycerol), mixed gently and stored at −80°C until further use. This aliquot serves as a back-up batch of nuclei.
-
Chromatin is isolated from the second aliquot by resuspending the nuclei in 700 µl hypotonic Buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, 1× protease inhibitors, 5 mM NEM) by gently tapping the tubes or mixing with a wide-bore tip and incubation on ice for 30 min. The chromatin is collected by centrifugation at 1,700g, 4°C for 5 min, resuspended in 700 µl Buffer B, incubated on ice for 15 min with periodic mixing and centrifuged again. Chromatin amounts are normalized based on histone or DNA amounts as described below. At this stage, 400 µl SUME buffer (0.5% SDS, 8 M Urea, 10 mM MOPS, pH 6.8, 10 mM EDTA, protease inhibitors, 5 mM NEM) is added to the frozen spheroplast pellet set aside in Step 2 for whole cell extract preparation, vortexed vigorously, briefly sonicated in a Branson Sonifier 450 (30% output, 50% duty cycle, 6 pulses each, two times) to reduce viscosity and centrifuged at maximum speed in a table-top centrifuge. Protein concentration of the clarified supernatant is measured using BioRad DC Protein Reagent and equal total protein amounts (60 µg) are used in Western blotting to determine the steady state levels of a protein. As a loading control, the global level of 3-phosphoglycerate kinase is assessed in Western blotting using an antibody (α-Pgk1, 1:5000 dilution; Molecular Probes).
Technical Note: Nuclei isolation and chromatin preparation can be scaled down using only 40 × 107 cells per strain. The crude nuclear pellet obtained in step 3 is resuspended in 700 µl Buffer A, mixed with 700 µl 66% Percoll and transferred to a polyallomer microfuge tube (Beckman). Nuclei are collected by centrifugation at 24,000 rpm, 4°C for 40 min using a TLA100.3 rotor and a Beckman TL-100 ultracentrifuge.
- Normalization of chromatin amounts
- Based on histone amounts:
- The chromatin-containing pellet is resuspended in 400 µl SUME buffer and a chromatin extract is prepared essentially as described for the whole cell extract preparation. To measure the chromatin-bound level of a protein, 15–20 µg of chromatin extract is subjected to Western blotting.
- To determine whether equal amounts of chromatin have been subjected to the analysis, the amount of histone H3 is also assessed by Western blotting. Histone H3 serves as a loading control for the chromatin fraction for the following reasons: first, H3 and H4 form a stable tetramer core within the nucleosome. Second, an antibody raised against yeast H3 is commercially available and permits a highly sensitive detection.
- To ensure that the signals are in the linear range of detection, less amounts of chromatin extract can be used for checking the histone levels (<5 µg), which are measured by densitometric analysis of the Western blots. If H3 levels are not equal, then the amounts of chromatin extract prepared from different strains are made equal based on the H3 amounts, and normalization is confirmed by Western blotting. Alternatively, for more accurate measurement, initially a four-point, two-fold serial dilution of the chromatin extract is prepared (10, 5, 2.5 and 1.25 µg), subjected to Western blotting using α-H3 and the blots are used in densitometry. The four point serial dilution provides the linear range for detection and quantitation.
- For detecting the chromatin-bound level of a protein, the chromatin extract is then normalized based on H3 amounts, resolved in a SDS-PAGE prior to Western blotting using a protein-specific or an epitope tag-specific antibody.
- Based on DNA amounts:
- The chromatin-containing pellet is resuspended in 400 µl PB and sonicated as described above (section 2.1 step 5).
- DNA is isolated from an aliquot of this soluble chromatin as follows: 25 µl of soluble chromatin is mixed with 75 µl sterile water and incubated at 37°C for 30 min following the addition of 2 µl of RNase A (10 mg/ml, Qiagen). The sample is incubated at 42°C for 30 min following the addition of 10 µl 10× Proteinase K buffer (PNK) (100 mM Tris.Cl, pH 8.0, 50 mM EDTA, 5% SDS) and 1 µl Proteinase K (9–11 mg/ml, Sigma). Prior to DNA purification using QIAquick® PCR purification kit (Qiagen), 10 µl 3M Sodium Acetate, pH 5.0 DNA is added to adjust the pH. DNA is eluted in 50 µl sterile water.
- The DNA concentration is estimated by measuring the OD260 using a spectrophotometer or alternatively, employing any other sensitive DNA concentration measuring approach (quantitative PCR in real time or fluorimetry using PicoGreen).
- The soluble chromatin obtained from different yeast strains are normalized based on their DNA amounts and equal amounts of normalized soluble chromatin are subjected to Western blotting using a protein-specific or an epitope tag-specific antibody to determine changes, if any, in the chromatin-bound levels of a protein. To show equal amounts of chromatin were used in the assay, DNA is isolated from an aliquot of the normalized soluble chromatin essentially as described above. A three point ten-fold serial dilution of the purified DNA is used as template in a multiplex PCR. The PCR products are resolved in a native 5% PAGE and stained with ethidium bromide for visualization.
2.2. ChIP assay for measuring occupancy of histones, their modifications and regulatory factors
2.2.1. ChIP assay for measuring the occupancy and distribution of H3K4 and -K79 methylation and their regulatory factors
-
A.Cell growth and crosslinking
- Single crosslinking
- Cultures grown overnight are used to re-inoculate fresh 50 ml medium at a starting OD600 0.2–0.3 and grown to a final OD600 0.8–1.2. Crosslinking is then initiated by adding 1.35 ml of 37% formaldehyde solution (Sigma) directly to the culture and incubation is performed at room temperature for 20 min with gentle mixing on a laboratory rotator.
-
To stop cross-linking, 2.5 ml of 2.5 M glycine is added and incubation is continued at room temperature for 10 min. Crosslinked cells (40 × 107) are harvested, washed twice with 25 ml sterile water, transferred to a 2 ml screw-cap tube and stored at −80°C until further use.Technical Note: Cultures of set1Δ or dot1Δ strain should be included as controls to determine non-specific precipitation and/or background signals for H3K4 or -K79 methylation-specific antibodies, respectively.
-
Double crosslinkingMany factors that do not bind DNA directly are not efficiently crosslinked to chromatin by formaldehyde alone (20, 29). Therefore, a protein-protein crosslinking agent in addition to formaldehyde is needed to increase the efficiency of crosslinking of these factors to chromatin and their DNA target sites in vivo.
- Cells (40 × 107) cells are harvested and washed twice with 25 ml phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4). Cells are resuspended in 15 ml crosslinking solution [10 mM dimethyl adipimidate dihydrochloride (DMA, Sigma) and 0.25% dimethylsulfoxide (DMSO) in PBS] and incubated at room temperature for 45 min with constant mixing on a laboratory rotator. DMSO is added to permeabilize the cells.
-
Subsequently, cells are washed once with PBS, resuspended in 37 ml PBS containing 1% formaldehyde and incubated at room temperature for 45 min. Crosslinking is stopped with 125 mM glycine as described above, cells are washed twice with PBS prior to harvesting and final storage at −80°C.Technical Note: If epitope-tagged versions of histones or regulatory factors are used, a culture of the yeast strain lacking the epitope tag should be included to determine non-specific precipitation and/or background signal for the epitope tag-specific antibody.
-
B.Soluble chromatin preparation
- The frozen, crosslinked cells are resuspended thoroughly by vortexing in 400 µl ice-cold FA140 buffer (50 mM HEPES.KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate and protease inhibitors). Acid-washed glass beads (Sigma) are added up to the top of the cell suspension.
- Cells are lysed in a Mini-beadbeater (Biospec) employing six bead-beating cycles, each involving a 30 sec agitation and a 5 min cooling period on ice between successive agitations. Lysate is collected by brief centrifugation into a 15 ml collection tube fitted with an adaptor after punching a hole at the bottom of the 2 ml screw-cap tube as described in section 1.1 step 5.
-
The lysate is transferred to a 1.5 ml microfuge tube and sonicated using a Branson Sonifier 450 fitted with a 3/16-inch microtip and set at 30% output, 90% duty cycle. A total of 12 sonication cycles is performed, each comprising of 6 sonication pulses and a brief cooling on dry ice for 10 sec. It is imperative to cool the samples well to prevent protein denaturation, which adversely affects the efficiency of immunoprecipitation.Technical Note: The average size of sheared DNA obtained using the cell number and sonication parameters described above is ~500 bp.
- Following sonication, soluble chromatin is obtained by centrifugation at 13,200 rpm for 5 min, followed by clarification of the resulting supernatant by centrifugation for an additional 15 min.
- Protein concentration of the soluble chromatin is estimated using BioRad DC protein assay kit. Protein concentrations of all the samples are made equal by diluting the samples with FA140 buffer (~15–20 mg/ml). A 10 µl sample of the diluted chromatin is set aside to isolate DNA that will serve as the template for “input” in PCR.
-
C.Immunoprecipitation (IP)
-
For IP of H3 and K4- or K79-methylated H3, 250–500 µg soluble chromatin is added to a 0.65 ml microfuge tube in a final reaction volume of 260 µl made with FA140 buffer.Technical Note: For factor IP, the amount of soluble chromatin to be used is estimated based on the abundance of a factor within the cell (molecules/cell) as determined by Ghaemmaghami et al. (30). The abundance of histones, Dot1 and Set1-COMPASS components can be retrieved from the Saccharomyces Genome Database (http://www.yeastgenome.org). For instance, Set1 is present at 172 molecules/cell and Spp1 (a Set1-COMPASS component) is present at 1680 molecules/cell. Generally, 1 mg of soluble chromatin is used for factors that are present at <1000 molecules/cell and 250–500 µg of chromatin is used for factors that are present at >1000 molecules/cell.
-
The diluted chromatin is pre-cleared with end-over-end mixing for 20 min at 4°C in a rotisserie rotator following the addition of 10 µl 1:1 slurry containing Protein A- or Protein G-conjugated magnetic beads in FA140 buffer or 25 µl 1:1 slurry of Protein A- or Protein G-conjugated agarose beads in FA140 buffer.Technical Note: The use of magnetic beads is preferred over agarose beads, as they reduce the background signal resulting from non-specific binding of chromatin to the beads and they are not lost during the numerous pipetting involved in the washing steps. Magnetic beads are washed extensively with FA140 buffer prior to their addition. The agarose beads are pre-equilibrated in FA140 buffer by end-over-end mixing for 20 min at 4°C in addition to the extensive washing.
-
Following pre-clearing, the soluble chromatin is transferred to a fresh 0.65 ml microfuge tube. Care should be taken to avoid carry over of the beads and equal amounts of soluble chromatin from different samples should be transferred. Antibody (0.5–1 µl) is added to the tubes and incubated overnight at 4°C with end-over-end mixing.Technical Notes:
-
Accounting for histone occupancyFor determining the occupancy and/or distribution of H3K4 or -K79 methylation over a gene, it is imperative to also account for the changes in H3 occupancy. Therefore, an equivalent amount of soluble chromatin from each sample used to IP H3K4 or -K79 methylation is also used to IP H3 using a pan-H3 antibody.
-
Quality of the antibody to be used.Well-characterized antibodies with high antigen specificity are critical for both consistency and accuracy of the data. Several antibodies validated to immunoprecipitate crosslinked antigens, and deemed “ChIP-grade”, are now commercially available.
-
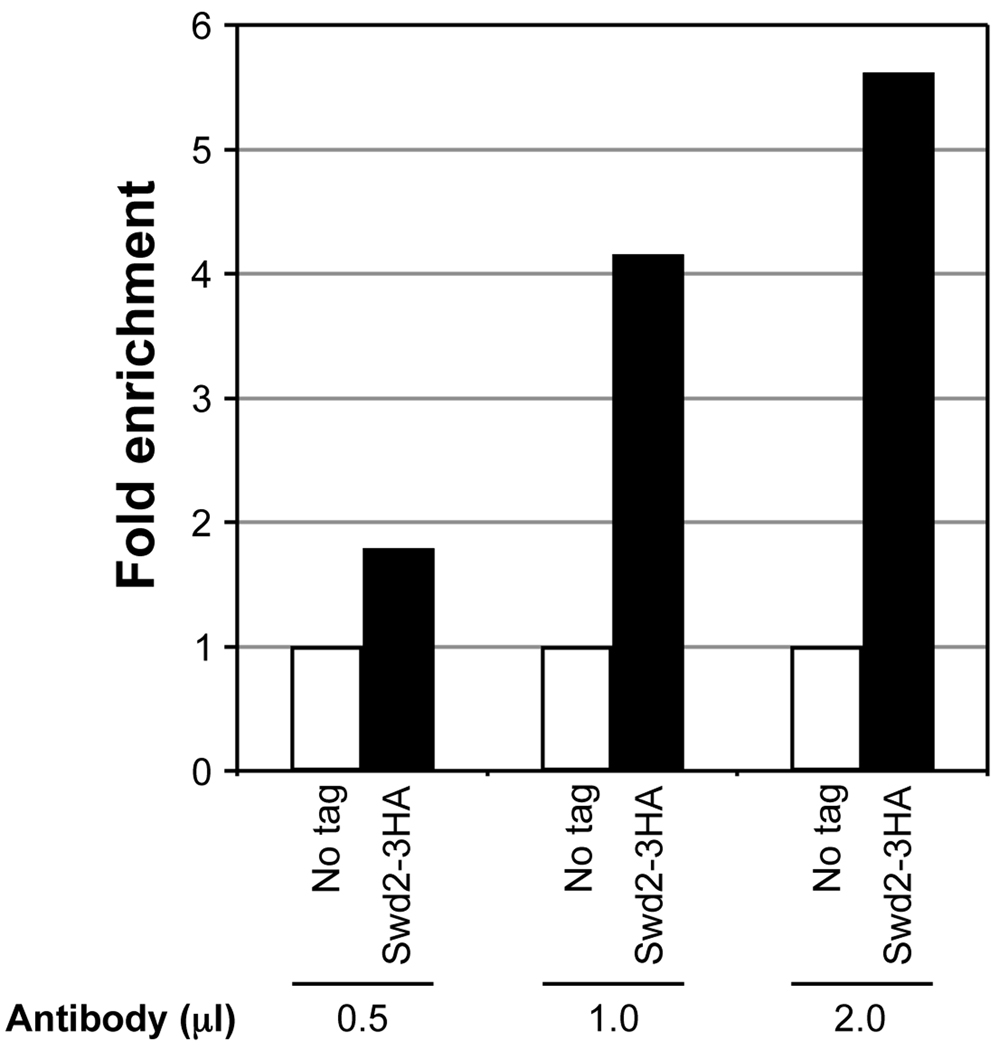
Amount of the antibody to be used.The amount of antibody to be used should be empirically determined, as excessive amounts tend to increase background signal from non-specific precipitation. Ideally, a pilot experiment using different amounts of the antibody should be conducted to determine minimal amount of antibody required to obtain significant enrichment over background (Fig. 2). In general, significant signal enrichment relative to the background (signal obtained from IP using chromatin from set1Δ) can be obtained using 0.5–1 µl of most commercially available antibody raised against the different forms of H3K4 methylation. For epitope-specific antibodies, significant signal enrichment can be obtained using 1–2 µl of the antibody, as shown for ChIP using α-HA (Fig. 2).
- Type of antibodies to be used.
- Any pan H3-specific antibody to be used should be unaffected by the H3K4 or -K79 methylation. Therefore, an antibody raised against the C-terminal region of H3 should be used. Furthermore, increased antigen specificity can also be obtained using an antibody raised against yeast H3.
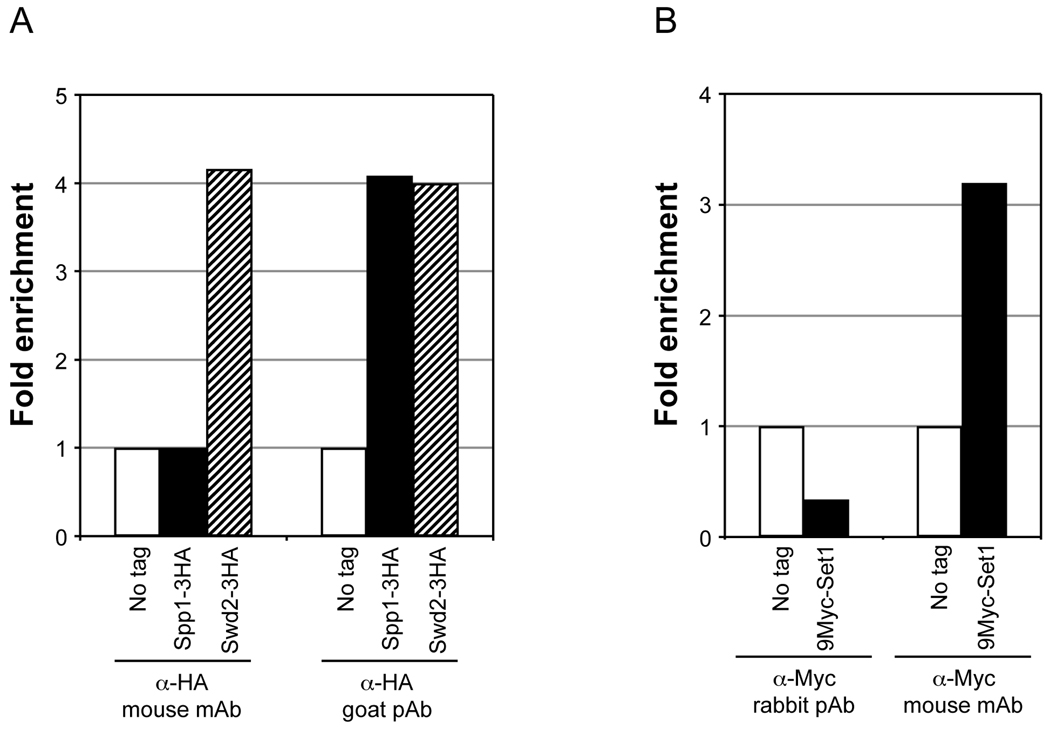
- Commercially available mouse monoclonal antibodies, α-Flag (M2, Sigma) and α-Myc (clone 9E10), can be used to IP proteins containing Flag or Myc epitope tags.
- Epitope masking from protein-protein interactions or due to chemical fixation can reduce IP efficiency. Therefore, a polyclonal antibody may be preferred over a monoclonal antibody. For Spp1 tagged with 3HA epitope, only a goat polyclonal antibody yielded 4-fold enrichment relative to background, whereas no enrichment was obtained using a mouse monoclonal antibody (Fig. 3A). However, in some cases, a mouse monoclonal might yield successful enrichment compared to a polyclonal antibody. For instance, successful enrichment (3.2-fold) in ChIP signal relative to the background was obtained for Set1 tagged with 9Myc epitope only using a mouse monoclonal antibody (Fig. 3B).
-
- The antigen-antibody complexes are captured by adding either 10 µl 1:1 slurry containing Protein A- or Protein G-conjugated magnetic beads in FA140 buffer or 25 µl 1:1 slurry of Protein A- or Protein G-conjugated agarose beads in FA140 buffer, and incubation is continued for 1 h at 4°C with end-over-end mixing. Magnetic beads are collected using a magnetic bead concentrator (Dyna-Mag™, Invitrogen) and agarose beads are collected by centrifugation at 1,000 rpm for 30 sec.
-
Prior to washing, the beads are transferred to a pre-lubricated 1.5 ml microfuge tube using FA140 buffer. The beads are washed sequentially twice with 1 ml of FA140 buffer, twice with 1 ml of FA500 buffer (50 mM HEPES.KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate) and once with 1 ml of LiCl/NP40 buffer (10 mM Tris.Cl, pH 8.0, 250 mM LiCl, 0.5% Igepal CA-630/NP40, 0.5% Sodium deoxycholate).Technical Notes:
- Pre-lubricated tubes are used to prevent adhesion of chromatin to the tubes, which can contribute to higher background signal.
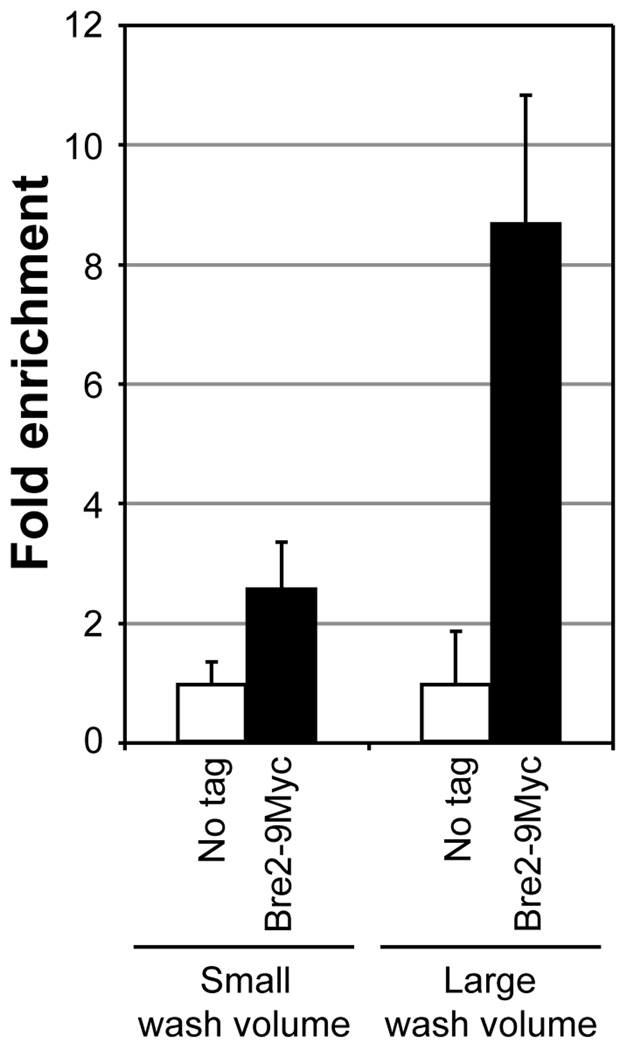
- Importantly, the modified washing process (step 5) performed in 1.5 ml tubes with a larger wash buffer volume (1 ml per wash) and an added high stringency wash result in significantly higher signal to noise ratio as compared to those obtained following the washing steps described in many standard ChIP protocols (500 µl buffer per wash performed in 0.65 ml tubes with a single wash using FA500 buffer). An 8.7-fold enrichment in ChIP signal is obtained relative to the background using the modified washing protocol as opposed to a 2.6-fold enrichment using the standard washing procedure (Fig. 4).
-
- D. DNA purification
- The beads are centrifuged at 2,000 rpm for 30 sec to remove any residual buffer prior to the addition of 400 µl elution solution (1% SDS, 100 mM sodium bicarbonate) and incubated at room temperature with end-over-end mixing for 40 min.
- The eluate is transferred to a fresh 0.65 ml tube, 16 µl 5M NaCl is added and mixed well. At this stage, 400 µl elution solution and 16 µl 5M NaCl is also added to the 10 µl soluble chromatin, set aside to isolate “input” DNA (section 2.2.1B step 5). Reverse crosslinking is done by incubation at 65°C for 5 h in a thermal cycler or a heat-block. The solution is transferred to a fresh 1.5 ml tube, 1 ml ethanol is added, mixed well and precipitation is allowed to occur overnight at −20°C.
- The samples are centrifuged at maximum speed in a tabletop centrifuge at 4°C. The pellet is washed with 800 µl 70% ethanol followed by centrifugation at high speed. Any residual ethanol is completely removed and the pellet is air-dried in a 37°C incubator for 10 min. The pellet is dissolved in 90 µl water, treated with 2 µl 10 mg/ml RNase A and followed by an incubation at 37°C for 30 min. Proteins are removed by adding 10 µl 10× PNK buffer (100 mM Tris.Cl, pH 8.0, 50 mM EDTA, 5% SDS), 1 µl Proteinase K (Sigma) and incubation is done at 42°C for 1h. DNA is purified using Qiaquick PCR Purification kit after addition of 10 µl 3M Sodium acetate to adjust the pH. DNA is eluted in 50 µl water and stored at −20°C.
Fig. 2.
Antibody titration to determine the optimal amount required for high enrichment in ChIP assay. Soluble chromatin was prepared using a yeast strain expressing Swd2 with 3 copies of the HA epitope tag at the C-terminus (Swd2-3HA) or lacking the tag (no tag) subjected to double crosslinking (DMA and HCHO). The soluble chromatin (500 µg) was subjected to immunoprecipitation (IP) using 0.5 µl, 1 µl or 2 µl of an antibody that recognizes the HA epitope (α-HA, GenScript, catalog no. A00168). Quantitative PCR in real time (qPCR) was used to measure DNA associated with Swd2 in the immunoprecipitate and the total DNA in the soluble chromatin subjected to IP (input). Relative enrichment (IP/input) was initially calculated. For no tag control, the relative enrichment obtained represents the background signal (set as 1). Fold enrichment relative to the no tag control is shown for Swd2-3HA using different amounts of the antibody.
Fig. 3.
Determining the type of antibody suitable for effective enrichment of chromatin-bound factors in ChIP assays. Yeast strains expressing either Spp1 or Swd2 with 3 copies of the HA epitope tag at the C-terminus (Spp1-3HA or Swd2-3HA, respectively), expressing Set1 containing 9 copies of Myc epitope tag at the N-terminus (9Myc-Set1) or strains lacking the tags (no tag) were subjected to double crosslinking (DMA and HCHO) prior to soluble chromatin preparation. For panel A, soluble chromatin (500 µg) was subjected to IP using 2 µl α-HA, which is either a mouse monoclonal antibody (mAb) (kindly provided by Ethan Lee) or goat polyclonal antibody (pAb) (GenScript, catalog no. A00168). For panel B, soluble chromatin (1 mg) was subjected to IP using 2 µl of a mouse monoclonal α-Myc (clone 9E10, kindly provided by Ethan Lee) or a rabbit polyclonal antibody α-Myc (GenScript, catalog no. A00172). Enrichment of ChIP DNA relative to input DNA was calculated following qPCR as described in Fig. 2. Fold enrichment using different types of antibody are shown for Spp1-3HA and Swd2-3HA (panel A) or 9Myc-Set1 (panel B) relative to the no tag control (set as 1).
Fig. 4.
Increased washing increases the enrichment of chromatin-bound factors in ChIP assays. Soluble chromatin was prepared using a yeast strain expressing Bre2 with 9 copies of the Myc epitope tag at the C-terminus (Bre2-9Myc) or lacking the tag (no tag) subjected to double crosslinking (DMA and HCHO). 500 µg soluble chromatin was used in IP along with 2 µl mouse monoclonal α-Myc. Following IP, bead-bound immunoprecipitates were washed either with a small volume of buffers [comprised of sequential washes with 500 µl each of FA140 buffer (two times), FA500 buffer and LiCl/NP40 buffer] or a large buffer volume [comprised of sequential washes with 1 ml each of FA140 buffer (two times), FA500 buffer (two times) and LiCl/NP40 buffer]. Enrichment of ChIP DNA relative to input DNA was calculated following qPCR as described in Fig. 2. Fold enrichment using different wash volumes are shown for Bre2-9Myc relative to the no tag control (set as 1).
2.2.2. Chromatin double immunoprecipitation assay for measuring occupancy of H2Bub1
Antibodies specific to mammalian (human, mouse and rat) and budding yeast H2Bub1 have been generated and used to perform ChIP assays (31–32). However, only the mammalian-specific H2Bub1 antibody is commercially available. Therefore, two sequential immunoprecipitations are needed to measure H2Bub1 occupancy in yeast and in any other species for which an H2Bub1-specific antibody is not commercially available. As described in this section, N-terminally Flag epitope-tagged H2B is isolated using α-Flag in the first IP followed by Flag peptide elution. The eluate is then used in a second round of IP using an antibody that recognizes mono- or poly-ubiquitinated proteins. For IP using α-Flag, chromatin from a yeast strain harboring H2B without the Flag tag (no tag control) should included as a control to confirm enrichment of Flag-H2B. Chromatin from a yeast strain harboring a mutation in the site of ubiquitination (Flag-H2B-K123R) should also be included as a control to measure and confirm enrichment of H2Bub1.
Soluble chromatin from formaldehyde-crosslinked cells is prepared as described in section 2.2.1 step B. 1.5–2.0 mg of soluble chromatin is added to 0.65 ml microfuge tube in a final reaction volume of 520 µl made with FA140 buffer. The chromatin is pre-cleared by with 50 µl 1:1 slurry of Protein G-agarose beads in FA140 buffer and incubation at 4°C with end-over-end mixing for 20 min. 2 µl α-Flag is added to precleared chromatin and incubated overnight until the end of following day at 4°C with end-over-end mixing. 50 µl 1:1 slurry of Protein G-agarose beads in FA140 buffer is then added and incubation is continued at 4°C for 1 h. The beads are collected by centrifugation at 1,000 rpm for 30 sec and four washes with 500 µl FA140 buffer each for 5 min at 4°C is then performed.
Bead-bound proteins are eluted by incubation at with 25 µl Flag peptide (4 mg/ml stock, 3× FLAG® Peptide, Sigma) in 500 µl reaction volume made with FA140 buffer and incubated overnight until the end of following day at 4°C with end-over-end mixing. Following centrifugation at 2,000 rpm for 30 sec, 500 µl eluate is transferred to a fresh tube and 50 µl is set aside to isolate DNA to be used as the template for “input” in PCR.
In the second round of IP, 400 µl of the eluate from first round of IP using α-Flag is added to a fresh 0.65 ml microfuge tube and incubated overnight at 4°C with end-over-end mixing along with 2 µl of an antibody that recognizes mono- or poly-ubiquitinated proteins (Enzo Life Sciences; 33). 50 µl 1:1 slurry of Protein G-agarose beads in FA140 buffer is then added and incubation is continued at 4°C for 1 h. Bead-bound antigen-antibody complexes are washed and eluted as described in section 2.2.1C step 5. At this stage, 350 µl elution solution and 16 µl 5M NaCl is also added to the 50 µl elutate from first round IP, set aside to isolate “input” DNA (section 2.2.2 step 2). Reverse crosslinking is done by incubation at 65°C for 5 h and DNA is purified following RNase A and Proteinase K treatments as described above (section 2.1 step 6b).
-
E.
Analysis and quantitation of ChIP DNA
An agarose gel and densitometry-based approach or quantitative PCR in real time (qPCR) using the ChIP DNA can be employed to determine and measure occupancy of histones, their modifications and regulatory factors on specific regions of chromatin. Details of qPCR can be obtained from previous reports (34–35).
Concluding Remarks
We have adopted and refined many of the standard and fundamental procedures used in studying the regulation and functions of histone modifications. Although the procedures described here have been tailored to investigate the crosstalk between H2Bub1 and H3K4/-K79 methylation in budding yeast, they are in general applicable to any histone or non-histone proteins and their modifications that might occur in yeast or in other model organisms. Advantages of the improved methodologies are as listed below.
For total cell lysate preparation (section 1.1), the use of SWBNG buffer increases overall protein solubility under native conditions. The presence of denaturants (urea and SDS) in SUME buffer not only increases protein solubility, but also inactivates proteases and any enzymatic activities that can remove protein modifications.
Preparation of nuclear extracts in SUME buffer offers an easy and rapid approach to obtain high enrichment for better detection of nuclear proteins or their modified forms by Western blotting, especially those present in low abundance or detected using antibodies with low avidity (section 1.2.3). Moreover, spheroplast lysis achieved using Triton X-100 in the modified nuclei isolation procedure obviates the need for the ergonomically challenging Dounce homogenizer used in many published protocols.
The boil procedure preserves highly labile modifications, such as ubiquitination and sumoylation (section 1.3). It is a much simpler alternative to the time-consuming TCA lysis-IP method used to enrich and detect H2Bub1 (25). Furthermore, use of a modified Laemmli sample buffer (lacking reducing agents and bromophenol blue) and the availability of a detergent-compatible protein assay kit enable estimation of the protein concentration of the lysate. In turn, this permits the use of equal amounts of lysates from different strains to accurately measure changes in the global, steady state levels of histone ubiquitination and sumoylation in Western blotting (18).
A chromatin fraction, without any contamination from unlysed spheroplasts and/or the cytoplasmic proteins, can be obtained by isolating nuclei using a Percoll gradient (section 2.1). The isolated contaminant-free nuclei can also be used to prepare nuclear/chromatin extracts to detect or evaluate changes in protein-protein interactions using Co-IP assays or to investigate changes in chromatin structure using nucleases (e.g., micrococcal nuclease, DNase1). Since protein-protein interactions are electrostatic in nature, isolated nuclei can be incubated in solutions with different salt concentrations to assess changes in the solubility of a protein(s), which might result from any change in its interaction with other proteins or with chromatin (e.g., salt-dependent nucleosome disruption assays) (18).
-
Since the antibody that specifically recognizes yeast H2Bub1 is not commercially available, our ChDIP assay provides a simple method to directly measure changes in the occupancy and distribution of H2Bub1 on chromatin (section 2.2.2). Additionally, it circumvents the inducible expression of an epitope-tagged ubiquitin needed for ChDIP assays (36–39). Chromatin-bound H2Bub1 and other ubiquitinated proteins can be isolated using the antibody that recognizes mono- or poly-ubiquitinated conjugates. True enrichment of only H2Bub1 can then be determined using the chromatin isolated from a yeast mutant lacking H2Bub1 as a negative control in the ChDIP assays (e.g., H2B-K123R).
Employing this rationale, it is also conceivable that the occupancy and distribution of any protein of interest modified by ubiquitin or ubiquitin-like moieties (e.g., SUMO, Urm1, Nedd8) on chromatin can be easily assessed using an antibody that specifically recognizes these modifications and using the chromatin obtained from a yeast strain harboring a mutation in the site of modification as the negative control. We recently showed that sumoylation could be induced to occur on the H2B C-terminal region by inserting SUMO consensus sites (18). Importantly, using an antibody that recognizes yeast SUMO (α-smt3) and chromatin obtained from a yeast mutant lacking the engineered sumoylation, we confirmed that the induced sumoylation occurs on chromatin similar to H2Bub1.
-
Most protocols for ChIP assays available in the literature can be used with great success to determine changes in H3K4 and -K79 methylation. However, determining the occurrence and distribution of chromatin-bound factors by ChIP assays can be challenging. Several parameters affect the effective enrichment and detection of chromatin bound factors by ChIP assays, such as, factor abundance within the cell and on chromatin, crosslinking efficiency (affected by whether the factor associates directly with DNA or is bridged by other interacting proteins) and the availability of an antibody with high avidity that specifically recognizes the protein of interest.
In some cases, use of a protein-protein crosslinking agent (such as, DMA) in addition to formaldehyde can increase the crosslinking efficiency (29). While the ease of epitope tagging factors in yeast circumvents the need to raise antibodies that specifically recognize a factor, epitope masking and avidity of the epitope-tag specific antibody can still hinder ChIP efficiency. As shown in Fig. 3, the type of an epitope tag and the epitope tag-specific antibody to be used varies from one factor to another and should be experimentally determined. Nevertheless, extensive washing with large volumes of wash buffer and including an additional high stringency wash increases the ChIP efficiency for many subunits of the Set1-COMPASS complex (Fig. 4; 20). Using these modified washing steps, we have also successfully obtained significant enrichment over the background for Jhd2 (Fu and Sun, unpublished observations), a protein shown to be recalcitrant in ChIP assays (40).
-
In general, ChIP assays are used to determine changes in chromatin-bound levels of a protein at a single gene or a subset of genes and at different regions of a gene in yeast mutants or in strains subjected to different treatments. Since only a small set of genes are investigated, any conclusion pertaining to the effect of a mutation or an experimental condition on the chromatin association of a protein is restricted to the local changes at the specific gene(s) or region(s) of a gene. A comprehensive view of the global chromatin bound levels of a protein can be obtained using genome-wide association studies employing ChIP-chip or ChIP-seq (41). While these approaches provide high information content and allow one to draw broad conclusions, they significantly impinge on financial resources. Therefore, the chromatin association assay described here using the chromatin obtained from isolated nuclei is a cost-effective and easy alternative, and it provides a quick snapshot of the global chromatin-bound levels of a nuclear protein.
Congruency of the data obtained from ChIP and chromatin association assays would confirm that a mutation or an experimental regimen does truly affect the chromatin binding of a protein of interest. However, any discrepancy or difference in the chromatin-bound levels of a protein using these assays also provides important clues regarding changes in the interactions of a given protein with other proteins or its interaction with chromatin (Fig. 5). In the chromatin association assay, weakened interactions of a protein might result in its dissociation from chromatin, culminating in its reduced levels in the chromatin fraction (Fig. 5 ii–iii). In ChIP assays, DMA and formaldehyde-mediated double crosslinking can stably tether the affected protein to chromatin and thereby, prevent its loss from chromatin in the mutant (Fig. 5 v–vi). Indeed, similar to the scenario depicted in Fig. 5, we found that the H2B levels on chromatin were reduced in yeast mutants lacking H2Bub1 using chromatin association assay, but ChIP assays showed that the H2B occupancy on chromatin in these mutants was similar to the wild type (18). These differences might be due to weakened interactions of H2B with other histones and/or with DNA in absence of H2Bub1. Since protein-protein and protein-DNA interactions are electrostatic in nature, these interactions can be sensitive to changes in the ionic strength of the surrounding environment. Indeed, salt-dependent nucleosome disruption assays showed that histones are easily extractable into low salt solutions in the absence of H2Bub1 and high salt solutions are required to solubilize H2Bub1-containing chromatin, confirming that H2Bub1 stabilizes nucleosome (18). Therefore, the combined approach of using ChIP and a chromatin association assay is likely to provide valuable insights into any subtle or profound changes in the protein-protein interactions that might occur on chromatin, for instance, interactions between various subunits of histone-modifying or chromatin-remodeling complexes.
Fig. 5.
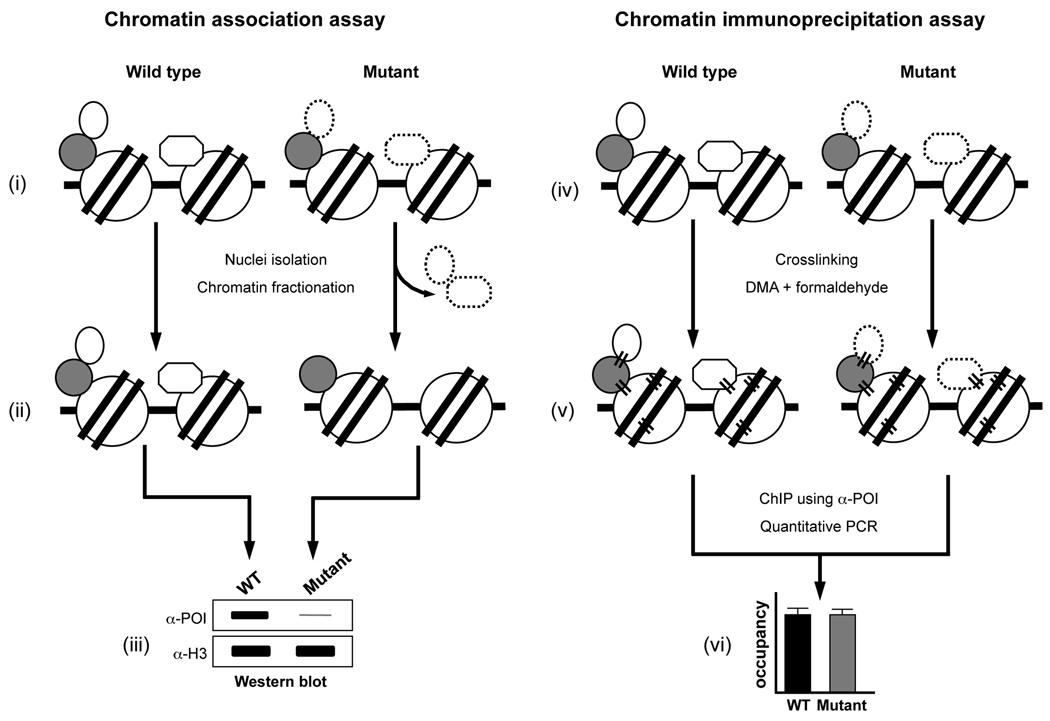
A combinatorial approach employing chromatin association and ChIP assays can reveal defects in protein-protein interactions on chromatin. Schematic representations of the chromatin association assay and ChIP assay are shown. A speculative scenario of destabilization in the interactions of a chromatin-bound protein under certain experimental condition (mutation or treatment) compared to the control (wild type or untreated) and discrepancy/difference in the detection of this destabilized interaction between the two assays are depicted. In (i) and (iv), a protein of interest (POI, oval or octagon) is shown stably associated with the wild type chromatin either by its interactions with a partner or bridging protein (gray circle) or via its direct interaction with the nucleosome (histone octamer, circle; DNA, solid line). These direct and indirect interactions of the POI may be destabilized or weakened in the mutant without any reduction in its overall levels on chromatin (oval or octagon with dashed line). (ii) The destabilized/weakened interactions might render the POI susceptible to dissociation from the chromatin during the multiple steps, with solutions of different ionic strengths, involved in the chromatin association assay (nuclei isolation and chromatin fractionation) and resulting in the reduced levels of POI on the chromatin in the mutant. (iii) Normalized soluble chromatin is subjected to Western blotting and an antibody (α-POI) is used to measure changes in the chromatin-bound levels of POI in the wild type or mutant. The amount of H3 is used to confirm that equal amounts of chromatin were used in the assay (detected using α-H3, loading control). As shown in the mock-up Western blot, chromatin-bound levels of POI in the mutant might be highly reduced (thin gray line) due to its dissociation as compared to the wild type. (v) Addition of a bifunctional crosslinking agent (DMA) and formaldehyde causes protein-protein and protein-DNA crosslinking (parallel black lines) and results in stable tethering of the POI to chromatin. Crosslinking stabilizes the “loose” or weakened interactions of POI in the mutant and prevents its dissociation during the several incubation and wash steps (including highly stringent conditions, such as the exposure to 500 mM NaCl and detergents) involved in the ChIP assay. (vi) Soluble chromatin is prepared from the wild type and mutant strains subjected to double crosslinking and IP is performed using α-POI. Changes in the chromatin-bound levels of POI (occupancy) in the wild type and mutant are determined using qPCR analysis of input and ChIP DNA. As depicted in the model graph, owing to crosslinking, occupancy of the POI on chromatin in the mutant might be similar to that in the wild type, unlike in chromatin association assay (iii).
Table 1.
Saccharomyces cerevisiae strains
| Strain | Genotype | Source |
|---|---|---|
| Y131 |
Mat a hta1-htb1Δ::LEU2, HTA2-GAL1/GAL10-HTB2, leu2-2,-112, ura3-1, trp1-1, his3–11,-15 ade2-1, can1-100, ssd1, HTA1-HTB1 (2µ, URA3) |
[42] |
| YZS276 | HTA1-Flag-HTB1 (CEN, HIS3); derived from Y131 | [17] |
| YZS277 | HTA1-Flag-htb1-K123R (CEN, HIS3); derived from Y131 | [17] |
| YZS383 | HTA1-HTB1 (2µ, URA3), bre1Δ::KanMX6; derived from Y131 | This study |
| YZS385 | HTA1-Flag-HTB1 (CEN, HIS3), bre1Δ::KanMX6; derived from YZS383 | This study |
| YZS440 | HTA1-HTB1 (2µ, URA3), BRE2-9Myc::TRP1; derived from Y131 | This study |
| YZS518 | HTA1-HTB1 (2µ, URA3), 9Myc-SET1::TRP1; derived from Y131 | [20] |
| YZS537 | HTA1-HTB1 (2µ, URA3), SPP1-3HA::KanMX4, 9Myc-SET1::TRP1; derived from Y518 | [20] |
| YZS631 | HTA1-HTB1 (2µ, URA3), SWD2-3HA::KanMX4, 9Myc-SET1::TRP1; derived from Y518 | [20] |
Acknowledgements
We thank Vincent Geli for the Set1 epitope-tagging plasmids; Ethan Lee for the HA and Myc mouse monoclonal antibodies; and Mary Ann Osley, Brian Strahl and Tony Weil for some of the yeast strains used in this study. We also thank Ya-Ting Chung and Yi-Chun Chen for their valuable contributions in the standardization of the experimental procedures. This work was supported by funds from The Vanderbilt-Ingram Cancer Center, The Robert J. and Helen C. Kleberg Foundation, and National Institutes of Heath (RO1CA109355).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell. Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campos EI, Reinberg D. Histones: annotating chromatin. Annu. Rev. Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 6.Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bähler J, Green RD, Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Latham JA, Dent SYR. Cross-regulation of histone modifications. Nat. Struct. Mol. Biol. 2007;14:1017–1024. doi: 10.1038/nsmb1307. [DOI] [PubMed] [Google Scholar]
- 8.Fingerman IM, Du HN, Briggs SD. Controlling histone methylation via transhistone pathways. Epigenetics. 2008;35:237–242. doi: 10.4161/epi.3.5.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol. Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 10.Osley MA. H2B ubiquitylation: The end is in sight. Biochim. Biophys. Acta. 2004;1677:74–79. doi: 10.1016/j.bbaexp.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 11.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 12.Henry KW, Berger SL. Trans-tail histone modifications: wedge and bridge? Nature Struc. 2002;9:565–566. doi: 10.1038/nsb0802-565. [DOI] [PubMed] [Google Scholar]
- 13.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 14.Shahbazian MD, Zhang K, Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell. 2005;19:271–277. doi: 10.1016/j.molcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 16.Vitaliano-Prunier A, Menant A, Hobeika M, Géli V, Gwizdek C, Dargemont C. Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat. Cell Biol. 2008;10:1365–1371. doi: 10.1038/ncb1796. [DOI] [PubMed] [Google Scholar]
- 17.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 18.Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of Histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc. Natl. Acad. Sci. USA. 2009;106:16686–16691. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fingerman IM, Li HC, Briggs SD. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway. Genes Dev. 2007;21:2018–2029. doi: 10.1101/gad.1560607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandrasekharan MB, Huang F, Chen YC, Sun ZW. Histone H2B C-terminal helix mediates the trans-histone H3K4 methylation independent of H2B ubiquitination. Mol. Cell. Biol. 2010;30:3216–3232. doi: 10.1128/MCB.01008-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardner RG, Nelson ZW, Gottschling DE. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol. Cell. Biol. 2005;25:6123–6139. doi: 10.1128/MCB.25.14.6123-6139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kizer KO, Xiao T, Strahl BD. Accelerated nuclei preparation and methods for analysis of histone modifications in yeast. Methods. 2006;40:296–302. doi: 10.1016/j.ymeth.2006.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piñeiro M, Puerta C, Palacián E. Yeast nucleosomal particles: structural and transcriptional properties. Biochemistry. 1991;30:5805–5810. doi: 10.1021/bi00237a025. [DOI] [PubMed] [Google Scholar]
- 24.Dong L, Xu CW. Carbohydrates induce mono-ubiquitination of H2B in yeast. J. Biol. Chem. 2004;279:1577–1580. doi: 10.1074/jbc.C300505200. [DOI] [PubMed] [Google Scholar]
- 25.Kao CF, Osley MA. In vivo assays to study histone ubiquitylation. Methods. 2003;31:59–66. doi: 10.1016/s1046-2023(03)00088-4. [DOI] [PubMed] [Google Scholar]
- 26.Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amati BB, Gasser SM. Chromosomal ARS and CEN elements bind specifically to the yeast nuclear scaffold. Cell. 1988;54:967–978. doi: 10.1016/0092-8674(88)90111-0. [DOI] [PubMed] [Google Scholar]
- 28.Ruiz-Garcí AB, Sendra R, Galiana M, Pamblanco M, Pérez-Ortín JE, Tordera V. HAT1 and HAT2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. J. Biol. Chem. 1998;273:12599–12605. doi: 10.1074/jbc.273.20.12599. [DOI] [PubMed] [Google Scholar]
- 29.Kurdistani SK, Grunstein M. In vivo protein-protein and protein-DNA crosslinking for genomewide binding microarray. Methods. 2003;31:90–95. doi: 10.1016/s1046-2023(03)00092-6. [DOI] [PubMed] [Google Scholar]
- 30.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 31.Minsky N, Shema E, Field Y, Schuster M, Segal E, Ore M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 32.Schulze JM, Jackson J, Nakanishi S, Gardner JM, Hentrich T, Haug J, Johnston M, Jaspersen SL, Kobor MS, Shilatifard A. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol. Cell. 2009;35:626–641. doi: 10.1016/j.molcel.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujimuro H, Yokosawa H. Production of antipolyubiquitin monoclonal antibodies and their use for characterization and isolation of polyubiquitinated proteins. Methods Enzymol. 2005;399:75–86. doi: 10.1016/S0076-6879(05)99006-X. [DOI] [PubMed] [Google Scholar]
- 34.Aparicio O, Geisberg JV, Struhl K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr. Protoc. Cell Biol. 2004 doi: 10.1002/0471143030.cb1707s23. Unit 17.7 (Chapter 17) [DOI] [PubMed] [Google Scholar]
- 35.Tsukuda T, Trujillo KM, Martini E, Osley MA. Analysis of chromatin remodeling during formation of a DNA double-strand break at the yeast mating type locus. Methods. 2009;48:40–45. doi: 10.1016/j.ymeth.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geng F, Tansey WP. Polyubiquitylation of histone H2B. Mol. Biol. Cell. 2008;19:3616–3624. doi: 10.1091/mbc.E08-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ingvarsdottir K, Edwards C, Lee MG, Lee JS, Schultz DC, Shilatifard A, Shiekhattar R, Berger SL. Histone H3 K4 demethylation during activation and attenuation of GAL1 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:7856–7864. doi: 10.1128/MCB.00801-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat. Rev. Genet. 2008;6:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]