Abstract
Components of the planar cell polarity (PCP) pathway are required for the caudal tangential migration of facial branchiomotor (FBM) neurons, but how PCP signaling regulates this migration is not understood. In a forward genetic screen, we identified a new gene, nhsl1b, required for FBM neuron migration. nhsl1b encodes a WAVE-homology domain-containing protein related to human Nance-Horan syndrome (NHS) protein and Drosophila GUK-holder (Gukh), which have been shown to interact with components of the WAVE regulatory complex that controls cytoskeletal dynamics and with the polarity protein Scribble, respectively. Nhsl1b localizes to FBM neuron membrane protrusions and interacts physically and genetically with Scrib to control FBM neuron migration. Using chimeric analysis, we show that FBM neurons have two modes of migration: one involving interactions between the neurons and their planar-polarized environment, and an alternative, collective mode involving interactions between the neurons themselves. We demonstrate that the first mode of migration requires the cell-autonomous functions of Nhsl1b and the PCP components Scrib and Vangl2 in addition to the non-autonomous functions of Scrib and Vangl2, which serve to polarize the epithelial cells in the environment of the migrating neurons. These results define a role for Nhsl1b as a neuronal effector of PCP signaling and indicate that proper FBM neuron migration is directly controlled by PCP signaling between the epithelium and the migrating neurons.
Keywords: Facial branchiomotor neuron, Nance-Horan syndrome-like 1b, Planar cell polarity, Neuron migration, Zebrafish
INTRODUCTION
In the developing vertebrate brain, neurons frequently migrate considerable distances from the proliferative zone where they are born to the location where they carry out their specialized functions. Cell migration in general involves complex interactions between the migrating cell and its environment. Examples of such interactions within the central nervous system are those between migrating cortical neurons and their radial glia substrates (Marin and Rubenstein, 2003) and between neurons of the rostral migratory stream and the astrocytic tubes through which they migrate to the olfactory bulb (Kaneko et al., 2010). An in vivo genetic approach is required to understand the interactions between migrating neurons and their environment and to identify the genes involved in these interactions.
Accumulating evidence has indicated that directed cell migration is impacted by activity of the non-canonical Wnt/planar cell polarity (PCP) signaling pathway. In vertebrates, as in Drosophila, PCP signaling coordinates the orientation of cellular structures within the plane of an epithelium, such as the orientation of stereocilia bundles in the inner ear (Kelly and Chen, 2007) and the asymmetric localization of motile cilia in epithelia (Park et al., 2008; Borovina et al., 2010). The PCP pathway is also known to be active in controlling directed cell motility in convergent extension (CE) movements during gastrulation and the polarized cell behaviors required for neural tube closure (Heisenberg and Tada, 2002; Wallingford, 2006), neural crest migration (De Calisto et al., 2005; Carmona-Fontaine et al., 2008) and epidermal wound healing (Caddy et al., 2010). The involvement of PCP genes in neuronal migration comes from the study of facial branchiomotor (FBM) neurons in the segmented hindbrain of vertebrates. FBM neurons are a subset of cranial branchiomotor neurons that are generated ventrally in rhombomere (r)4 and undergo a highly stereotyped caudal migration into r6 and r7 in the zebrafish. There, they form the facial motor nucleus from which axons exit the hindbrain in r4 and innervate muscles in the head derived from the second branchial arch (Chandrasekhar, 2004). During this migration, FBM neurons move through the neuroepithelium adjacent to the floor plate, in contact both with the basement membrane and with other migrating FBM neurons (Grant and Moens, 2010). Zygotic loss-of-function of the core PCP components Vang-like 2 (Vangl2), Prickle (Pk1a and Pk1b), Frizzled (Fzd3a) and Celsr (Celsr2) in zebrafish all lead to a specific failure of FBM neuron migration (Bingham et al., 2002; Jessen et al., 2002; Carreira-Barbosa et al., 2003; Wada et al., 2005; Wada et al., 2006; Rohrschneider et al., 2007). This role for PCP in directing FBM neuron migration is evolutionarily conserved, as similar phenotypes are observed in mouse mutants for Vangl2, Fzd3 and Celsr (Vivancos et al., 2009; Qu et al., 2010).
Although this genetic evidence implicates the PCP pathway in FBM migration, it is unclear how PCP components regulate migration. In epithelia, PCP core components function to communicate subcellular differences in polarized information between neighboring cells in a cell-cell contact-dependent manner (Vladar et al., 2009). This molecular polarity is then transferred into context-dependent morphological asymmetries through the activity of cell type-specific downstream effector molecules that link polarity information to changes in the actin cytoskeleton (Strutt et al., 1997; Lee and Adler, 2002; Strutt and Warrington, 2008). In FBM neuron migration, previous chimeric analyses have suggested that the PCP components Vangl2, Fzd3a and Celsr2 act primarily non cell-autonomously, as wild-type neurons fail to migrate through a mutant neuroepithelium and mutant neurons do migrate through a wild-type environment, albeit incompletely (Jessen et al., 2002; Wada et al., 2005; Wada et al., 2006). Because the neuroepithelium through which FBM neurons migrate displays aspects of planar polarity and expresses PCP components (Ciruna et al., 2006; Borovina et al., 2010), it has been suggested that this environment shapes the trajectory of FBM neuron migration indirectly, by providing a permissive route for migration. However, other evidence suggests a more direct role for PCP signaling, as the core component Pk1b is required cell-autonomously for FBM neuron migration (Rohrschneider et al., 2007; Mapp et al., 2011). Importantly for this work, no PCP effectors for migration have been identified to date that could help to elucidate how the PCP pathway regulates neuronal migration in vivo.
The large PDZ-domain containing protein Scribble (Scrib) is also required for FBM neuron migration (Wada et al., 2005; Vivancos et al., 2009). Scrib has diverse functions in cell polarity and migration. In addition to its well known function in defining apico-basal polarity in epithelial cells in Drosophila together with Discs large (Dlg; Dlg1 – FlyBase) and Lethal giant larvae [Lgl; L(2)gl – FlyBase] (Bilder et al., 2000; Bilder and Perrimon, 2000), Scrib functions as a PCP component in vertebrates, where it interacts with Vangl2 to control the orientation of ear sensory cells, convergent extension and neural tube closure (Montcouquiol et al., 2003; Murdoch et al., 2003; Wada et al., 2005; Montcouquiol et al., 2006). In migratory cells in culture, Scrib is localized to the leading edge, where it promotes cell protrusions by locally modulating the activity and localization of Rac and Cdc42 as a complex with the exchange factors βPIX and GIT1 (Osmani et al., 2006; Dow et al., 2007; Nola et al., 2008). Thus, Scrib represents a polarity protein with well characterized cell-autonomous functions in migration; however, until now, its function in FBM neuron migration, like that of other PCP components, has been described as largely non-autonomous (Wada et al., 2005).
In a zebrafish forward genetic screen for mutants with defective FBM neuron migration, we identified a mutation in Nance-Horan syndrome-like 1b (nhsl1bfh131). Nhsl1b is related to the human Nance-Horan syndrome (NHS) protein and to the Drosophila Scrib-interacting protein Guk-holder (Gukh). We show that nhsl1b is required cell-autonomously in FBM neuron migration and show that Nhsl1b protein localizes to the edge of membrane protrusions in FBM neurons in vivo. In cell transplantation experiments, we show that scrib and vangl2 have a previously unappreciated cell-autonomous role in FBM neuron migration in addition to their non-cell-autonomous role, and that nhsl1b interacts genetically with scrib in this process. We hypothesize that Nhsl1b is a neuronal PCP effector, the first in this system, which functions in migrating neurons to execute directed cell movements. Our results thus support a model whereby PCP signaling between FBM neurons and their environment functions to control directly the trajectory of migration. Furthermore, our chimeric analyses revealed that FBM neurons have an alternative, collective mode of migration that requires interactions between migrating FBM neurons themselves and occurs independently of Nhsl1b or PCP proteins in the migrating neurons. We propose a model in which PCP-dependent and collective modes together drive directed migration of FBM neurons in vivo.
MATERIALS AND METHODS
Zebrafish husbandry, screening and positional cloning
Zebrafish (Danio rerio) were maintained according to standard procedures and staged as previously described (Kimmel et al., 1995). The Isl1:GFP transgenic line, registered as Tg(isl1:GFP)rw0 at The Zebrafish International Resource Center (ZIRC) (Higashijima et al., 2000) was maintained in the *AB background. The isl1:membRFP transgenic line, designated Tg(isl1CREST-hsp70l:mRFP)fh1, was described previously (Grant and Moens, 2010). The WIK strain was used for positional cloning (Shimoda et al., 1999). The scrib mutant was originally described as landlocked, llkrw468 (Wada et al., 2005). The vangl2/trilobite mutant was originally described as trim209 (Jessen et al., 2002).
To induce point mutations in premeiotic germ cells, male Isl1-GFP fish were treated with the chemical mutagen n-ethyl-n-nitrosourea (ENU, Sigma) according to standard methods (van Eeden et al., 1999). Mutants with defects in the migration of FBM neurons were isolated by screening gynogenetic diploid zebrafish embryos produced using the early pressure method (Beattie et al., 1999; Walker et al., 2009). Putative mutants were outcrossed to the wild-type (WT) WIK strain and mutants and carriers identified by random crosses between siblings. Bulk segregant analysis was performed on mutant progeny and phenotypically WT animals collected from incrosses (Bahary et al., 2004).
In our screen, a single allele of nhsl1b (nhsl1bfh131) was identified. Two further non-complementing alleles, nhsl1bfh280 and nhsl1bfh281, were identified by screening nhsl1b exon 6 (1923 bp) on a library of 8600 F1 ENU-mutagenized fish by TILLING (Draper et al., 2004).
Morpholino injections
Antisense morpholinos (MO) were injected at the 1-cell stage. Morpholinos were as follows: Nhsl1b: (MO E4I4 5′-CTAAAAGTTTAACTTCTCACCCGTG-3′; MO exon1 ATG 5′-CGGGAAACGGCATTTTAAATCCTGT-3′), 5 ng; Hoxb1a: (Cooper et al., 2003), 2 ng; Pk1b: MO1 + MO2 (Rohrschneider et al., 2007), 2 ng each; Scrib MO: (Wada et al., 2005), 5 ng; Vangl2 MO (Park et al., 2008), 3 ng.
Plasmids and mRNA injections
Plasmids encoding the scrib gene and the psd95 gene (Meyer et al., 2005; Wada et al., 2005) were subcloned into pCS2 expression vectors as GST- or GFP-N-terminal fusion proteins using the gateway system (Villefranc et al., 2007). GFP-prickle mRNA was used as described (Ciruna et al., 2006). Sense-capped mRNA was synthesized using mMessage mMachine (Ambion). Approximately 1 nl of mRNA was injected into one-cell- or eight-cell-stage embryos at concentrations ranging from 0.1 to 0.5 ng/nl in nuclease-free water (Ambion).
In situ hybridization and whole-mount immunohistochemistry
RNA in situ hybridization was carried out as previously described (Feng et al., 2010; Moens et al., 1998). Whole-mount immunostaining was performed as described previously (Grant and Moens, 2010) with the following antibodies: anti-islet1 (1:50, Developmental Studies Hybridoma Bank); anti-GFP (1:2000, Torrey Pines), anti-ZO-1 (1:1000, Zymed), anti-γ-tubulin (1:500, Sigma), anti-Arl13b (1:200, gift from Z. Sun, Department of Genetics, Yale University School of Medicine). The anti-Nhsl1b antibody is a rabbit polyclonal antibody directed against the C-terminus of zebrafish Nhsl1b (1:200, AnaSpec).
Cell culture, transfections and immunoprecipitations
For protein-protein interaction studies, HEK293T cells were grown in DMEM (Gibco), 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco). Cells were transfected using Lipofectamine 2000 (Invitrogen) using standard protocols. Cells were washed twice in cold PBS, and lysed in NP40 lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% NP40, 0.01 M EDTA) with complete protease cocktail inhibitor (Roche) and 1 mM PMSF. Lysates were cleared and incubated with anti-GST (Abcam), anti-myc (9E10) or anti-GFP (Torrey Pines) for 2 hours at 4°C. Immunocomplexes were precipitated by the addition of protein A-, or G-conjugated Dynabeads (Invitrogen) for 1 hour at 4°C. Beads were washed three times in NP40 lysis buffer and resuspended in 2× SDS sample buffer. Immunoprecipitates were separated by SDS-PAGE and analyzed by immunoblotting with the indicated antibodies.
Cell transplantation
Chimeric embryos were made by transplantation at the early gastrula stage as described (Carmany-Rampey and Moens, 2006; Kemp et al., 2009). To track transplanted cells, donor embryos carrying the isl1:GFP transgene were injected with cascade blue-dextran or rhodamine dextran (10,000 mw, Molecular Probes). In some experiments, host embryos carried the Tg(isl1CREST-hsp70l:mRFP)fh1 transgene so that host motorneurons could be visualized in live embryos. Alternatively, the position of host motorneurons was visualized by immunostaining with anti-islet1 antibody. Embryos were imaged on a Zeiss Pascal or Zeiss 510 confocal microscope.
RESULTS
A forward genetic screen yields a mutant with specific disruption in migration of facial branchiomotor neurons
To identify novel genes required for the tangential caudal migration of facial branchiomotor (FBM) neurons, we conducted a forward genetic screen using Tg(isl1:GFP)rw0 transgenic zebrafish, which express GFP in branchiomotor neurons (Fig. 1A) (Higashijima et al., 2000). We screened clutches from 355 independent F1 females using the early pressure method (Beattie et al., 1999; Walker et al., 2009). From this screen, we identified new mutant alleles of known PCP components in which the migration of FBM neurons is perturbed, including scrib (Wada et al., 2005) and fzd3a (Wada et al., 2006) (data not shown). We also isolated a novel mutant, designated as fh131, which displays a similar specific impairment in the migration of FBM neurons previously seen in other zebrafish mutants for PCP components (Fig. 1B,C). Experiments in this study explore the basis of the neuronal migration defect in the fh131 mutant.
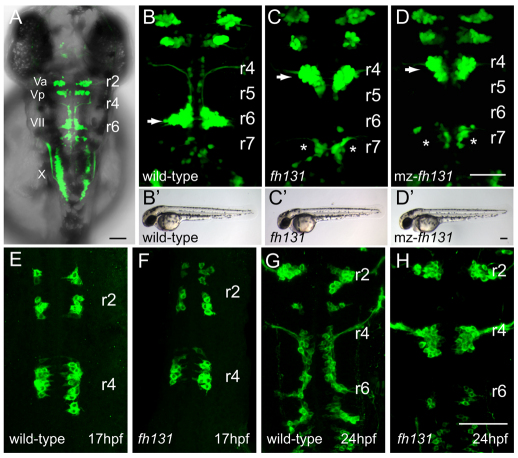
Fig. 1.
The fh131 mutant disrupts facial branchiomotor (FBM) neuron migration. Confocal images showing dorsal views of the hindbrain of Tg(isl1:GFP)rw0 transgene expression in embryos. Anterior is to the top. (A) Cranial motorneurons are easily visible in whole-mount zebrafish embryos at 48 hours post-fertilization (hpf). Va and Vp, anterior and posterior trigeminal nuclei, respectively, in hindbrain rhombomere (r)2 and r3; VII, facial branchiomotor neurons in r6 with axons exiting the hindbrain in r4; X, vagal motorneurons. (B) Wild-type embryo at 48 hpf with FBM neurons fully migrated into r6 (arrow). (C,D) Zygotic fh131 mutant (B) and maternal-zygotic (mz) fh131 mutant (C) with similarly unmigrated FBM neurons in r4 (arrows). Asterisks mark the cell bodies of the glossopharyngeal (cranial nerve IX) neurons in r7. (B′-D′) Low power transmitted light images of embryos with the genotypes shown in B-D showing otherwise normal morphology at 48 hpf. (E-H) FBM neurons in wild type (E,G) and fh131 mutants (F,H) at the onset of migration at 17 hpf (E,F) and at 24 hpf (G,H) showing that fh131 mutant FBM neurons never leave r4. Scale bars: 50 μm.
In wild-type embryos, FBM neurons begin to differentiate and can first be visualized by GFP fluorescence in rhombomere (r)4 at 16 hours post-fertilization (hpf; Fig. 1E). Almost immediately, FBM neurons begin to migrate ventrally and posteriorly, reaching the basement membrane near the r4-r5 boundary, at which point they accelerate and migrate posteriorly to r6 (Chandrasekhar et al., 1997; Higashijima et al., 2000; Wada et al., 2005; Grant and Moens, 2010). A subset of earliest-born FBM neurons migrate to r7 (P.K.G. and C.B.M., unpublished). The first FBM neurons reach their target by 24 hpf (Fig. 1G) and the migration of later-born FBM neurons is complete by 48 hpf (Fig. 1B). In fh131 mutants, GFP-expressing neurons appear normally beginning at 16 hpf in r4 (Fig. 1F); however, none of the r4-derived GFP-expressing neurons migrate posteriorly and they instead remain in r4 (Fig. 1C,F,H). The location of other branchiomotor neurons in the hindbrain is normal, including neurons of the trigeminal, glossopharnygeal and vagal nucleus (Fig. 1A-C). Despite their abnormal positioning in r4, FBM neurons in fh131 mutant embryos extend axons to the correct target muscles in the second branchial arch (see Fig. S1 in the supplementary material). Therefore, fh131 mutant embryos have a specific impairment in the caudal migration of FBM neurons.
fh131 mutant embryos are morphologically normal, and adult homozygous mutants are viable and fertile (Fig. 1B′,C′). Because the maternal functions of other genes involved in FBM neuron migration such as vangl2 and scrib are required for convergent extension movements and neural tube morphogenesis, we tested whether fh131 functions more broadly in PCP processes by generating embryos that lack both maternal and zygotic fh131 function (mz mutants). mz-fh131 were identical to zygotic fh131 mutant embryos, indicating that its role in neuronal migration is the earliest detectable function for this gene (Fig. 1D,D′).
Correct segmental patterning of r4 is required for FBM neuron migration (Studer et al., 1996; Cooper et al., 2003). We determined that segmental patterning is normal in fh131 mutants, as is the expression of genes required to initiate a migratory transcriptional program in FBM neurons (see Fig. S2A-H in the supplementary material) (Coppola et al., 2005; Song et al., 2006). Expression of tag-1 (cntn2 – Zebrafish Information Network), which encodes a cell adhesion molecule specifically expressed by FBM neurons, was also normal (see Fig. S2I,J in the supplementary material) (Sittaramane et al., 2009). Taken together, these results suggest that the overall patterning of the hindbrain and differentiation of FBM neurons was unaffected by the mutation in fh131 embryos, and that fh131 functions more directly in the migratory process.
fh131 encodes Nance-Horan syndrome-like 1b (Nhsl1b)
Using high-resolution mapping and positional cloning, we found that the fh131 mutation disrupts the Nance-Horan syndrome-like 1b (nhsl1b) gene. Briefly, we used standard positional cloning and recombination mapping to place the fh131 mutation within a defined interval on chromosome 20 (Fig. 2A). This interval contained 13 genes, including nhsl1b, a member of the Nance-Horan syndrome (NHS) family of genes, which in mammals includes NHS, NHSL1 and NHSL2 (Brooks et al., 2004; Brooks et al., 2010). In humans, mutations in the founding member of this family, NHS, cause X-linked cataracts, dental anomalies and partially penetrant mental retardation (Brooks et al., 2004). The zebrafish genome encodes four NHS-related genes, two orthologs of NHS (nhsa and nhsb) and two orthologs of NHSL1 (nhsl1a and nhsl1b) (Fig. 2D). No NHSL2 orthologs have been identified to date. Using a bioinformatics approach, Katoh (Katoh, 2004) suggested that vertebrate NHS genes are orthologs of Drosophilia guanylate kinase holder (Gukh), which was isolated based on its physical interaction with the polarity proteins Discs large (Dlg) and Scribble (Mathew et al., 2002). Given the known requirement for zebrafish Scrib in FBM neuron migration, we pursued nhsl1b as a likely candidate.
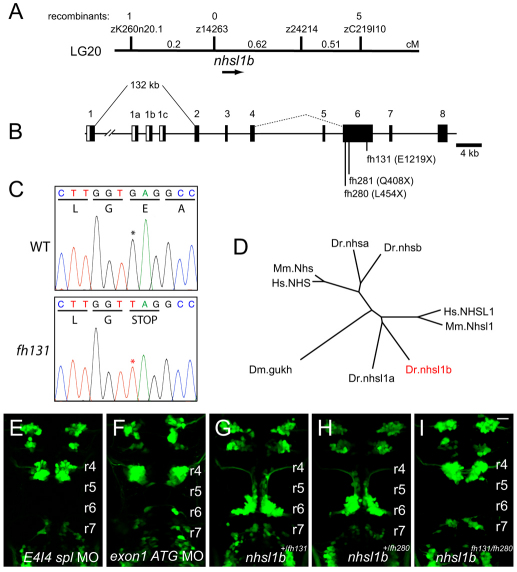
Fig. 2.
nhsl1b is disrupted in fh131 mutants. (A) Genetic mapping of 1444 fh131 mutant zebrafish embryos identifies a genetic interval on chromosome 20 containing nhsl1b. (B) Genomic structure of nhsl1b. Black boxes mark exons 1-8. (C) Sequence trace of a nonsense mutation in nhsl1b in fh131 mutants. (D) Phylogram of the NHS protein family. Mm, mouse; Dr, zebrafish; Hs, human; Dm, Drosophila. (E,F) Tg(isl1:GFP)rw0 expression in an embryo injected with a splice-blocking morpholino targeted to the nhsl1b exon 4-intron 4 boundary (E) or a translation-blocking morpholino targeted to the ATG of exon 1 (F). (G-I) Tg(isl1:GFP)rw0 expression in PCR-genotyped embryos heterozygous for nhsl1bfh131 (G) or for the nhs1bfh280 nonsense allele generated by TILLING (H) and in nhsl1bfh131/280 trans-heterozygotes (I). Note the strong block to FBM migration in the trans-heterozygotes indicating that fh131 and fh280 are alleles of the same gene. r4-r7, rhombomeres 4-7. Scale bar: 20 μm.
Sequence analysis of nhsl1b exons revealed that the fh131 allele carries a nonsense mutation (E1219X) resulting in a premature stop codon in exon 6 (Fig. 2C) that co-segregated with the fh131 mutant phenotype (n=72/72). Injection of an antisense morpholino oligonucleotide (MO) targeted to the exon 4-intron 4 splice junction caused a mis-splicing of the nhsl1b transcript leading to the retention of intron 4 and resulted in a strong block in FBM neuron migration (Fig. 2E and see Fig. S3 in the supplementary material). Furthermore, two additional nonsense alleles, nhsl1bfh280 (Q408X) and nhsl1bfh281 (L454X), identified by TILLING (Draper et al., 2004), failed to complement the fh131 allele originally found in our forward genetic screen (Fig. 2G-I). Taken together, these findings demonstrate that Nhsl1b function is necessary for the caudal migration of FBM neurons. Hereafter, we refer to the fh131 mutant as nhsl1bfh131.
RACE (3′ and 5′ rapid amplification of cDNA ends) indicate that nhsl1b is composed of eight exons, with an alternatively spliced fifth exon and four alternative translational start sites encoded from four alternative first exons (exon 1, exon 1a, exon 1b and exon 1c) (Fig. 2B). Exon 1 is the largest of these first exons and is located 132 kb upstream of exon 2, a genomic structure that is highly conserved in human NHSL1 (Brooks et al., 2010). Similar to the human NHS homologs, exon 1 of zebrafish nhsl1b encodes an N-terminal WAVE homology domain (WHD) found in WAVE (Wiskott-Aldrich syndrome protein family Verprolin-homologous) proteins (Brooks et al., 2010). Injection of a translation-blocking morpholino targeted specifically to the ATG of exon 1 also caused a complete block in FBM neuron migration indicating that the exon 1-encoded WHD domain is essential for the function in migration of Nhsl1b (Fig. 2F).
Nhsl1b interacts genetically and physically with Scrib to regulate FBM neuron migration
We crossed scrib+/rw468 with nhsl1b+/fh131 heterozygotes together to create double heterozygous embryos. We observed that 62% (n=88) of double heterozygous scrib+/rw468; nhsl1b+/fh131 embryos exhibited an almost complete loss of FBM migration, compared with much milder migration defects in only 8% (n=85) and 18% (n=69) of single nhsl1b+/fh131 or single scrib+/rw468 heterozygotes, respectively (Fig. 3A-C). This strong genetic interaction was not observed in double heterozygotes with nhsl1bfh131 and vangl2m209, fz3arw689 or celsrrw71 (data not shown).
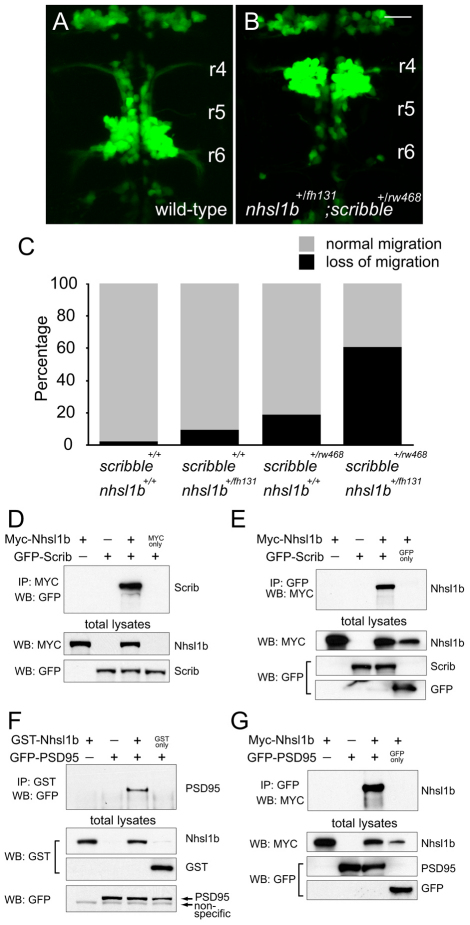
Fig. 3.
Nhsl1b interacts genetically and physically with Scrib in the regulation of facial branchiomotor (FBM) neuron migration. (A,B) Wild-type (A) and double heterozygous nhsl1bfh131/+; scribrw468/+ zebrafish embryos (B) at 48 hours post-fertilization (hpf). Double heterozygotes have unmigrated FBM neurons in rhombomere (r)4, indicative of a strong genetic interaction between the two genes. (C) Histogram of phenotypes in the genotypic classes arising from a nhsl1bfh131/+ x scribrw468/+ cross. (D-G) Nhsl1b associates with Scrib and Psd95 (Dlg4). cDNA constructs were transfected into HEK293T cells as indicated. Whole cell lysates were immunoprecipitated (IP) and western-blotted (WB) with the indicated antibodies. Scale bar: 20 μm.
Guanylate-kinase holder (gukh), the single Drosophila homolog of the vertebrate NHS family, encodes a scaffold protein bridging Dlg and Scrib at the neuromuscular synapse (Mathew et al., 2002). Our genetic studies linking Nhsl1b and Scrib in FBM migration prompted us to investigate whether the zebrafish proteins interact biochemically. We observed that immunoprecipitation of Myc-tagged Nhsl1b, but not the Myc epitope alone, co-precipitated GFP-Scrib and vice versa when the two proteins were expressed in HEK293T cells (Fig. 3D,E). Nhsl1b also co-immunoprecipitated with a zebrafish ortholog of Dlg, PSD95 (Dlg4) (Fig. 3F,G). These findings indicate that, like Drosophila GukH, vertebrate Nhsl1b can exist in a protein complex with both Scrib and PSD95.
nhsl1b is expressed in FBM neurons and Nhsl1b protein localizes to membrane protrusions during migration.
nhsl1b is expressed at low levels maternally and at higher levels zygotically (Fig. 4A). RNA in situ hybridization revealed that nhsl1b is expressed in somitic mesoderm as well as weakly in progenitor cells throughout the nervous system at 14 hpf (Fig. 4B). At 24 hpf, when FBM neurons are migrating, nhsl1b was expressed weakly throughout the neuroepithelium but was specifically upregulated in branchiomotor neurons, including FBM neurons (Fig. 4C-E). The nhsl1b paralog nhsl1a and the more distantly related nhsa gene were also expressed in neural progenitors and somitic mesoderm; however, neither were expressed in migrating FBM neurons (see Fig. S4 in the supplementary material).
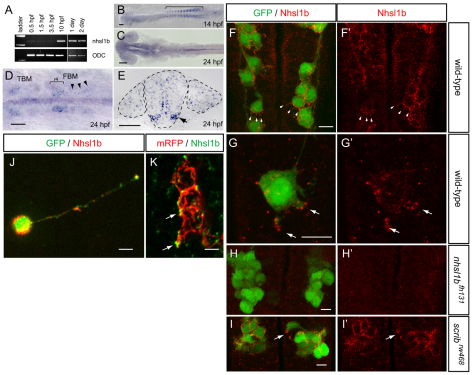
Fig. 4.
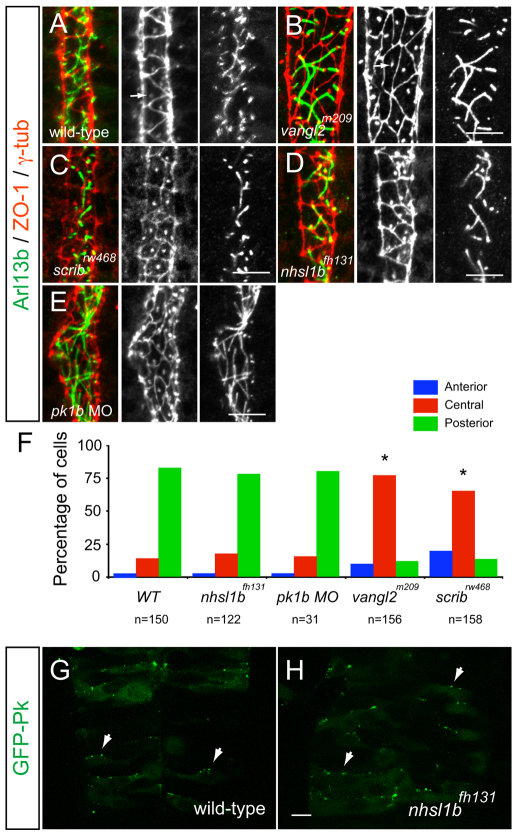
nhsl1b is expressed in facial branchiomotor (FBM) neurons and localizes to membrane protrusions. (A) RT-PCR from fertilization to 2 days old shows onset of zygotic nhsl1b expression at the end of epiboly [10 hours post-fertilization (hpf)]. ODC, ornithine decarboxylase control. (B-E) mRNA in situ hybridization with nhsl1b in whole mount (B-D) and in cross section at the level of r5 (E) showing widespread, low-level expression in somites (B) and CNS (C) and specific upregulation in cranial motorneurons (D,E). nhsl1b is expressed in FBM neurons in rhombomere (r)4 at the onset of migration (bracket in D) and in r5 and r6 during migration (arrowheads in D, arrow in E) and in trigeminal branchiomotor neurons in r2 (TBM). (F-I′) Whole-mount immunocytochemistry with anti-Nhsl1b (red). Isl1:GFP marks FBM neurons (green). Nhsl1b is localized to the membrane surface of FBM neurons, particularly to membrane protrusions (arrowheads) in wild type (F,G) but not Nhsl1b mutant embryos (H). Nhsl1b is similarly localized in FBM neurons in Scrib mutants (I). I′-J′ show Nhsl1b immunostaining alone. (J) Primary cultures of FBM neurons isolated from Tg(isl1:GFP) fish immunostained for Nhsl1b (red) and GFP (green) shows colocalization of Nhsl1b in motorneurons. (K) Anti-Nhsl1b staining in Tg(isl1CREST-hsp70l:mRFP)fh1 fish showing clear colocalization of Nhsl1b with the cell membrane (arrows). Scale bars: 50 μm for B-E; 7 μm for F-K.
Using an antibody directed against the C-terminus of zebrafish Nhsl1b, we observed, similar to our RNA in situ results, that Nhsl1b protein was detectable at low levels in neuroepithelial progenitors and more strongly in migrating FBM neurons, where it localized as foci at the membrane and was abundant at the edges of membrane protrusions (Fig. 4F,G). This immunolocalization was absent in nhsl1b mutant embryos, as mutant Nhsl1bfh131 protein is predicted to have a C-terminal truncation due to the premature stop codon (E1219X), demonstrating the specificity of the antibody for Nhsl1b (Fig. 4H). To confirm that the Nhsl1b immunolocalization was motorneuron-derived, we generated primary neuronal cultures from Tg(isl1:GFP)rw0 transgenic zebrafish (Fassier et al., 2010). We found that Nhsl1b colocalized with GFP-expressing motorneurons (Fig. 4J). Membrane localization of Nhsl1b was confirmed by staining Tg(isl1CREST-hsp70l:mRFP)fh1 transgenic embryos, in which mRFP localizes to membranes of FBM neurons (Fig. 4K). Nhsl1b was similarly localized on trigeminal motorneurons, the segmental homologs of the FBM neurons in hindbrain r2 that do not undergo posterior migration, and on the unmigrated FBM neurons in scribrw468 mutants (Fig. 4I; data not shown), indicating that Nhsl1b is required but not sufficient for posterior-directed migration, and that Scrib is not required for the membrane localization of Nhsl1b.
Nhsl1b functions cell-autonomously in migrating FBM neurons
FBM neurons migrate through a complex cellular milieu in the ventral neural tube, amongst neural progenitors and adjacent to floorplate cells (Grant and Moens, 2010; Mapp et al., 2010). Neuroepithelial cells are polarized along the anterior-posterior axis in a PCP-dependent manner. For instance, maternal and zygotic vangl2 function is required for the anterior membrane localization of GFP-tagged Prickle (GFP-Pk) on neuroepithelial progenitors (Ciruna et al., 2006) and the asymmetric positioning of cilia and basal body at the posterior surface of floorplate cells (Borovina et al., 2010). We observed that in zygotic mutants of both vangl2 and scrib, which lack motorneuron migration but have milder convergent extension defects than the maternal-zygotic mutants, planar polarity of neuroepithelial progenitor and floorplate was disrupted (compare Fig. 5B,C with 5A and 5F, χ2 test, P<0.0001; data not shown). This is consistent with a function for core PCP components in the migratory environment, as suggested by previous chimeric analysis (Jessen et al., 2002; Wada et al., 2005; Wada et al., 2006). By contrast, nhsl1b mutants had normal neuroepithelial and floor plate planar polarity (Fig. 5D,F-H). Apicobasal polarity of progenitor cells was also normal in nhsl1b mutants (see Fig. S5 in the supplementary material). Together with the localization of Nhsl1b protein described above, these results indicate that Nhsl1b functions in the FBM neurons and not in their environment.
Fig. 5.
Scrib and Vangl2, but not Nhsl1b or Pk1b, are required for neuroepithelial cell polarity. (A-E) Confocal images showing floorplate planar polarity in 33 hours post-fertilization (hpf) zebrafish embryos. Anterior is to the top. ZO-1 marks subapical tight junctions (red), γ-tubulin marks basal bodies (red, indicated by arrows in A,B) and Arl13b marks the axonemes of primary cilia (green). Whereas basal bodies are localized to the posterior side of floorplate cells in wild type (A), nhsl1bfh131 mutants (D) and pk1b morphants (E) they are centrally located in zygotic vangl2m209 mutants (B) which have a widened floorplate due to defective neural tube convergence and in zygotic scribrw468 mutants (C), which have only a mild neural tube convergence defect. (F) Quantification of the percentage of cells displaying an anterior, central or posterior position of basal bodies in floorplate cells. Asterisk indicates statistically significant difference from wild type (WT) as determined by χ2 test, P<0.0001. (G,H) Live confocal imaging (dorsal view, anterior to the top) of mosaically expressed GFP-Pk marking anterior membranes (arrows) of neuroepithelial progenitors in 16 hpf wild-type (A) and nhsl1bfh131 mutant (B) embryos. Scale bars: 10 μm.
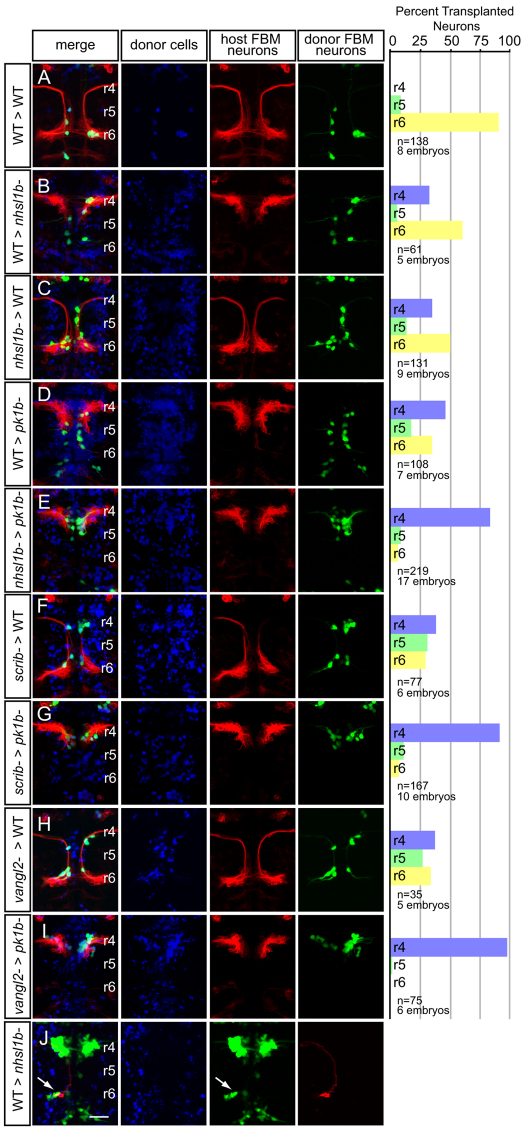
We confirmed a cell-autonomous function for Nhsl1b by chimera analysis. We transplanted Cascade Blue-dextran (CB)-labeled cells from donor embryos into the presumptive ventral hindbrain territory of gastrula stage hosts, such that donor-derived cells contributed mosaically to FBM neurons as well as to other ventral hindbrain cells (Cooper et al., 2003). In these experiments, donor embryos expressed the Tg(isl1:GFP)rw0 transgene and host embryos expressed the Tg(isl1CREST-hsp70l:mRFP)fh1 transgene, both marking FBM neurons. In control experiments, 90% of wild-type FBM neurons migrated normally from r4 into r6 in a wild-type environment (Fig. 6A). 60% of wild-type FBM neurons were similarly capable of migrating into r6 in an nhsl1b morphant or nhsl1b mutant environment, albeit not as well as in a wild-type environment (Fig. 6B). This observation is consistent with a cell-autonomous function for nhsl1b and is similar to the behavior of wild-type cells in a pk1b or hoxb1a morphant environment (61% and 64%, respectively; Fig. 6D and see Fig. S6B in the supplementary material), both of which are known to act cell-autonomously in FBM neuron migration (Cooper et al., 2003; Rohrschneider et al., 2007). This is different from the complete failure of wild-type FBM neurons to migrate in vangl2 or scrib mutant hosts (Jessen et al., 2002; Wada et al., 2005), consistent with a non-cell-autonomous role for these PCP proteins in polarizing the environment.
Fig. 6.
A cell-autonomous role for Nhsl1b, Scrib and Vangl2 in migration. (A-J) Live confocal images at 48 hours post-fertilization (hpf) of chimeric zebrafish embryos with anterior to the top. Cascade blue marks donor-derived cells (blue), Tg(isl1CREST-hsp70l:mRFP)fh1 marks host motorneurons (red) and Tg(isl1:GFP) marks donor-derived motorneurons (green). Histograms on the right indicate the percent of donor-derived FBM neurons in rhombomere (r)4 (unmigrated), r5 and r6 (fully migrated) under the transplantation conditions indicated on the far left, which are written as Donor>Host. n refers to the total number of FBM neurons scored in each condition. Pk1b MOs were used in D, E, G and I to prevent host FBM neurons from migrating by a cell-autonomous mechanism. J shows the rescue of host nhsl1b mutant FBM neurons expressing Tg(isl1:GFP) (green) (arrow) by wild-type donor FBM neurons expressing Tg(isl1CREST-hsp70l:mRFP)fh1 (red). Scale bar: 50 μm.
In reciprocal transplants with nhsl1b, vangl2 or scrib FBM neurons transplanted into wild-type hosts, the majority of mutant FBM neurons migrated out of r4 (65% for nhsl1b, 61% for scrib and 63% for vangl2; Fig. 6C,F,H). This result has been interpreted as proof of a non-autonomous function for vangl2 and scrib (Jessen et al., 2002; Wada et al., 2005); however, observing it for nhsl1b, which otherwise appeared to function cell-autonomously, led us to explore this finding further.
In addition to neuroepithelial progenitor cells and floorplate cells, FBM neurons contact one another during migration, and we considered the possibility that mutant FBM neurons might be rescued in their migration via interactions with neighboring wild-type FBM neurons. To test this, we made use of the fact that the PCP component Prickle1b (Pk1b) is expressed specifically in FBM neurons and is required strictly cell-autonomously for their migration (Rohrschneider et al., 2007). We reasoned that if nhsl1b, vangl2 or scrib mutant FBM neurons fail to migrate in pk1b-depleted hosts, this would mean that the rescue of their migration that we observed in a wild-type environment was mediated by the host FBM neurons themselves. First, we confirmed that planar polarity was normal in the pk1b morphant neuroepithelium and that wild-type FBM neurons could successfully migrate into r6 in a pk1b morphant environment, indicating that the environmental cues to support FBM neuron migration were present even though the host neurons failed to migrate (Fig. 5E,F and Fig. 6D). In this pk1b-morphant environment, the vast majority of mutant neurons failed to migrate out of r4 (84% for nhsl1b, 91% for scrib, 97% for vangl2; Fig. 6E,G,I). Identical results were observed when nhsl1b and scrib mutant cells were placed into a host lacking hoxb1a, which is also required cell-autonomously for FBM neuron migration (Cooper et al., 2003) (see Figs S6 and S7 in the supplementary material). Thus, FBM neurons that lack nhsl1b, scrib or vangl2 can be ‘rescued’ in their migration by an alternative, collective mode that depends on the presence of wild-type migrating neurons. Indeed, we found that transplantation of a small number of wild-type FBM neurons into an nhsl1bfh131 mutant host could rescue the migration of a subset of nhsl1b mutant motorneurons (19/20 nhsl1bfh131 hosts exhibit rescue by wild-type donor cells) (Fig. 6J).
The fact that FBM neurons lacking scrib and vangl2 failed to migrate in a pk1b morphant host, which has the environmental cues to support wild-type FBM neuron migration, reveals an essential cell-autonomous requirement for these core PCP components in addition to their function in the polarized environment. This cell-autonomous function was obscured by collective migration in previous studies (Jessen et al., 2002; Wada et al., 2005). A cell-autonomous role for Scrib is consistent with its physical and genetic interaction with Nhsl1b, as we discuss further below.
DISCUSSION
We have identified a new gene, nhsl1b, required for FBM neuron migration. Nhsl1b encodes one of four zebrafish NHS family proteins, all of which have an N-terminal WAVE homology domain (WHD) encoded by an alternatively spliced first exon (Brooks et al., 2010). WAVE proteins, members of the larger Wiskott-Aldrich syndrome protein (WASP) family, exist in an inhibitory heteropentameric WAVE complex that is activated by Rac to promote actin polymerization in protrusive membrane structures via interaction with the Arp2/3 complex (Takenawa and Suetsugu, 2007; Insall and Machesky, 2009; Derivery and Gautreau, 2010). Human NHS binds components of the hetero-pentameric WAVE complex, but lacks the other domains required for interaction with actin and Arp2/3, suggesting a model in which NHS family proteins regulate actin polymerization by controlling the assembly of the WAVE complex (Brooks et al., 2010). We find that Nhsl1b protein is localized at the membrane and is often abundant in protrusive structures of migrating FBM neurons in vivo, consistent with a role for Nhsl1b in modulating cytoskeletal-membrane rearrangements in migrating cells, downstream of PCP signaling.
In Drosophila, the single NHS family homolog Gukh interacts physically with Scribble and is required for Scribble localization at the neuromuscular junction (Mathew et al., 2002). Consistent with this, we observed that, in zebrafish, Nhsl1b and Scrib interact physically and exhibit a strong genetic interaction. Interestingly, Scrib has also been implicated in directed migration in other cellular contexts. Scrib is required for polarization and migration of astrocytes and mammary epithelial cells in an in vitro scratch ‘wound healing’ assay and in transwell cultures (Osmani et al., 2006; Dow et al., 2007; Nola et al., 2008). In these cells, Scrib is recruited to the leading edge where it is required for the localized activation of Rac and Cdc42 via a direct interaction with the Rac/Cdc42 GEF, βPIX (Audebert et al., 2004; Osmani et al., 2006; Dow et al., 2007; Nola et al., 2008). Given that Rac is known to activate the WAVE complex (Derivery and Gautreau, 2010), our finding that Nhsl1b and Scrib physically and genetically interact raises the possibility that Scrib could function as a scaffold that brings together components that regulate assembly (via Nhsl1b) and activation (via Rac) of the WAVE complex in migrating FBM neurons.
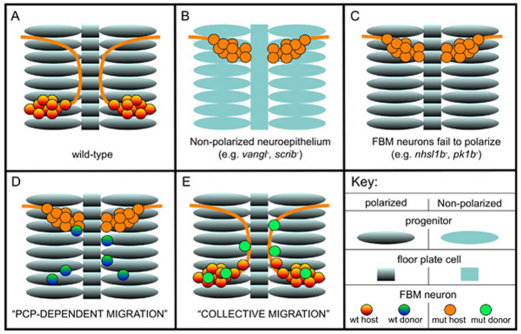
Previous work has shown that the PCP components Scrib and Vangl2 function non-cell-autonomously in FBM neuron migration, and suggested that a planar polarized epithelium shapes the trajectory of this migration (Jessen et al., 2002; Wada et al., 2005; Wada et al., 2006). Consistent with this idea, we have shown that the zygotic functions of Scrib and Vangl2 are required for planar polarization of neuroepithelial progenitors and floorplate cells across the anterior-posterior axis of the neural tube at a time when FBM neurons are migrating (see also Borovina et al., 2010). By contrast, our investigation of Nhsl1b function supports a cell-autonomous role for Nhsl1b within migrating FBM neurons: (1) Nhsl1b is not required for planar polarity in the surrounding neuroepithelial progenitors or in the nearby floorplate, (2) wild-type neurons can migrate in an nhsl1b mutant environment, and (3) nhsl1b mutant neurons fail to migrate through a wild-type environment if host neurons are unmigrated. Importantly, our chimeric analysis also uncovered essential cell-autonomous functions for the PCP components Scrib and Vangl2 in this migration. Taken together, our data support a model in which FBM neuron migration depends both on planar polarization of the epithelium/floorplate, which requires Vangl2 and Scrib (Fig. 7B), and on the ability of FBM neurons to be polarized in response to it, which requires Vangl2, Scrib, Nhsl1b as well as Pk1b (Mapp et al., 2011) in the neurons themselves (Fig. 7C). In this scenario, extrinsic planar polarity in neuroepithelial cells is translated into intrinsic neuronal polarity to control the direction of migration. The dual requirement for PCP components in the FBM neurons and their environment is reminiscent of the cell-autonomous and non-cell-autonomous functions of core PCP components in the fly wing (Lawrence et al., 2007; Wu and Mlodzik, 2009) and suggests the intriguing possibility that FBM neuron migration involves direct PCP signaling between the planar polarized neuroepithelium/floorplate and the migrating neurons. We refer to this as ‘PCP-dependent migration’ (Fig. 7D). The precise molecular mechanism by which polarity is communicated in this context remains to be determined.
Fig. 7.
A model for facial branchiomotor (FBM) neuron migration. (A-C) FBM neuron migration requires the planar polarization of both the neurons and the surrounding neuroepithelium. Neurons fail to migrate either owing to lack of neuroepithelial polarity, e.g. in a vangl2 or scrib mutant (B) or owing to the inability of the neurons to be polarized in response to this environment, e.g. in an nhsl1b or pk1b mutant (C). (D,E) Chimeric analysis reveals that FBM neurons can migrate by one of two distinct mechanisms: one which requires the function of PCP proteins both within FBM neurons and the neuroepithelium (D), or collectively, independent of these functions in the ‘rescued’ neurons but requiring the presence of other normally migrated neurons (E). The incomplete migration of donor-derived neurons observed in D or E when only one of these two mechanisms is available indicates that both mechanisms are functioning during normal migration.
PCP effectors are cell type-specific proteins that function cell-autonomously downstream of PCP signals to link planar polarity to changes in cytoskeletal networks (Strutt et al., 1997; Lee and Adler, 2002; Strutt and Warrington, 2008). For example, the most downstream PCP effector Multiple Wing Hairs was recently shown to encode a Formin Homology 3-domain containing protein that regulates actin polymerization at the apical surface of fly wing cells in a PCP-dependent manner (Strutt and Warrington, 2008). The cell-autonomous function of Nhsl1b specifically in FBM neuron migration and not in other PCP-dependent processes, its localization to cell protrusions, and the known role of NHS family members in regulating WAVE complex activity (Brooks et al., 2010) together argue that Nhsl1b functions as a neuron-specific PCP effector, the first in this system.
Analysis of our transplantation experiments also distinguishes an alternate form of migration that depends on interactions between FBM neurons themselves. We observed that vangl2, scrib and nhsl1b mutant FBM neurons, which are unable to migrate using the ‘PCP-dependent’ mode of migration, can be ‘rescued’ in their migration if they are in the presence of neighboring wild-type FBM neurons. We refer to this as ‘collective migration’ (Fig. 7E). This is analogous to the collective migration of cells in the zebrafish lateral line primordium, where cells lacking the receptor for the chemokine Sdf1 (Cxcl12a – Zebrafish Information Network) are nevertheless able to migrate if they are in the presence of wild-type cells that can detect the signal, or to the fly egg chamber where border cells lacking the transcription factor slbo can migrate in the presence of wild-type border cells (Rorth et al., 2000; Haas and Gilmour, 2006). A collective mode of FBM neuron migration, demonstrated in this paper, can explain previous observations that not all wild-type neurons efficiently migrate in environments where host neurons are unmigrated but epithelial polarity is normal (Cooper et al., 2003; Rohrschneider et al., 2007). The ability of one FBM neuron to direct the migration of another is presumably mediated through cell-cell contact-mediated signaling. Although the molecular mechanism of collective migration remains to be explored, our data argue that it is genetically distinguishable from PCP-dependent migration because it does not require the function of vangl2, scrib or nhsl1b in the ‘rescued’ neurons.
PCP-dependent and collective modes of migration are likely to both be active during wild-type FBM neuron migration, as neither mode alone is sufficient for complete migration. We hypothesize that initial migration out of r4 might predominantly be driven by the first, PCP-and-Nhsl1b-dependent mode, whereas later migrating cells might use the collective mode. However, the same neuron might use the two modes at different times during their migration, or the two modes might even be active in different parts of a cell at the same time. High-resolution live imaging of chimeric embryos in which one or the other mode is unavailable will help to elucidate the relative contributions of PCP-dependent and collective modes of FBM neuron migration.
Supplementary Material
Acknowledgements
K. Cooper, A. Carmany-Rampey and D. Tobin participated in the forward genetic screen, and E. Wolf-Saxon and T. Ma identified non-complementing nhsl1b alleles by TILLING. M. Zigman assessed planar polarity defects in epithelial progenitors in scrib and vangl2 mutants. The many essential contributions of these colleagues to this work are gratefully acknowledged. We also thank H. Wada, H., Okamoto, Z. Sun, V. Prince and J. Cooper for generously providing reagents and resources. S. Rhodes and Y. Rabena provided excellent zebrafish care. Finally, we thank M. Zigman, S. Parkhurst, A. Miller, R. Bachmann and C. Davey for their comments on the manuscript. This work was supported by NIH grant RO1 HD037909 and NIH RO1 HG002995 (TILLING) to C.B.M. P.K.G. was supported by the University of Washington CMB Training Grant. G.S.W. was supported by a Human Frontier Science Program Long Term Fellowship. C.B.M. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.063842/-/DC1
References
- Audebert S., Navarro C., Nourry C., Chasserot-Golaz S., Lecine P., Bellaiche Y., Dupont J. L., Premont R. T., Sempere C., Strub J. M., et al. (2004). Mammalian Scribble forms a tight complex with the betaPIX exchange factor. Curr. Biol. 14, 987-995 [DOI] [PubMed] [Google Scholar]
- Bahary N., Davidson A., Ransom D., Shepard J., Stern H., Trede N., Zhou Y., Barut B., Zon L. I. (2004). The Zon laboratory guide to positional cloning in zebrafish. Methods Cell Biol. 77, 305-329 [DOI] [PubMed] [Google Scholar]
- Beattie C. E., Raible D. W., Henion P. D., Eisen J. S. (1999). Early pressure screens. Methods Cell Biol. 60, 71-86 [PubMed] [Google Scholar]
- Bilder D., Perrimon N. (2000). Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature 403, 676-680 [DOI] [PubMed] [Google Scholar]
- Bilder D., Li M., Perrimon N. (2000). Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science 289, 113-116 [DOI] [PubMed] [Google Scholar]
- Bingham S., Higashijima S., Okamoto H., Chandrasekhar A. (2002). The Zebrafish trilobite gene is essential for tangential migration of branchiomotor neurons. Dev. Biol. 242, 149-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovina A., Superina S., Voskas D., Ciruna B. (2010). Vangl2 directs the posterior tilting and asymmetric localization of motile primary cilia. Nat. Cell Biol. 12, 407-412 [DOI] [PubMed] [Google Scholar]
- Brooks S. P., Ebenezer N. D., Poopalasundaram S., Lehmann O. J., Moore A. T., Hardcastle A. J. (2004). Identification of the gene for Nance-Horan syndrome (NHS). J. Med. Genet. 41, 768-771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks S. P., Coccia M., Tang H. R., Kanuga N., Machesky L. M., Bailly M., Cheetham M. E., Hardcastle A. J. (2010). The Nance-Horan syndrome protein encodes a functional WAVE homology domain (WHD) and is important for co-ordinating actin remodelling and maintaining cell morphology. Hum. Mol. Genet. 19, 2421-2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caddy J., Wilanowski T., Darido C., Dworkin S., Ting S. B., Zhao Q., Rank G., Auden A., Srivastava S., Papenfuss T. A., et al. (2010). Epidermal wound repair is regulated by the planar cell polarity signaling pathway. Dev. Cell 19, 138-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmany-Rampey A., Moens C. B. (2006). Modern mosaic analysis in the zebrafish. Methods 39, 228-238 [DOI] [PubMed] [Google Scholar]
- Carmona-Fontaine C., Matthews H. K., Kuriyama S., Moreno M., Dunn G. A., Parsons M., Stern C. D., Mayor R. (2008). Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature 456, 957-961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira-Barbosa F., Concha M. L., Takeuchi M., Ueno N., Wilson S. W., Tada M. (2003). Prickle 1 regulates cell movements during gastrulation and neuronal migration in zebrafish. Development 130, 4037-4046 [DOI] [PubMed] [Google Scholar]
- Chandrasekhar A. (2004). Turning heads: development of vertebrate branchiomotor neurons. Dev. Dyn. 229, 143-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekhar A., Moens C. B., Warren J. T., Jr, Kimmel C. B., Kuwada J. Y. (1997). Development of branchiomotor neurons in zebrafish. Development 124, 2633-2644 [DOI] [PubMed] [Google Scholar]
- Ciruna B., Jenny A., Lee D., Mlodzik M., Schier A. F. (2006). Planar cell polarity signalling couples cell division and morphogenesis during neurulation. Nature 439, 220-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper K. L., Leisenring W. M., Moens C. B. (2003). Autonomous and nonautonomous functions for Hox/Pbx in branchiomotor neuron development. Dev. Biol. 253, 200-213 [DOI] [PubMed] [Google Scholar]
- Coppola E., Pattyn A., Guthrie S. C., Goridis C., Studer M. (2005). Reciprocal gene replacements reveal unique functions for Phox2 genes during neural differentiation. EMBO J. 24, 4392-4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Calisto J., Araya C., Marchant L., Riaz C. F., Mayor R. (2005). Essential role of non-canonical Wnt signalling in neural crest migration. Development 132, 2587-2597 [DOI] [PubMed] [Google Scholar]
- Derivery E., Gautreau A. (2010). Generation of branched actin networks: assembly and regulation of the N-WASP and WAVE molecular machines. BioEssays 32, 119-131 [DOI] [PubMed] [Google Scholar]
- Dow L. E., Kauffman J. S., Caddy J., Zarbalis K., Peterson A. S., Jane S. M., Russell S. M., Humbert P. O. (2007). The tumour-suppressor Scribble dictates cell polarity during directed epithelial migration: regulation of Rho GTPase recruitment to the leading edge. Oncogene 26, 2272-2282 [DOI] [PubMed] [Google Scholar]
- Draper B. W., McCallum C. M., Stout J. L., Slade A. J., Moens C. B. (2004). A high-throughput method for identifying N-ethyl-N-nitrosourea (ENU)-induced point mutations in zebrafish. Methods Cell Biol. 77, 91-112 [DOI] [PubMed] [Google Scholar]
- Fassier C., Hutt J. A., Scholpp S., Lumsden A., Giros B., Nothias F., Schneider-Maunoury S., Houart C., Hazan J. (2010). Zebrafish atlastin controls motility and spinal motor axon architecture via inhibition of the BMP pathway. Nat. Neurosci. 13, 1380-1387 [DOI] [PubMed] [Google Scholar]
- Feng L., Yelon D., Waxman J. S., Hernandez R. E., Moens C. B. (2010). Dhrs3a regulates retinoic acid biosynthesis through a feedback inhibition mechanism. Dev. Biol. 338, 1-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P. K., Moens C. B. (2010). The neuroepithelial basement membrane serves as a boundary and a substrate for neuron migration in the zebrafish hindbrain. Neural Dev. 5, 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas P., Gilmour D. (2006). Chemokine signaling mediates self-organizing tissue migration in the zebrafish lateral line. Dev. Cell 10, 673-680 [DOI] [PubMed] [Google Scholar]
- Heisenberg C. P., Tada M. (2002). Zebrafish gastrulation movements: bridging cell and developmental biology. Semin. Cell Dev. Biol. 13, 471-479 [DOI] [PubMed] [Google Scholar]
- Higashijima S., Hotta Y., Okamoto H. (2000). Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J. Neurosci. 20, 206-218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insall R. H., Machesky L. M. (2009). Actin dynamics at the leading edge: from simple machinery to complex networks. Dev. Cell 17, 310-322 [DOI] [PubMed] [Google Scholar]
- Jessen J. R., Topczewski J., Bingham S., Sepich D. S., Marlow F., Chandrasekhar A., Solnica-Krezel L. (2002). Zebrafish trilobite identifies new roles for Strabismus in gastrulation and neuronal movements. Nat. Cell Biol. 4, 610-615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko N., Marin O., Koike M., Hirota Y., Uchiyama Y., Wu J. Y., Lu Q., Tessier-Lavigne M., Alvarez-Buylla A., Okano H., et al. (2010). New neurons clear the path of astrocytic processes for their rapid migration in the adult brain. Neuron 67, 213-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. (2004). Identification and characterization of human GUKH2 gene in silico. Int. J. Oncol. 24, 1033-1038 [PubMed] [Google Scholar]
- Kelly M., Chen P. (2007). Shaping the mammalian auditory sensory organ by the planar cell polarity pathway. Int. J. Dev. Biol. 51, 535-547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp H. A., Carmany-Rampey A., Moens C. (2009). Generating chimeric zebrafish embryos by transplantation. J. Vis. Exp. 29, 1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310 [DOI] [PubMed] [Google Scholar]
- Lawrence P. A., Struhl G., Casal J. (2007). Planar cell polarity: one or two pathways? Nat. Rev. Genet. 8, 555-563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Adler P. N. (2002). The function of the frizzled pathway in the Drosophila wing is dependent on inturned and fuzzy. Genetics 160, 1535-1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapp O. M., Wanner S. J., Rohrschneider M. R., Prince V. E. (2010). Prickle1b mediates interpretation of migratory cues during zebrafish facial branchiomotor neuron migration. Dev. Dyn. 239, 1596-1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapp O. M., Walsh G. S., Moens C. B., Tada M. S., Prince V. E. (2011). Zebrafish Prickle1b mediates facial branchiomotor neuron migration via a farnesylation-dependent nuclear activity. Development 138, 2121-2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O., Rubenstein J. L. (2003). Cell migration in the forebrain. Annu. Rev. Neurosci. 26, 441-483 [DOI] [PubMed] [Google Scholar]
- Mathew D., Gramates L. S., Packard M., Thomas U., Bilder D., Perrimon N., Gorczyca M., Budnik V. (2002). Recruitment of scribble to the synaptic scaffolding complex requires GUK-holder, a novel DLG binding protein. Curr. Biol. 12, 531-539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M. P., Trimmer J. S., Gilthorpe J. D., Smith S. J. (2005). Characterization of zebrafish PSD-95 gene family members. J. Neurobiol. 63, 91-105 [DOI] [PubMed] [Google Scholar]
- Moens C. B., Cordes S., Giorgianni M. W., Barsh G., Kimmel C. B. (1998). Equivalence in the genetic control of hindbrain segmentation in fish and mice. Development 125, 381-391 [DOI] [PubMed] [Google Scholar]
- Montcouquiol M., Rachel R. A., Lanford P. J., Copeland N. G., Jenkins N. A., Kelley M. W. (2003). Identification of Vangl2 and Scrb1 as planar polarity genes in mammals. Nature 423, 173-177 [DOI] [PubMed] [Google Scholar]
- Montcouquiol M., Sans N., Huss D., Kach J., Dickman J. D., Forge A., Rachel R. A., Copeland N. G., Jenkins N. A., Bogani D., et al. (2006). Asymmetric localization of Vangl2 and Fz3 indicate novel mechanisms for planar cell polarity in mammals. J. Neurosci. 26, 5265-5275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch J. N., Henderson D. J., Doudney K., Gaston-Massuet C., Phillips H. M., Paternotte C., Arkell R., Stanier P., Copp A. J. (2003). Disruption of scribble (Scrb1) causes severe neural tube defects in the circletail mouse. Hum. Mol. Genet. 12, 87-98 [DOI] [PubMed] [Google Scholar]
- Nola S., Sebbagh M., Marchetto S., Osmani N., Nourry C., Audebert S., Navarro C., Rachel R., Montcouquiol M., Sans N., et al. (2008). Scrib regulates PAK activity during the cell migration process. Hum. Mol. Genet. 17, 3552-3565 [DOI] [PubMed] [Google Scholar]
- Osmani N., Vitale N., Borg J. P., Etienne-Manneville S. (2006). Scrib controls Cdc42 localization and activity to promote cell polarization during astrocyte migration. Curr. Biol. 16, 2395-2405 [DOI] [PubMed] [Google Scholar]
- Park T. J., Mitchell B. J., Abitua P. B., Kintner C., Wallingford J. B. (2008). Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat. Genet. 40, 871-879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y., Glasco D. M., Zhou L., Sawant A., Ravni A., Fritzsch B., Damrau C., Murdoch J. N., Evans S., Pfaff S. L., et al. (2010). Atypical cadherins Celsr1-3 differentially regulate migration of facial branchiomotor neurons in mice. J. Neurosci. 30, 9392-9401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrschneider M. R., Elsen G. E., Prince V. E. (2007). Zebrafish Hoxb1a regulates multiple downstream genes including prickle1b. Dev. Biol. 309, 358-372 [DOI] [PubMed] [Google Scholar]
- Rorth P., Szabo K., Texido G. (2000). The level of C/EBP protein is critical for cell migration during Drosophila oogenesis and is tightly controlled by regulated degradation. Mol. Cell 6, 23-30 [DOI] [PubMed] [Google Scholar]
- Shimoda N., Knapik E. W., Ziniti J., Sim C., Yamada E., Kaplan S., Jackson D., de Sauvage F., Jacob H., Fishman M. C. (1999). Zebrafish genetic map with 2000 microsatellite markers. Genomics 58, 219-232 [DOI] [PubMed] [Google Scholar]
- Sittaramane V., Sawant A., Wolman M. A., Maves L., Halloran M. C., Chandrasekhar A. (2009). The cell adhesion molecule Tag1, transmembrane protein Stbm/Vangl2, and Lamininalpha1 exhibit genetic interactions during migration of facial branchiomotor neurons in zebrafish. Dev. Biol. 325, 363-373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. R., Shirasaki R., Cai C. L., Ruiz E. C., Evans S. M., Lee S. K., Pfaff S. L. (2006). T-Box transcription factor Tbx20 regulates a genetic program for cranial motor neuron cell body migration. Development 133, 4945-4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt D., Warrington S. J. (2008). Planar polarity genes in the Drosophila wing regulate the localisation of the FH3-domain protein Multiple Wing Hairs to control the site of hair production. Development 135, 3103-3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt D. I., Weber U., Mlodzik M. (1997). The role of RhoA in tissue polarity and Frizzled signalling. Nature 387, 292-295 [DOI] [PubMed] [Google Scholar]
- Studer M., Lumsden A., Ariza-McNaughton L., Bradley A., Krumlauf R. (1996). Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature 384, 630-634 [DOI] [PubMed] [Google Scholar]
- Takenawa T., Suetsugu S. (2007). The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8, 37-48 [DOI] [PubMed] [Google Scholar]
- van Eeden F. J., Granato M., Odenthal J., Haffter P. (1999). Developmental mutant screens in the zebrafish. Methods Cell Biol. 60, 21-41 [DOI] [PubMed] [Google Scholar]
- Villefranc J. A., Amigo J., Lawson N. D. (2007). Gateway compatible vectors for analysis of gene function in the zebrafish. Dev. Dyn. 236, 3077-3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivancos V., Chen P., Spassky N., Qian D., Dabdoub A., Kelley M., Studer M., Guthrie S. (2009). Wnt activity guides facial branchiomotor neuron migration, and involves the PCP pathway and JNK and ROCK kinases. Neural Dev. 4, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar E. K., Antic D., Axelrod J. D. (2009). Planar cell polarity signaling: the developing cell's compass. Cold Spring Harb. Perspect. Biol. 1, a002964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H., Iwasaki M., Sato T., Masai I., Nishiwaki Y., Tanaka H., Sato A., Nojima Y., Okamoto H. (2005). Dual roles of zygotic and maternal Scribble1 in neural migration and convergent extension movements in zebrafish embryos. Development 132, 2273-2285 [DOI] [PubMed] [Google Scholar]
- Wada H., Tanaka H., Nakayama S., Iwasaki M., Okamoto H. (2006). Frizzled3a and Celsr2 function in the neuroepithelium to regulate migration of facial motor neurons in the developing zebrafish hindbrain. Development 133, 4749-4759 [DOI] [PubMed] [Google Scholar]
- Walker C., Walsh G. S., Moens C. (2009). Making gynogenetic diploid zebrafish by early pressure. J. Vis. Exp. 28, 1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford J. B. (2006). Planar cell polarity, ciliogenesis and neural tube defects. Hum. Mol. Genet. 15 Spec No 2, R227-R234 [DOI] [PubMed] [Google Scholar]
- Wu J., Mlodzik M. (2009). A quest for the mechanism regulating global planar cell polarity of tissues. Trends Cell Biol. 19, 295-305 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.