Abstract
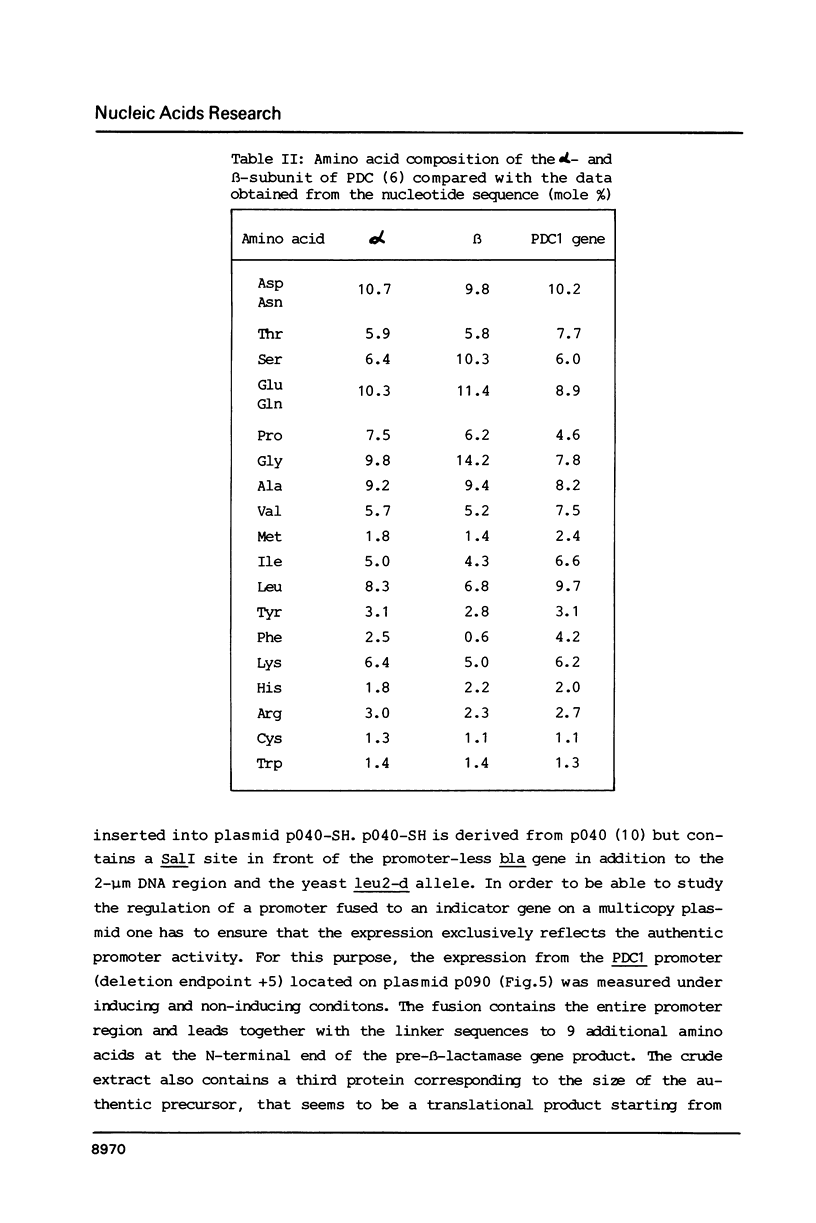
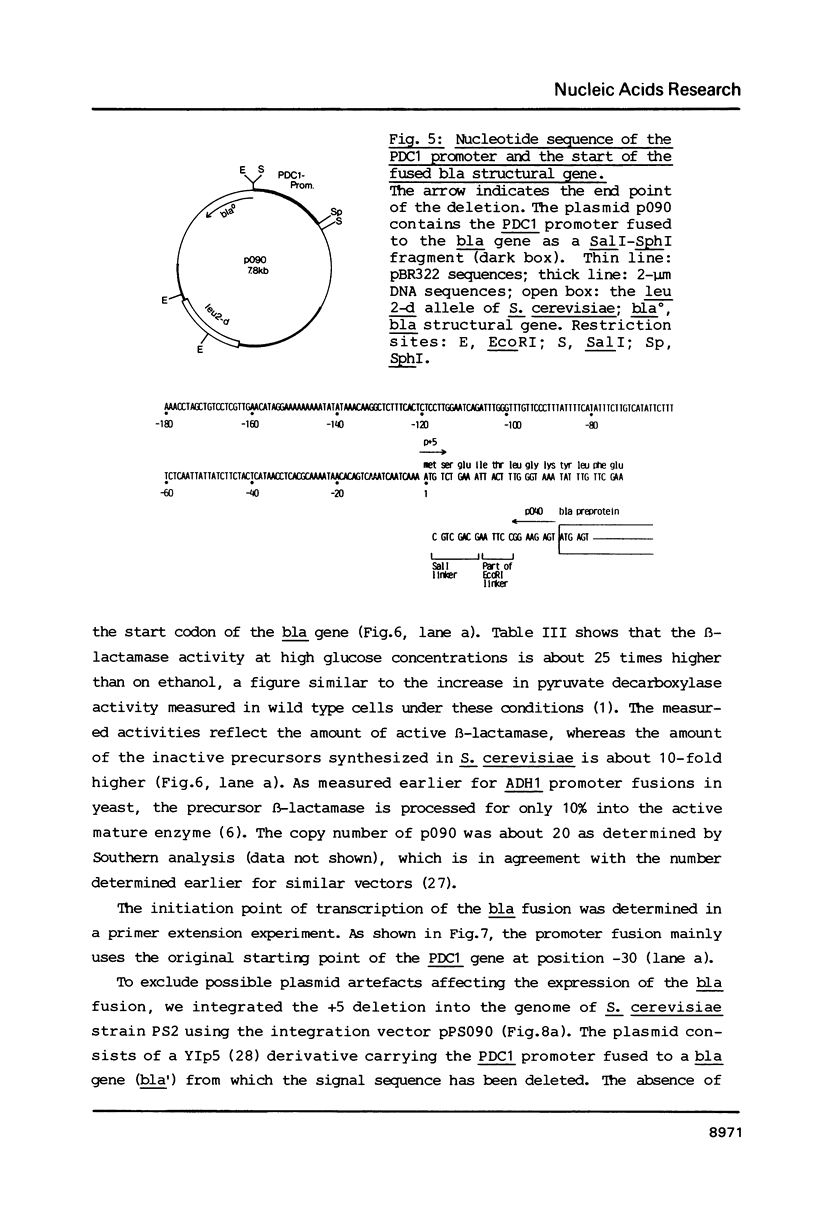
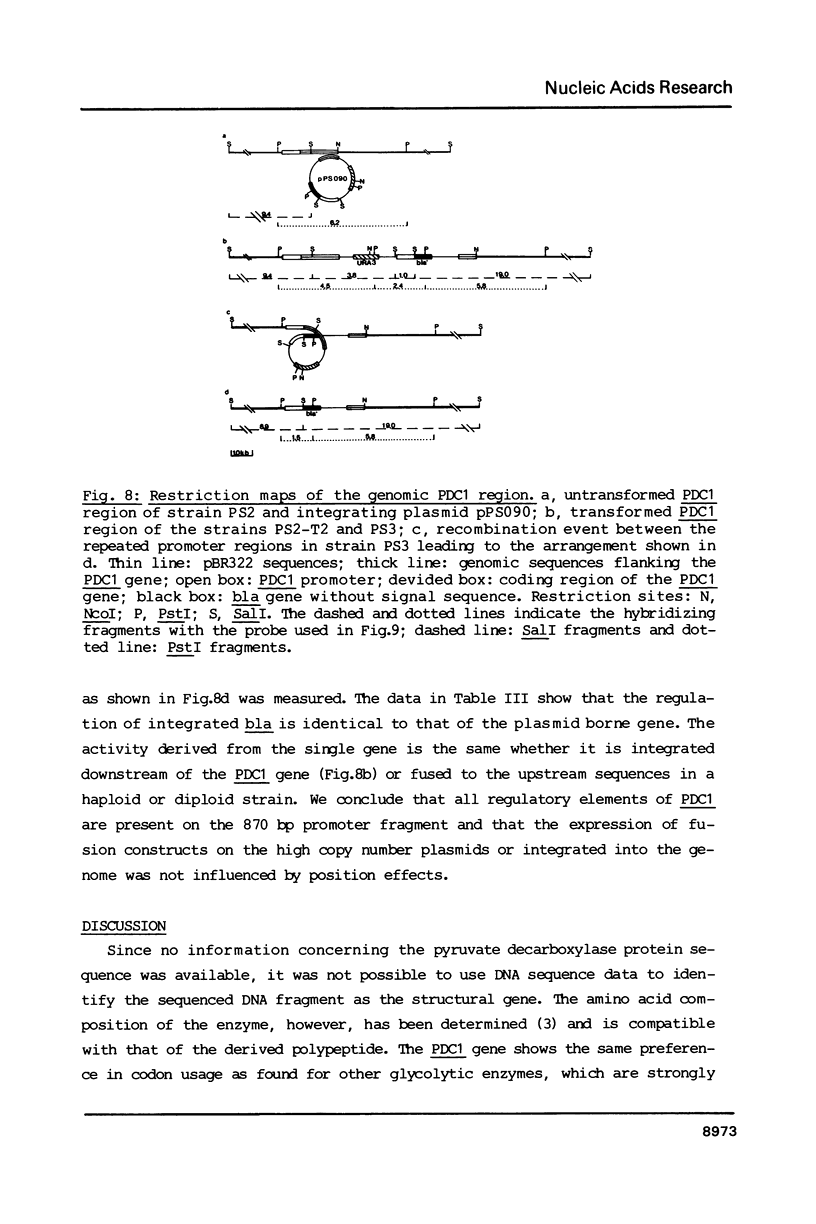
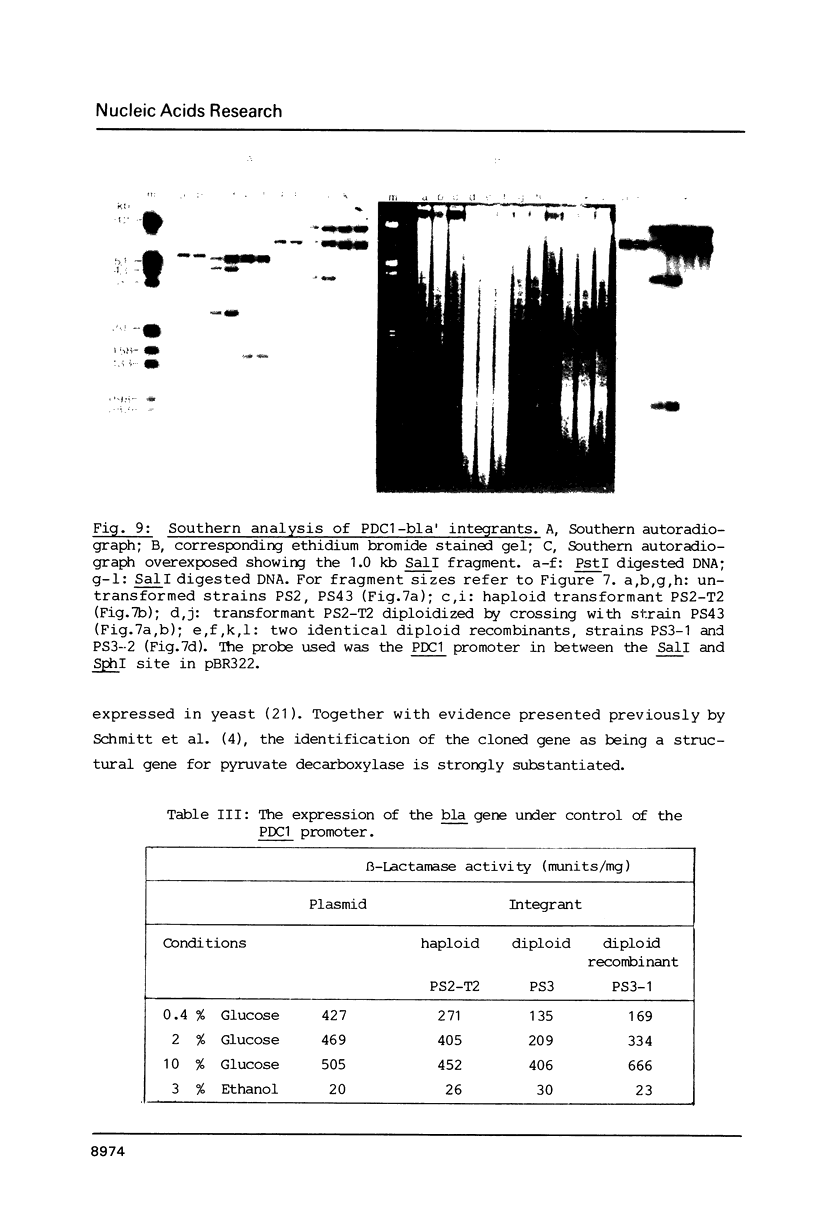
The PDC1 gene of Saccharomyces cerevisiae, encoding pyruvate decarboxylase was sequenced. The gene contains an open reading frame of 1647 base pairs. The codon usage shows the same strong bias as found for some other glycolytic enzymes. Transcription starts mainly at -30 and terminates 100 base pairs downstream of the termination codon. In some strains a second termination site, 46 base pairs upstream of the stop codon was observed. The function of the promoter region was analyzed by fusion to the bacterial structural gene encoding beta-lactamase (bla). On multicopy plasmid or integrated in the genome, the expression of the bla gene showed the regulation of the authentic PDC1 gene.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achstetter T., Emter O., Ehmann C., Wolf D. H. Proteolysis in eukaryotic cells. Identification of multiple proteolytic enzymes in yeast. J Biol Chem. 1984 Nov 10;259(21):13334–13343. [PubMed] [Google Scholar]
- Beggs J. D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978 Sep 14;275(5676):104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig K. D., Mackedonski V., Hollenberg C. P. Transcription of the bacterial beta-lactamase gene in Saccharomyces cerevisiae. Gene. 1982 Nov;20(1):1–10. doi: 10.1016/0378-1119(82)90082-8. [DOI] [PubMed] [Google Scholar]
- Ciriacy M. Isolation and characterization of further cis- and trans-acting regulatory elements involved in the synthesis of glucose-repressible alcohol dehydrogenase (ADHII) in Saccharomyces cerevisiae. Mol Gen Genet. 1979 Nov;176(3):427–431. doi: 10.1007/BF00333107. [DOI] [PubMed] [Google Scholar]
- Dobson M. J., Tuite M. F., Roberts N. A., Kingsman A. J., Kingsman S. M., Perkins R. E., Conroy S. C., Fothergill L. A. Conservation of high efficiency promoter sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 1982 Apr 24;10(8):2625–2637. doi: 10.1093/nar/10.8.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhart E., Hollenberg C. P. The presence of a defective LEU2 gene on 2 mu DNA recombinant plasmids of Saccharomyces cerevisiae is responsible for curing and high copy number. J Bacteriol. 1983 Nov;156(2):625–635. doi: 10.1128/jb.156.2.625-635.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Swinscoe J., Lebowitz P., Weissman S. M. Heterogeneity and 5'-terminal structures of the late RNAs of simian virus 40. J Mol Biol. 1978 Dec 25;126(4):813–846. doi: 10.1016/0022-2836(78)90022-0. [DOI] [PubMed] [Google Scholar]
- Gounaris A. D., Turkenkopf I., Civerchia L. L., Greenlie J. Pyruvate decarboxylase III. Specificity restrictions for thiamine pyrophosphate in the protein association step, sub-unit structure. Biochim Biophys Acta. 1975 Oct 20;405(2):492–499. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Roggenkamp R., Kustermann-Kuhn B., Hollenberg C. P. Expression and processing of bacterial beta-lactamase in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4466–4470. doi: 10.1073/pnas.78.7.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt H. D., Ciriacy M., Zimmermann F. K. The synthesis of yeast pyruvate decarboxylase is regulated by large variations in the messenger RNA level. Mol Gen Genet. 1983;192(1-2):247–252. doi: 10.1007/BF00327674. [DOI] [PubMed] [Google Scholar]
- Schmitt H. D., Zimmermann F. K. Genetic analysis of the pyruvate decarboxylase reaction in yeast glycolysis. J Bacteriol. 1982 Sep;151(3):1146–1152. doi: 10.1128/jb.151.3.1146-1152.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich J. Structure-function relationships in pyruvate decarboxylase of yeast and wheat germ. Ann N Y Acad Sci. 1982;378:287–305. doi: 10.1111/j.1749-6632.1982.tb31203.x. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]