Abstract
Xanthones are a class of molecules that bind to a number of drug targets and possess a myriad of biological properties. An understanding of xanthone biosynthesis at the genetic level should facilitate engineering of second-generation molecules and increasing production of first-generation compounds. The filamentous fungus Aspergillus nidulans has been found to produce two prenylated xanthones, shamixanthone and emericellin, and we report the discovery of two more, variecoxanthone A and epishamixanthone. Using targeted deletions that we created, we determined that a cluster of 10 genes including a polyketide synthase gene, mdpG, is required for prenyl xanthone biosynthesis. mdpG was shown to be required for the synthesis of the anthraquinone emodin, monodictyphenone, and related compounds, and our data indicate that emodin and monodictyphenone are precursors of prenyl xanthones. Isolation of intermediate compounds from the deletion strains provided valuable clues as to the biosynthetic pathway, but no genes accounting for the prenylations were located within the cluster. To find the genes responsible for prenylation, we identified and deleted seven putative prenyltransferases in the A. nidulans genome. We found that two prenyltransferase genes, distant from the cluster, were necessary for prenyl xanthone synthesis. These genes belong to the fungal indole prenyltransferase family that had previously been shown to be responsible for the prenylation of amino acid derivatives. In addition, another prenyl xanthone biosynthesis gene is proximal to one of the prenyltransferase genes. Our data, in aggregate, allow us to propose a complete biosynthetic pathway for the A. nidulans xanthones.
INTRODUCTION
Xanthones are polyphenolic compounds produced by higher plants, lichens, and fungi. In common they share a 9H-xanthen-9-one scaffold, but the class is highly diverse owing to functionalization with a range of substituents at various positions.1 As a consequence, the xanthone core has been described as a “privileged structure,”2 with members of this group exhibiting the potential to bind to a variety of targets. Indeed, xanthones have proven to be an important class of secondary metabolites. Over 250 of them have been shown to possess biological activities, including antimicrobial, antioxidant, cytotoxic, and neuropharmacological activities.1
In many instances xanthones are functionalized with prenyl groups, commonly the five carbon dimethylallyl moieties. Examples of prenylated xanthones include mangostanin and γ-mangostin, which are strongly inhibitory to both sensitive and methicillin-resistant strains of Staphylococcus aureus.3 γ-Mangostin also displays potent inhibitory activity against mediators of prostaglandin release, COX-1 and COX-2.4 Other prenylated xanthones have been found to be strongly active against Bacillus subtilis, Staphylococcus faecalis, Staphylococcus typhi, and Candida glabrata.5 By themselves, prenyl groups are important contributors to the outstanding structural diversity of natural products.6 Prenylated secondary metabolites isolated from plants, bacteria, and fungi display a large variety of medicinal properties, including antitumor, antiretroviral, and psychotrophic activities, that are often distinct from their nonprenylated precursors.7
Molecular genetic analysis of the biosynthesis of prenylated and nonprenylated xanthones, as well as prenylations in general, would greatly advance our understanding of fungal secondary metabolite biosynthesis and, by relating gene sequence to function, facilitate prediction of the function of homologous genes. Moreover, identification of the genes involved in the production of particular secondary metabolites offers the possibility of enhanced production through replacement of their native promoters with strong or inducible promoters. In addition, semisynthesis, mutasynthesis, and chemoenzymatic synthesis that lead to second-generation compounds with improved pharmacodynamic and pharmacokinetic properties should greatly benefit from identification and manipulation of the genes involved in secondary metabolite production.
The filamentous fungus Aspergillus nidulans is, in principle, an excellent organism in which to study the biosynthesis of prenylated xanthones. It is known to produce two prenylated xanthones, shamixanthone (2) and emericellin (3) (Figure 1).8,9 Its genome has been sequenced and is reasonably well annotated,10,11 and development of an efficient gene targeting system12,13 has greatly facilitated the targeted deletion of secondary metabolite genes.14,15 We have taken advantage of these attributes to identify and characterize the prenyl xanthone biosynthesis pathway in A. nidulans.
Figure 1.
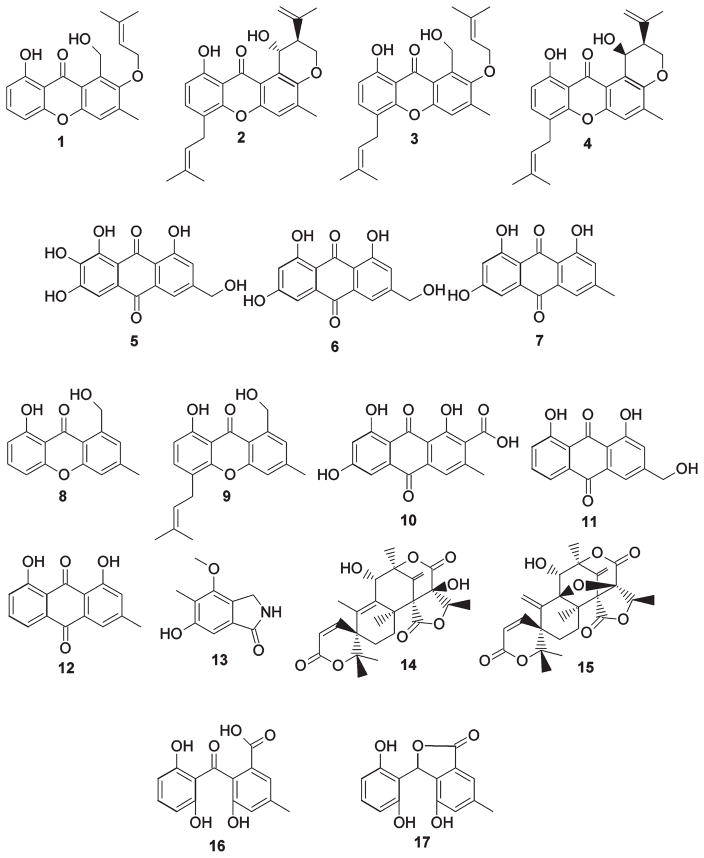
Prenyl xanthones and compounds that emerged from the study of targeted gene deletions. The compounds are as follows: variecoxanthone A, 1; shamixanthone, 2; emericellin, 3; epishamixanthone, 4; 2,ω-dihydroxyemodin, 5; ω-hydroxyemodin, 6; emodin, 7; 9H-xanthen-9-one, 8-hydroxy-1-(hydroxymethyl)-3-methyl-, 8; paeciloxanthone, 9; endocrocin, 10; aloe-emodin, 11; chrysophanol, 12; cichorine, 13; austinol, 14; dehydroaustinol, 15; monodictyphenone, 16; and 1(3H)-isobenzofuranone, 3-(2,6-dihydroxyphenyl)-4-hydroxy-6-methyl-, 17.
We previously identified, using a set of deletions of the aromatic, nonreducing PKS genes in a chromatin remodeling mutant background, a PKS gene, mdpG, that is responsible for synthesis of the related polyketides monodictyphenone and emodin.16,17 Here, we employed the same set of deletions in the investigation of prenyl xanthone synthesis and found that, again, mdpG is responsible. Deletion of genes flanking mdpG revealed that the prenylated xanthone biosynthetic pathway is complex, involving the products of nine genes that cluster with mdpG. Identification of intermediate compounds that accumulate in the deletion strains provides important clues as to the steps in the xanthone biosynthesis pathway. Many of these intermediates have not previously been identified from A. nidulans and are of potential medical interest. Surprisingly and interestingly, however, the prenyltransferase genes required for prenyl xanthone biosynthesis are not clustered with the PKS gene. Bioinformatic analysis of the A. nidulans genome allowed us to identify putative prenyltransferase genes elsewhere in the genome whose products might be required for prenylation of the xanthones. By deleting each of these candidate genes, we found two genes that are required for prenylation of xanthones. They are distant from the mdpG cluster, and it is unlikely that they could have been identified by traditional approaches. Interestingly, the products are members of a family of prenyltransferases that are recognized to prenylate amino acids and their derivatives, as opposed to polyketides. Lastly, we discovered a gene proximal to one of the prenyltransferase genes that, upon deletion, leads to the loss of two of the four prenyl xanthones that we identified. Our data, in aggregate, allowed us to unravel the complex xanthone biosynthesis pathway in A. nidulans, and they highlight the utility of genomics coupled with powerful molecular genetic methods for determining biosynthetic pathways, especially when the cluster is complex and critical genes are distant from the primary cluster.
RESULTS
Characterization of Prenylated Xanthones from A. nidulans
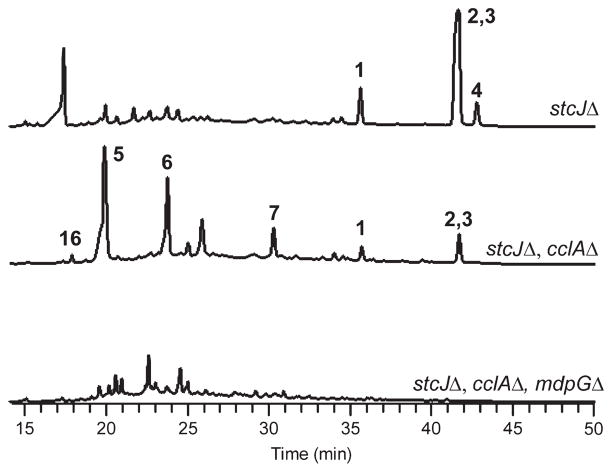
LO2026, an A. nidulans strain carrying a deletion of the stcJ gene (stcJΔ), which is required for the biosynthesis of the carcinogenic secondary metabolite sterigmatocystin,18 was cultivated on Yeast Extract Sucrose (YES) agar for 5 days at 37 °C. The rationale for the use of an stcJΔ strain is that elimination of sterigmatocystin frees up the common polyketide precursor malonyl-CoA and facilitates detection and isolation of other metabolites. The strain also carries nkuAΔ to facilitate gene targeting.12 Reverse-phase LC/MS analysis of the crude organic extract revealed the presence of several late-eluting, nonpolar metabolites (Figure 2). For full characterization, we cultivated the strain on YES plates at a larger scale and purified the compounds using flash chromatography, HPLC, and preparatory TLC when necessary. NMR characterization revealed the molecules to be shamixanthone (2) and emericellin (3), as well as, in lesser amounts, two prenylated xanthones not previously observed from this species, variecoxanthone A (1)19 and epishamixanthone (4).20 (Figure 1; NMR data available in the Supporting Information, Structural Characterization and Figures S3–S15). We also found that other solid media including Yeast Agar Glucose (YAG) and shredded wheat are conductive to xanthone production.
Figure 2.
HPLC profiles of extracts from stcJΔ; stcJΔ, cclAΔ; and stcJΔ, cclAΔ, mdpGΔ strains, as detected by UV absorbance at 254 nm. Numbering of peaks correspond to the compounds in Figure 1. Shamixanthone 2 and emericellin 3 elute at the same retention time.
Analysis of Prenyl Xanthone Biogenesis through Targeted Deletions
The structural similarity of the prenyl xanthones (Figure 1) suggests a common biosynthetic origin, and the presence of aromatic rings is indicative of formation catalyzed by a nonreduced polyketide synthase (NR-PKS).21 Analysis of the sequenced genome of A. nidulans suggests there are 12 NR-PKS genes in this species. The products of two of the genes, stcA encoding the sterigmatocystin PKS and wA, a spore pigment PKS, were already determined,22,23 leading us to focus on the 10 remaining NR-PKSs. In a previous project, we discovered that deletion of cclA, a bre2 homologue orchestrating histone H3 lysine 4 methylation, resulted in the synthesis of related aromatic compounds including monodictyphenone and emodin under conditions in which these molecules were normally not observed.16 For that project we deleted the 10 annotated NR-PKS genes in a cclAΔ background and discovered that deletion of one NR-PKS gene, AN0150.4 (using the nomenclature of the Central Aspergillus Data Repository, CADRE, http://www.cadre-genomes.org.uk/, and the Aspergillus genome database, http://www.aspgd.org/) in a cclAΔ background, resulted in the elimination of these compounds.17 We designated AN0150.4 mdpG and the PKS it encodes MdpG. (See Table 1 for our gene designations and the corresponding annotations from CADRE/AspGD and the Broad Institute Aspergillus Comparative Database.) For the current project we utilized the same 10 NR-PKS deletant strains to learn whether any of them were required for the synthesis of the prenyl xanthones in A. nidulans. We were surprised to discover that, in addition to monodictyphenone and its related molecules, the xanthones disappeared in the mdpGΔ strain (Figure 2) and not in the other deletant strains (data not shown). This reveals that MdpG is the PKS responsible for the synthesis of not only emodin, monodictyphenone, and related compounds but also the xanthones.
Table 1.
Gene Designations of the Text and the Corresponding Annotations from Two Websitesa
| gene designation | putative function | CADRE/AspGD annotation | Broad annotation |
|---|---|---|---|
| AN10039 | histidine acid phosphatase | AN10039.4 | unannotated |
| mdpA | regulatory gene | AN10021.4 | ANID_10021.1 |
| mdpB | dehydratase | AN10049.4 | ANID_10049.1 |
| mdpC | ketoreductase | AN0146.4 | ANID_00146.1 |
| mdpD | monooxygenase | AN0147.4 | ANID_00147.1 |
| mdpE | regulatory gene | AN0148.4 | ANID_00148.1 |
| mdpF | Zn-dependent hydrolase | AN0149.4 | ANID_00149.1 |
| mdpG | polyketide synthase | AN0150.4 | ANID_00150.1 |
| mdpH | hypothetical protein | AN10022.4 | ANID_11847.1 + ANID_11848.1 |
| mdpI | acyl-CoA synthase | AN10035.4 | ANID_10035.1 |
| mdpJ | glutathione S transferase | AN10038.4 | ANID_10038.1 |
| mdpK | oxidoreductase | AN10044.4 | ANID_10044.1 |
| mdpL | Baeyer–Villiger oxidase | AN10023.4 | ANID_10023.1 |
| AN0153 | MYB DNA binding protein | AN0153.4 | ANID_00153.1 |
| xptA | prenyltransferase | AN6784.4 | ANID_06784.1 |
| xptB | prenyltransferase | unannotated | ANID_12402.1 + ANID_12430.1 |
| xptC | GMC oxidoreductase | AN7998.4 | ANID_07998.1 |
CADRE (http://www.cadre-genomes.org.uk) which uses the same designations as the Aspergillus genome database (AspGD, http://www.aspgd.org) and the Broad Institute Aspergillus Comparative Database (http://www.broadinstitute.org/annotation/genome/aspergillus_group/MultiHome.html). Putative functions are from BLAST searches performed previously16 or for this study.
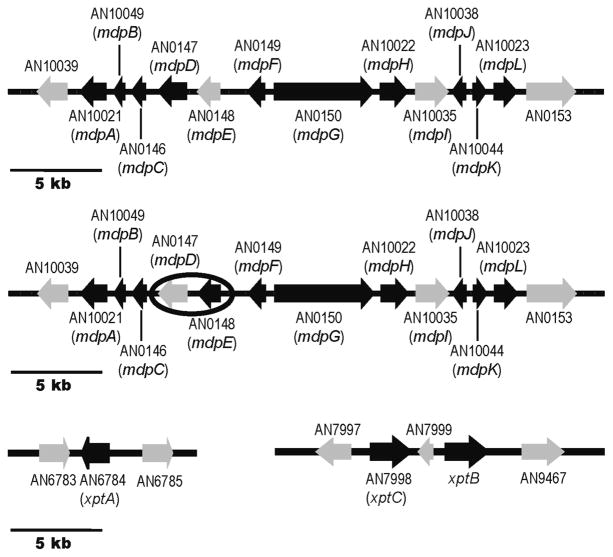
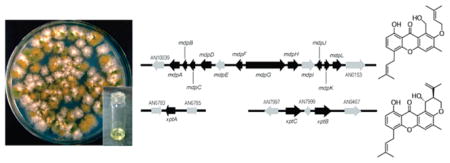
Next, we tried to identify additional genes involved in A. nidulans prenyl xanthone biosynthesis. Facilitating the search, secondary metabolism genes in A. nidulans are usually clustered, prompting us to focus on the genes surrounding mdpG (AN10039 to AN0153) (Figure 3). To study the synthesis of the xanthones as it would occur naturally, without the influence of a potentially powerful chromatin remodeling mutation, we deleted these genes in the LO2026 background carrying the wild-type cclA gene (cclA+). All deletions were verified by diagnostic PCR (using three different primer sets for each gene) (see Materials and Methods).
Figure 3.
(Top) Organization of the genes surrounding the PKS mdpG involved in prenyl xanthone biosynthesis. Each arrow indicates gene size and direction of transcription. On the basis of a set of deletions we created and analyzed, genes shown in black are involved in prenyl xanthone biosynthesis while those in gray are not. (Middle) Organization of the genes for monodictyphenone biosynthesis in a cclAΔ background (based on ref 17). AN0147 and AN0148 are circled to emphasize that AN0147 is unnecessary for monodictyphenone generation but required for xanthone synthesis, whereas AN0148 is necessary for monodictyphenone generation but not required for xanthone synthesis. (Bottom) Organization of the genes outside of the mdpG cluster that are involved in prenyl xanthone synthesis. The genes that were successfully deleted were AN10039 through mdpL, xptA, xptC, AN7999, and xptB.
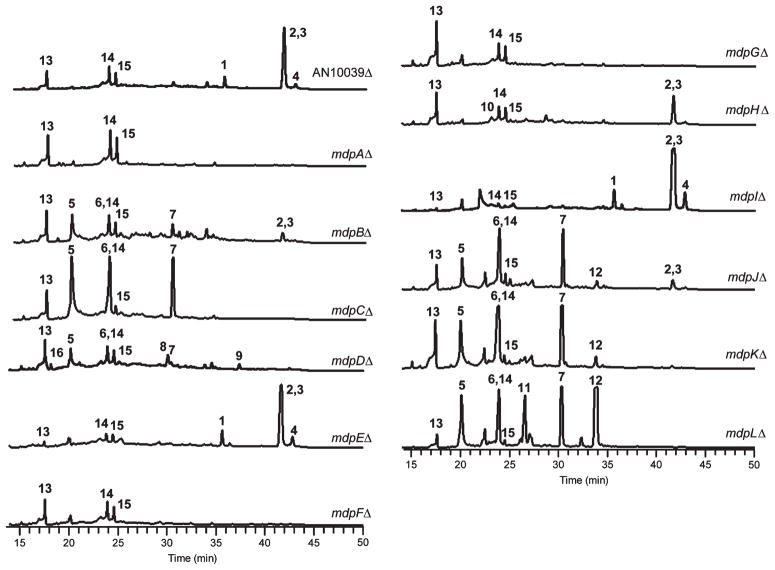
Attempted deletion of AN0153, homologous to a DNA-binding protein, failed to yield any viable transformants, in agreement with previous observations.17 This gene appears to be essential for viability and unrelated to secondary metabolism and was thus excluded from further study here. Deletion of the other genes significantly reduced or eliminated prenyl xanthone formation except for AN10039, mdpEΔ, and mdpIΔ (Figures 3 and 4). Two differences were observed in relation to our investigation using cclAΔ: In the cclAΔ background, deletion of the transcriptional regulator gene mdpE eliminates the synthesis of monodictyphenone and related compounds; however, for the prenyl xanthones this gene does not appear consequential (Figure 3). Additionally, the monooxygenase gene mdpD was not found to be important for the production of monodictyphenone and analogous products in the cclAΔ background, whereas it plays a crucial role in prenyl xanthone formation.
Figure 4.
HPLC extracts of strains in the cluster as detected by UV absorbance at 254 nm. ω-Hydroxyemodin 6 and the unrelated metabolite austinol 14 elute at the same retention time.
The deletant strains that reduced prenyl xanthone production were examined for the presence of any new metabolites that might represent intermediates in the biosynthetic pathway of the xanthones. Extracts from strains carrying deletions of mdpG, mdpA, and mdpF contained no obvious intermediates from the prenyl xanthone biosynthesis pathways. A strain carrying a deletion of mdpH did not display new metabolites in significant amounts, but a compound with the mass and retention time of endocrocin (10), recently identified from an mdpH/cclA double-deletant strain,17 was observed.
The mdpLΔ strain, the extract of which displayed the greatest number of significant peaks in the chromatogram, was grown in large scale on YES plates, and the major metabolites were isolated by flash chromatography and HPLC and characterized primarily using NMR. The compounds were determined to be 2,ω-dihydroxyemodin (5), ω-hydroxyemodin (6), emodin (7), aloe-emodin (11), and chrysophanol (12) (Figure 4; NMR data shown in the Supporting Information). Also, mdpCΔ, mdpJΔ, and mdpKΔ extracts each exhibited 5, 6, and 7, as determined by their masses and retention times in comparison to the mdpLΔ extract, but little or no 11 or 12. Chromatograms of mdpBΔ extracts similarly displayed the three metabolites, along with a number of small, indistinct peaks.
The mdpDΔ extract also yielded 5, 6, and 7 but with addition of monodictyphenone (16), a core xanthone structure (8), and the C-prenylated analog paeciloxanthone (9). Finally, the polyketide cichorine (13) and the meroterpenoids austinol (14) and dehydroaustinol (15) could be detected in all strains tested, indicating they are unrelated to prenyl xanthone biosynthesis.
Identification of Two Genes Required for Prenylation of the Xanthone Scaffold
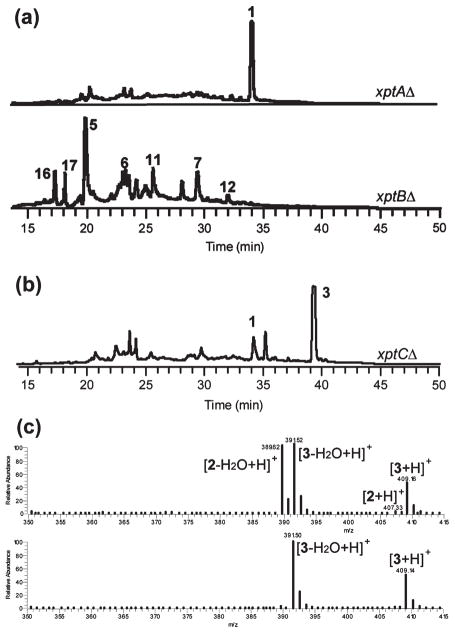
Although our results revealed a gene cluster responsible for synthesis of A. nidulans prenyl xanthones, the lack of a prenyltransferase gene in this cluster made it clear that the required prenyltransferase gene(s) must be located elsewhere in the genome. On the basis of homology to known genes of this class,24 we were able to identify several putative prenyltransferase genes in the A. nidulans genome: AN6784.4, AN8514.4 [tdiB25], AN10289.4, AN11080.4, AN11194.4, and AN11202.4. We deleted each of these genes, and the deletant strains were cultured on YES plates. LC/MS analysis revealed that deletion of AN6784.4, but not the other prenyltransferase genes, resulted in the elimination of 2–4 (Figure 5a) and accumulation of a compound which upon large-scale isolation and characterization was confirmed to be variecoxanthone A (1). Because AN6784.4 is a prenyltransferase gene required for synthesis of the xanthones, we hereby designate it xptA (xanthone prenyltransferase A).
Figure 5.
(a) HPLC extracts of the prenyltransferase deletant strains xptAΔ and xptBΔ as detected by UV absorption at 254 nm. (b) HPLC extract of xptCΔ. (c) Mass spectra of the major LC/MS peak from (top) stcJΔ control extract and (bottom) xptCΔ extract.
Since the A. nidulans xanthones contain two prenyl groups (except for variecoxanthone A, which only has one), it was apparent that an additional prenyltransferase gene was required. We hypothesized that the genome must contain an additional, unannotated prenyltransferase gene. We performed a local BLAST search of the Broad Institute Aspergillus Comparative Database (http://www.broadinstitute.org/annotation/genome/aspergillus_group/MultiHome.html) using AN6784.4 as a query sequence. We identified two annotated partial gene fragments designated ANID_12402.1 and ANID_12430.1. Inspection of the two fragments revealed that the 3′-end of ANID_12402.1 overlaps with the 5′-end of ANID_12430.1 by ~600 bp. Further, the Web site predicted that the sequence of ANID_12402 is prematurely truncated at its 3′-end. For these reasons we concluded that the nonoverlapping portion of ANID_12430 was in fact the continuation of ANID_12402 and that the 5′-end of ANID_12402 and the 3′-end of ANID_12430 are the boundaries of a single gene. Indeed, the sequence of this putative gene is found intact in CADRE/AspGD but without annotation. Deletion of this gene led to the elimination of the xanthones and accumulation of several metabolites, including 7, 12, 16, and a putative monodictyphenone derivative (17) (Figure 5a). We designate this new gene xptB (xanthone prenyltransferase B).
Identification of an Additional Prenyl Xanthone Biosynthesis Gene
The data were suggestive of a biosynthesis in which emericellin (3) is a precursor to shamixanthone (2) and epishamixanthone (4) (see Discussion). Even after identification of xptA and xptB it was clear that a gene encoding the enzyme responsible for the transformation from emericellin to shamixanthone and epishamixanthone was required but not yet identified. Given the tendency of fungal secondary metabolite genes to exist in clusters, we reasoned that the missing gene might reside proximate to xptA or xptB. An analysis of the putative function of the genes neighboring xptA revealed none that were likely involved in secondary metabolism; however, AN7998.4 and AN7999.4 upstream of xptB were possible candidates. Deletion of AN7998.4 yielded a metabolite profile that continued to display variecoxanthone A (1) and 3 but lacked 2 and 4 (Figure 5b and 5c), revealing that AN7998.4 is the missing gene in the biosynthesis of the two prenyl xanthones. We designate AN7998.4 as xptC. The amino acid sequence indicates that it is a member of the glucose–methanol–choline (GMC) oxidoreductase superfamily, a broad class that includes cellobiose dehydrogenase, choline dehydrogenase, and methanol oxidase.26 Deletion of AN7999.4 had no obvious effect on prenyl xanthone biosynthesis.
DISCUSSION
We used a combination of genomics, efficient gene targeting, and natural products chemistry to elucidate the complex prenyl xanthone biosynthesis pathway in A. nidulans, the first such pathway to be deciphered in a fungus. This pathway is complex, involving the products of 10 clustered genes and three genes located apart from the main cluster. The genes mdpA through mdpL are located on the right arm of Chromosome VIII, whereas xptA is located on the right arm of Chromosome I and xptB and xptC are found on the left arm of Chromosome II. In all cases the genes are ~0.5 Mb from a telomere. Other uncommon examples of separated fungal secondary metabolism genes include the genes for T-toxin biosynthesis in Cochliobolus heterostrophus, with 9 genes in two unlinked loci, and the nonclustered acetyl-transferase gene Tri101 involved in trichothecene biosynthesis in Fusarium species, having a different evolutionary history from other trichothecene genes.27,28 Without genomics and efficient gene targeting it would have been difficult, if not impossible, to identify all of the genes in the pathway. In particular, it would have probably been extremely difficult to identify the three genes that are separated in the genome from the bulk of the xanthone biosynthesis genes in the absence of the ability to identify candidate genes and to delete them en masse.
The two prenyltransferases, encoded by the xptA and xptB genes, are particularly interesting. They are both homologous to members of a class of enzymes known as fungal indole prenyltransferases.29 For example, XptA is 27% identical at the amino acid level to SirD from Leptosphaeria maculans30 and 24% identical to FgaPT2 from Aspergillus fumigatus.31 XptB is 29% and 27% identical to these enzymes, respectively, and XptA and XptB are 37% identical to each other. Unlike many other prenyltransferase enzymes, the fungal indole prenyltransferases are soluble, lack (N/D)DxxD motifs, and continue to be active in the absence of Mg2+.29 The fungal indole prenyltransferases are capable of regular and reverse C-prenylations as well as N- and O-prenylations. Their solubility may be useful for chemoenzymatic synthesis in terms of ease of manipulation and efficiency of catalysis. All members of this class studied in detail to date prenylate compounds of amino acid origin, whereas our data reveal that XptA and XptB clearly prenylate polyketides. There were hints from the literature that this class might have a range of substrates extending to polyketides. For example, the X-ray structure of dimethylallyl tryptophan synthase (DMATS) from A. fumigatus revealed a common architecture (but no significant primary amino acid similarity) with the bacterial enzyme NphB that catalyzes addition of a 10-carbon geranyl moiety to a polyketide-based aromatic scaffold.32 In addition, a gene in Penicillium aethiopicum that is homologous to indole prenyl-transferases is proximal to a polyketide gene necessary for the formation of viridicatumtoxin and was proposed to contribute to addition of a geranyl group to that metabolite.33 Our findings provide the first direct evidence, however, that this class of prenyltransferases can prenylate polyketides, and the possibility must be considered that XptA and XptB may prenylate polyketide substrates beyond those reported in this study.
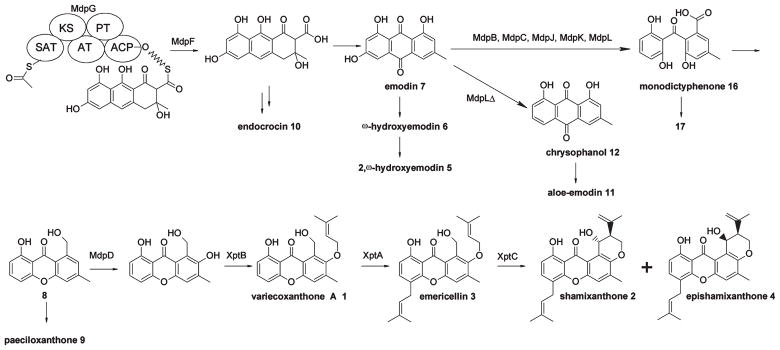
Our deletions reveal the genes required for prenyl xanthone synthesis, but correlation of the deleted genes with the intermediates that accumulate in the deletion strains allows us to propose a biosynthetic pathway for the A. nidulans prenyl xanthones (Figure 1). We previously detailed the synthesis of monodictyphenone (16) in a cclAΔ background, which was dependent on many of the same secondary metabolite genes also responsible for the prenyl xanthones.17 We propose that 16 is an intermediate en route to prenyl xanthones (Figure 6). One new observation, however, is that from the mdpLΔ strain we have now isolated chrysophanol (12).
Figure 6.
Proposed biosynthetic pathway of prenyl xanthones.
Earlier work by others used isotopic precursors to propose the biosynthesis of 2 and a similar metabolite, tajixanthone, in Aspergillus variecolor.34 The labeling patterns from [1-13C]- and [2-13C]-acetate incorporation suggested an octaketide precursor, which could condense into an anthrone and possibly autooxidize to an anthraquinone. Interestingly, the earlier work also found that isotopically labeled 12 was incorporated into tajixanthone; it was proposed that xanthone formation proceeds from an octaketide to 7 and further to 12. Because our earlier analysis17 indicates that 12 is not in the direct biosynthetic pathway from 7 to 16, we propose that it is a shunt metabolite; more specifically, the mdpL knockout may yield an unstable intermediate that transforms to chrysophanol. 7 might not be converted to 12 when the biosynthesis is uninterrupted. An alternative explanation for incorporation of 12 into tajixanthone is that 12 is metabolized to 7 at a low rate through an endogenous mechanism. Aloe-emodin (11), the ω-hydroxylated analog of 12, may be a side product arising from oxidation by an endogenous cytochrome P450 enzyme.35
Elucidating the enzymatic conversion of 7 to 16 is complicated by the fact that the process appears to be dependent on five of the clustered genes, mdpB, mdpC, mdpJ, mdpK, and mdpL, and that the metabolite profiles from each of the corresponding deletion strains are similar. As a further complication, the presence of several metabolites in a deletant strain (for example, five major compounds from mdpLΔ) makes it difficult to ascribe their roles in the biosynthesis and their relation to each other. We refer the reader to ref 17 for a discussion of the possible functions of mdpB, mdpC, mdpJ, mdpK, and mdpL in monodictyphenone biosynthesis. Our data allow us, however, to deduce the remainder of the prenyl xanthone biosynthesis pathway with considerable confidence.
In this study we show conclusively that four genes, mdpD, xptA, xptB, and xptC are, at a minimum, responsible for conversion of monodictyphenone to the prenyl xanthones. It is easiest to deduce the steps of the biosynthetic pathway by working back from the final products, shamixanthone (2) and epishamixanthone (4). The xptC deletion does not accumulate 2 or 4 but does accumulate emericellin (3) and variecoxanthone A (1). This allows us to deduce that XptC is required for conversion of 3 or 1 to 2 and 4. The xptA deletion strains accumulate 1 but not 3, 2, or 4. This result allows us to deduce that XptA is required for the C-prenylation of 1 to form 3 and also allows us to deduce that 3 is the compound that is converted by XptC to 2 and 4. Given the similarity of XptC to oxidative enzymes, we suggest that oxidation of the primary alcohol of 3 to an aldehyde, followed by ene cyclization,19 may yield 2 and its epimer 4. From the facts that XptA catalyzes the C-prenylation of 1 and no prenylated xanthones are found in xptB deletion strains, we deduce that XptB catalyzes the O-prenylation required for formation of 1 and that the O-prenylation occurs before the C-prenylation and is a prerequisite for the C-prenylation. From the mdpD deletant we isolated compound 8, which contains the xanthone core but does not bear functionality at the 2 position, at which the final products feature the O-prenyl group. Oxidation of the xanthone core at the C-2 position is necessary for O-prenylation, so we postulate that MdpD, homologous to known monooxygenases, catalyzes oxidation prior to O-prenylation by XptB. In the absence of the C-2 oxygen, XptA is able to yield the C-prenylated product 9 in small amounts. In the xptB deletant strain, we were able to detect compound 17, which we suspect to be a shunt metabolite arising from reduction of the carbonyl group of monodictyphenone (16) and condensation with the carboxylic acid.
In addition to assisting in elucidating the biosynthetic pathway, many of the shunt products and intermediate molecules are bioactive. For instance, 11 is cytotoxic to neuroblastoma, Ewing sarcoma, and pPNET cell lines and inhibits neuroblastoma tumors in vivo but is not toxic to normal cells nor animal models.36 9 displays antimicrobial and antiacetylcholineesterase activity and is also cytotoxic against hepG2 cells.37 Compound 8 has antifungal and antibacterial activities.38 These findings indicate that a benefit of targeted deletions of genes is the accumulation of useful quantities of biosynthetic intermediates that may themselves possess noteworthy biological properties.
CONCLUSION
Previous work demonstrated that a cluster of 10 genes was responsible for several related compounds that emerged in a chromatin deletant strain of A. nidulans, including mono-dictyphenone.17 The current work reveals that four prenyl xanthones rely on most of these genes for their synthesis but in addition require a monooxygenase gene within the cluster and three genes outside of the cluster, including two belonging to a family of indole prenyltransferase genes and one homologous to GMC oxidoreductase genes. The combined data allow us to propose that these latter genes are involved in the later stages of prenyl xanthone formation to complete a biosynthesis in which monodictyphenone is a precursor.
MATERIALS AND METHODS
Generation of Fusion PCR Fragments, A. nidulans Protoplasting, and Transformation
All gene deletions were carried out according to the gene targeting procedures of Szewczyk et al.13 Two ~1000 base pair fragments upstream and downstream of every targeted gene were amplified from A. nidulans genomic DNA using PCR. Primers used in this study are listed in Table S1, Supporting Information. The two amplified flanking sequences and an A. fumigatus pyrG selectable marker cassette were fused together by PCR using nested primers. A. nidulans strains in this study are displayed in Table S2, Supporting Information. Protoplast generation and transformation were performed as described.13 The strain LO2026 carrying a deletion of the stcJ gene that eliminates sterigmatocystin production was used as the recipient strain. Diagnostic PCR of the deletant strains was carried out using the external primers from the first round of PCR. The difference in the size between the gene replaced by the selective marker and the native gene allowed us to establish if the transformants carried correct gene replacements. For further verification, diagnostic PCR was performed two more times, using one of the external primers and a primer located inside the marker gene, then the other external primer and an internal primer. In these instances, the deletants yielded the PCR product of the expected size whereas no product was present in nondeletants.
Media and Cultivation of Strains
YES media was prepared by combining 20 g of yeast extract, 120 g of sucrose, 20 g of agar, and 2 mL of trace element solution15 in 1 L H2O. For LC/MS screening, spores of LO2026 (the control strain) and three strains of each gene deletant were individually cultivated (1 × 107 spores) on 10 × 150 mm Petri dishes containing YES agar and cultivated at 37 °C for 5 days. The agar was chopped into ~2 cm2 pieces, and the material was extracted using sonication with methanol, followed by 1:1 methanol:dichloromethane. The organic solvents were removed in vacuo, and the remaining material was partitioned between H2O (25 mL) and ethyl acetate (25 mL × 2). The combined ethyl acetate layers were evaporated, and the crude material was redissolved at a concentration of 20 mg/mL in DMSO and then diluted 5-fold in methanol.
LC/MS Analysis
LC/MS was carried out using a ThermoFinnigan LCQ Advantage ion trap mass spectrometer with an RP C18 column (Alltech Prevail; 2.1 × 100 mm with a 3 μm particle size) at a flow rate of 125 μL/min and monitored by a UV detector at 254 nm. The solvent gradient was 95% MeCN–H2O (solvent B) in 5% MeCN–H2O (solvent A) both containing 0.05% formic acid:0% B from 0 to 5 min, 0 to 100% B from 5 to 35 min, 100% B from 35 to 40 min, 100% B to 0% B from 40 to 45 min, and reequilibration with 0% B from 45 to 50 min.
Isolation of Metabolites
The LO2026 (stcJΔ), LO3387 (mdpLΔ), LO3337 (mdpDΔ), LO3896 (xptAΔ), and LO4178 (xptBΔ) strains were each cultivated in 25 × 150 mm Petri dishes containing 2 L YES media and extracted in the same manner as above. The crude material was subjected to silica gel column chromatography, using ethyl acetate and hexanes as the eluent. The materials were further separated by preparative HPLC [Phenomenex Luna 5 μm C18 (2), 250 × 21.2 mm] with a flow rate of 5.0 mL/min and measured by a UV detector at 250 nm. Shamixanthone (2), emericellin (3), epishamixanthone (4), and paeciloxanthone (9) required further purification using preparative TLC. See Supporting Information for more details about isolation.
Structural Characterization
See the Supporting Information under Structural Characterization and Figures S3–S15.
Supplementary Material
Acknowledgments
This work was supported by grant PO1GM084077 from the National Institute of General Medical Sciences and by the Kansas University Endowment Fund.
Footnotes
Supporting Information. A nidulans strains and primers used in this study, detailed structural characterization, purification methods, and complete refs 10 and 11. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.El-Seedi HR, El-Barbary MA, El-Ghorab DMH, Bohlin L, Borg-Karlson AK, Goransson U, Verpoorte R. Curr Med Chem. 2010;17:854–901. doi: 10.2174/092986710790712147. [DOI] [PubMed] [Google Scholar]
- 2.Lesch B, Brase S. Angew Chem, Int Ed. 2004;43:115–118. doi: 10.1002/anie.200352154. [DOI] [PubMed] [Google Scholar]
- 3.Panthong K, Pongcharoen W, Phongpaichit S, Taylor WC. Phytochemistry. 2006;67:999–1004. doi: 10.1016/j.phytochem.2006.02.027. [DOI] [PubMed] [Google Scholar]
- 4.Nakatani K, Nakahata N, Arakawa T, Yasuda H, Ohizumi Y. Biochem Pharmacol. 2002;63:73–79. doi: 10.1016/s0006-2952(01)00810-3. [DOI] [PubMed] [Google Scholar]
- 5.Saleem M, Nazir M, Ali MS, Hussain H, Lee YS, Riaz N, Jabbar A. Nat Prod Rep. 2010;27:238–254. doi: 10.1039/b916096e. [DOI] [PubMed] [Google Scholar]
- 6.Heide L. Curr Opin Chem Biol. 2009;13:171–179. doi: 10.1016/j.cbpa.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 7.Botta B, Vitali A, Menendez P, Misiti D, Delle Monache G. Curr Med Chem. 2005;12:713–739. doi: 10.2174/0929867053202241. [DOI] [PubMed] [Google Scholar]
- 8.(a) Marquez-Fernandez O, Trigos A, Ramos-Balderas JL, Viniegra-Gonzalez G, Deising HB, Aguirre J. Eukaryot Cell. 2007;6:710–720. doi: 10.1128/EC.00362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Marquez-Fernandez O, Trigos A, Ramos-Balderas JL, Viniegra-Gonzalez G, Deising HB, Aguirre J. Eukaryot Cell. 2007;6:710. doi: 10.1128/EC.00362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishida M, Hamasaki T, Hatsuda Y. Agric Biol Chem. 1975;39:2181–2184. [Google Scholar]
- 10.Galagan JE, et al. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 11.Wortman JR, et al. Fungal Genet Biol. 2009;46:S2–S13. doi: 10.1016/j.fgb.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR. Genetics. 2006;172:1557–1566. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. Nat Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- 14.Chiang YM, Szewczyk E, Nayak T, Davidson AD, Sanchez JF, Lo HC, Ho WY, Simityan H, Kuo E, Praseuth A, Watanabe K, Oakley BR, Wang CCC. Chem Biol. 2008;15:527–532. doi: 10.1016/j.chembiol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez JF, Chiang YM, Szewczyk E, Davidson AD, Ahuja M, Oakley CE, Bok JW, Keller N, Oakley BR, Wang CCC. Mol Biosyst. 2010;6:587–593. doi: 10.1039/b904541d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bok JW, Chiang YM, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, Lo HC, Watanabe K, Strauss J, Oakley BR, Wang CCC, Keller NP. Nat Chem Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiang YM, Szewczyk E, Davidson AD, Entwistle R, Keller NP, Wang CCC, Oakley BR. Appl Environ Microb. 2010;76:2067–2074. doi: 10.1128/AEM.02187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown DW, Adams TH, Keller NP. Proc Natl Acad Sci USA. 1996;93:14873–14877. doi: 10.1073/pnas.93.25.14873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chexal KK, Holker JSE, Simpson TJ, Young K. J Chem Soc, Perkin Trans. 1975;1:543–548. [PubMed] [Google Scholar]
- 20.Ishida M, Hamasaki T, Hatsuda Y, Fukuyama K, Tsukihara T, Katsube Y. Agric Biol Chem. 1976;40:1051–1052. [Google Scholar]
- 21.Cox RJ. Org Biomol Chem. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe A, Fujii I, Sankawa U, Mayorga ME, Timberlake WE, Ebizuka Y. Tetrahedron Lett. 1999;40:91–94. [Google Scholar]
- 23.Minto RE, Townsend CA. Chem Rev. 1997;97:2537–2555. doi: 10.1021/cr960032y. [DOI] [PubMed] [Google Scholar]
- 24.von Dohren H. Fungal Genet Biol. 2009;46:S45–S52. doi: 10.1016/j.fgb.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Bok JW, Hoffmeister D, Maggio-Hall LA, Murillo R, Glasner JD, Keller NP. Chem Biol. 2006;13:31–37. doi: 10.1016/j.chembiol.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 26.Cavener DR. J Mol Biol. 1992;223:811–814. doi: 10.1016/0022-2836(92)90992-s. [DOI] [PubMed] [Google Scholar]
- 27.Inderbitzin P, Asvarak T, Turgeon BG. Mol Plant-Microbe Interact. 2010;23:458–472. doi: 10.1094/MPMI-23-4-0458. [DOI] [PubMed] [Google Scholar]
- 28.O’Donnel K, Kistler HC, Tacke BK, Casper HH. Proc Natl Acad Sci USA. 2000;93:7905–7910. doi: 10.1073/pnas.130193297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steffan N, Grundmann A, Yin WB, Kremer A, Li SM. Curr Med Chem. 2009;16:218–231. doi: 10.2174/092986709787002772. [DOI] [PubMed] [Google Scholar]
- 30.Kremer A, Li SM. Microbiology (UK) 2010;156:278–286. doi: 10.1099/mic.0.033886-0. [DOI] [PubMed] [Google Scholar]
- 31.Unsold IA, Li SM. Microbiology (UK) 2005;151:1499–1505. doi: 10.1099/mic.0.27759-0. [DOI] [PubMed] [Google Scholar]
- 32.Metzger U, Schall C, Zocher G, Unsold I, Stec E, Li SM, Heide L, Stehle T. Proc Natl Acad Sci USA. 2009;106:14309–14314. doi: 10.1073/pnas.0904897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chooi YH, Cacho R, Tang Y. Chem Biol. 2010;17:483–494. doi: 10.1016/j.chembiol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmed SA, Bardshiri E, McIntyre CR, Simpson TJ. Aust J Chem. 1992;45:249–274. [Google Scholar]
- 35.Kelly DE, Krasevec N, Mullins J, Nelson DR. Fungal Genet Biol. 2009;46:S53–S61. doi: 10.1016/j.fgb.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 36.Pecere T, Gazzola MV, Mucignat C, Parolin C, Dalla Vecchia F, Cavaggioni A, Basso G, Diaspro A, Salvato B, Carli M, Palu G. Cancer Res. 2000;60:2800–2804. [PubMed] [Google Scholar]
- 37.Wen L, Lin YC, She ZG, Du DS, Chan WL, Zheng ZH. J Asian Nat Prod Res. 2008;10:133–137. doi: 10.1080/10286020701273783. [DOI] [PubMed] [Google Scholar]
- 38.Hein SM, Gloer JB, Koster B, Malloch D. J Nat Prod. 1998;61:1566–1567. doi: 10.1021/np9801918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.