Abstract
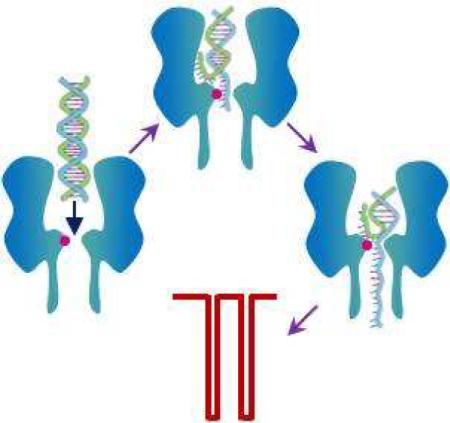
Biological protein α-hemolysin nanopore is under intense investigation as a potential platform for rapid and low-cost DNA sequencing. However, due to its narrow constriction, analysis of DNA in the α-hemolysin pore has long time been restricted to single strands. In this paper, we report that by introducing new surface functional groups into the α-hemolysin pore, facilitated unzipping of double-stranded DNA through the channel could be achieved. Since the mean residence time of the DNA events is dependent on the length of the duplex, and also varies with the nucleotide base composition, the modified protein pore approach offers the potential for rapid double-stranded DNA analysis, including sequencing.
Keywords: α-hemolysin, protein engineering, double-stranded DNA, nanopore sequencing, facilitated DNA duplex unzipping
The transmembrane protein α-hemolysin (αHL) pore has been used to develop ultrasensitive biosensors1–10, study biomolecular folding and unfolding11, investigate covalent and non-covalent bonding interactions12–14, and probe enzyme kinetics15. In particular, αHL pore is under intense investigation as a potential platform for rapid and low-cost DNA sequencing16–21, where a DNA strand is electrophoretically driven through the pore, which allows each individual base of the polymer to be potentially identified in sequence if the ionic current would be modulated to a different extent by each base. Since the αHL pore has a ~ 1.5 nm diameter constriction22, which is only slightly larger than the diameter (~ 1.3 nm) of a single-stranded DNA (ssDNA), analysis of DNA in the αHL pore has long time been restricted to single strands. In addition to the αHL pore, solid-state nanopores and several other biological nanopores including MspA and phi29 motor protein have been investigated for potential DNA sequencing23–28. Although the fabricated solid-state nanopores are robust and can function in a variety of extreme conditions, the difficulty in creating these synthetic nanopores with reproducible pore sizes and especially the low resolution of these pores due to the lack of surface functions have limited their practical sequencing application. Similar to the αHL pore, the biomedical application of the MspA channel has been mainly focused on the study of ssDNA due to its narrow constriction (~ 1 nm diameter)27. On the other hand, the phi29 motor protein nanopore is between 3.6 and 6.0 nm wide, and it has shown that double stranded DNA (dsDNA) could translocate through the phi29 nanopore28. However, the relative lack of results to date makes it impossible to gauge whether this protein pore could be used to measure the length of dsDNA at this point in time. It should be noted that DNA is rarely present in single-stranded form. The ability to direct detection of dsDNA without the need for denaturation has the potential to simplify sample preparation and bring down the sequencing cost. In this work, we systematically investigated the interaction between dsDNA and three types of mutant αHL pores, and demonstrated that it is feasible to utilize properly engineered αHL pores to analyze dsDNA to obtain useful information about its structure and length.
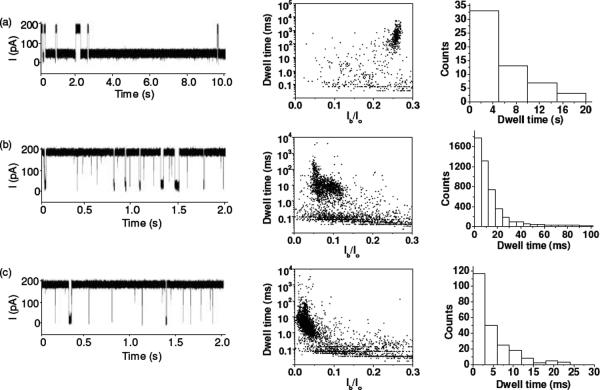
The initial experiments were performed in 1 M NaCl and 10 mM Tris·HCl (pH 7.5) at +200 mV with a 20 bp dsDNA with 0% GC content, d(A)20•d(T)20 (sequence: 5'-AAAAAAAAAAAAAAAAAAAA-3' and its complementary strand), in the wild-type and three mutant αHL pores, including (M113F)7, (M113E)7, and (M113K)7. The position 113 is close to the narrowest part of the lumen of the αHL channel, and has been used to design nanopore sensors for a wide variety of compounds2,3,5,8. The new amino acid residues in the mutant protein pores belonged to three major classes. The engineered (M113K)7 protein contains seven positively charged Lys side chains, providing an interaction site for negatively charged molecules. The mutant (M113E)7 pore presents an electrostatic interaction site (containing seven negatively charged Glu amino acid residues) for positively charged compounds. The (M113F)7 channel contains an aromatic binding site (consisting of seven aromatic Phe side chains) for aromatic analytes. In contrast, the wild-type αHL pore has seven Met residues at position 113, proving a hydrophobic interaction surface. It should be noted that hydrophobic interactions can also occur in the mutant pores, although the designed binding between the target analytes and the protein is predominantly due to the electrostatic or aromatic interactions. It is believed that the current modulations with the blockage amplitudes less than 70% of the open channel value are attributed to the brief residency of DNA polymers in the vestibule or their collision with the opening of the αHL pore.21 Since the residence times of these blockades are not polymer length dependent (Fig. S1), for convenience, only the modulations with at least 70% of the complete channel block were counted as events in this work. As shown in Fig. 1 and Fig. S2 (Supporting Information), with the exception of the (M113E)7 pore, in which no events were observed, the 20 bp dsDNA caused two significantly different types of events in the wild-type, (M113F)7, and (M113K)7 αHL channels. One type of events shows a small mean residence time and a wide range of current blockage amplitudes, while the other type of events presents a narrow range of current blockage amplitudes and a large mean duration value but with a broad distribution of residence times. Furthermore, we noticed that the mean residence times of the short-lived events were not significantly different in the three different αHL pores and the value (~30 μs) was similar to the mean duration of translocation of 20-mer ssDNA polymers through the αHL pore (data not shown). In contrast, although the mean residence times of the long-lived events varied greatly with different αHL pores (4840 ± 450 ms, 9.7 ± 1.2 ms, and 4.4 ± 0.6 ms for the wild-type, (M113F)7, and (M113K)7 αHL pores, respectively), they were at least two orders larger than those of the short-lived ones. Our results suggest that the short-lived events may be due to the translocation of ssDNA molecules, which were obtained after dsDNA unzipping, through the pore, while the long-lived events are probably attributed to the tangling of dsDNA with/near the constriction region of the channel. In the latter case, either DNA duplexes could escape backwards and exit from the cis mouth of the pore, or they would be unzipped by the electrical potential and one of the two strands translocate through the pore. This interpretation was supported by our observation that many of such long-lived events in the wild-type αHL pore began with a long duration mid-amplitude (~75%) channel block and then ended with a short-lived upward or downward terminal spike. In the (M113F)7 and (M113K)7 αHL pores, the 20 bp dsDNA produced events without such mid-amplitude channel blocks and with much smaller residence times than those in the wild-type αHL pore, suggesting that dsDNA molecules would not plug the constrictions of the mutant pores for a long time but instead they were rapidly unzipped and translocated through the channels (note that facilitated dsDNA unzipping in mutant protein pores has also been observed by Maglia et al29).
Figure 1.
Interaction of d(A)20•d(T)20 with various αHL pores: (a) wild-type; (b) (M113F)7 and (c) (M113K)7. (Left) Typical single channel current recording traces; (Middle) corresponding scatter plots of event amplitude vs. residence time; and (Right) residence time histogram of the long-lived events. Ir/Io in Fig. 1b is normalized blockage residual current, which was obtained by dividing the average blockage residual current of events by the average open channel current. The experiment was performed at +200 mV in 1 M NaCl buffered with 10 mM Tris-HCl (pH 7.5) in the presence of 1.0 μM d(A)20•d(T)20. The traces in Fig. 1 were low-pass filtered with a four-pole Bessel filter at 10 kHz.
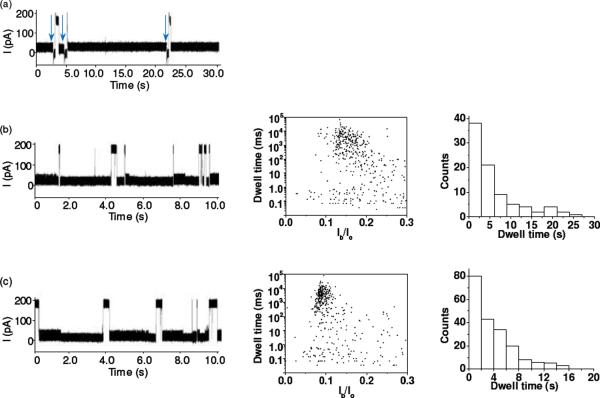
The experiments with a 20 bp dsDNA with 100% GC content, d(GC)10•d(GC)10 (sequence: 5'-GCGCGCGCGCGCGCGCGCGC-3' and its complimentary strand) in the wild-type, (M113F)7, and (M113K)7 pores provided further evidence that the long-lived events may be attributed to the unzipping or escape of dsDNA. Since the GC pair contains three hydrogen bonds, while the AT pair has only two hydrogen bonds, it could be expected that the two DNA strands in d(GC)10•d(GC)10 hold more tightly than those in d(A)20•d(T)20, and hence it would be more difficult to unzip d(GC)10•d(GC)10 in the αHL pores, leading to blockades with a larger mean residence time than those of d(A)20•d(T)20. This hypothesis was confirmed by our experimental results. For example, the residence times of d(A)20•d(T)20 in the (M113F)7 and (M113K)7 pores were 9.7 ± 1.2 ms and 4.4 ± 0.6 ms, respectively, while those of d(GC)10•d(GC)10 were 4100 ± 400 ms and 3200 ± 200 ms, respectively. Note that d(GC)10•d(GC)10 molecules permanently block the wild-type αHL pore, and even switching the polarity of the applied potential could not restore the open channel current (vertical arrows in Fig. 2a). Again, the long-lived events of d(GC)10•d(GC)10 in the (M113F)7 and (M113K)7 pores had significantly smaller residence times than those in the wild-type αHL pore, indicating that the surface functional groups at/near the constriction of the pore play a significant role in the unzipping of dsDNA. Furthermore, coinciding with the significantly larger duration of the long-lived events of d(GC)10•d(GC)10 over those of d(A)20•d(T)20, we noticed that no short-lived events were observed with d(GC)10•d(GC)10 in the wild-type αHL pore, and the short duration events of d(GC)10•d(GC)10 in the (M113F)7 and (M113K)7 pores were much less frequent than those of d(A)20•d(T)20. However, the percentages (~39% and ~45%) of the number of short-lived events to the total number of events for d(GC)10•d(GC)10 in the (M113F)7 and (M113K)7 pores were similar to those (~44% and ~38%) for d(A)20•d(T)20 in the (M113F)7 and (M113K)7 pores. This supports the interpretation that the short-lived events are caused by the translocation of ssDNA molecules, which are obtained after dsDNA unzipping. In addition, as an added importance, d(A)20•d(T)20 and d(GC)10•d(GC)10 produced events with significantly different blockage amplitudes (94 ± 1% vs. 85 ± 3%) in the (M113F)7 pore but not in the (M113K)7 pore (96 ± 2% vs. 94 ± 3%), suggesting that AT base pair and GC base pair could be possibly differentiated via blockage amplitudes in appropriately engineered pores. Indeed, events with a sequence of current modulations at different blockage levels (or substates) were observed in the experiment with the (M113F)7 pore and a 20 bp dsDNA with 50% GC content, d(CTAG)5•d(CTAG)5 (sequence: 5'-CTAGCTAGCTAGCTAGCTAG-3' and its complimentary strand) (Supporting Information, Fig. S3). The feasibility of utilizing these substate events to rapidly discriminate dsDNA sequences is currently in progress.
Figure 2.
Interaction of d(GC)10•d(GC)10 with various αHL pores: (a) wild-type; (b) (M113F)7; and (c) (M113K)7. (Left) Typical single channel current recording traces; (Middle) corresponding scatter plots of event amplitude vs. residence time; and (Right) residence time histogram of the long-lived events. The experiment was performed at +200 mV in 1 M NaCl buffered with 10 mM Tris-HCl (pH 7.5) in the presence of 1.0 μM d(GC)10•d(GC)10. The traces in Fig. 2 were low-pass filtered with a four-pole Bessel filter at 10 kHz.
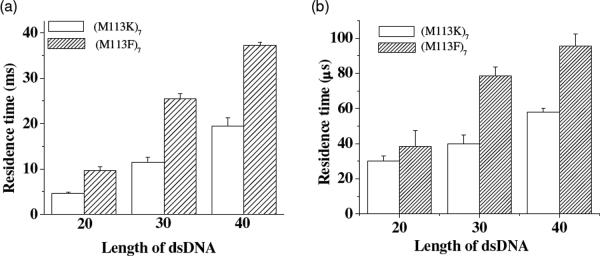
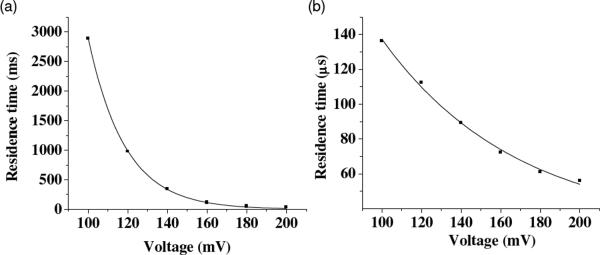
To further demonstrate that the long-lived events were caused by unzipping and then translocating of DNA in the αHL pores, length dependence and voltage dependence studies was performed. Our experimental results showed that the mean residence times for both the long-lived events and short-lived events increased with an increase in the length of dsDNA (Fig. 3), while decreased as the voltage increased (Fig. 4). In addition, the salt effect on dsDNA's interaction with the αHL (M113F)7 pore was investigated. We found that with a decrease in the salt concentration, the residence time of the long-lived DNA events decreased (Fig. S4, Supporting Information). Taken together, the combined results provided evidence that the long-lived events were indeed attributed to translocation.
Figure 3.
Plot of the event mean residence time as a function of the length of dsDNA polymers: (a) long-lived events; and (b) short-lived events. The experiments were performed at +200 mV in 1 M NaCl buffered with 10 mM Tris-HCl (pH 7.5). The 20-mer, 30-mer, and 40-mer double-stranded DNA samples used in this experiment were d(A)20•d(T)20, d(A)30•d(T)30, and d(A)40•d(T)40, respectively (see Supporting Information for their sequences).
Figure 4.
Effect of the applied voltage on the event mean residence time: (a) long-lived events; and (b) short-lived events. The experiment was performed with 1.0 μM d(A)40•d(T)40 and the (M113F)7 pore at +200 mV in 1 M NaCl buffered with 10 mM Tris-HCl (pH 7.5).
In conclusion, the interaction between dsDNA and various αHL pores with different surface functions were systematically investigated. By introducing positively charged or aromatic groups into the αHL pores, facilitated dsDNA unzipping by the protein pore could be achieved. The engineered protein pore approach offers the potential for rapid detection, differentiation, characterization, and even sequencing of double-stranded DNA.
Supplementary Material
Acknowledgements
This work was financially supported by the National Institutes of Health (1R011HG005095).
Footnotes
Supporting Information Experimental details, and additional table and figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Gu L-Q, Braha O, Conlan S, Cheley S, Bayley H. Stochastic Sensing of Organic Analytes by a Pore-Forming Protein Containing a Molecular Adapter. Nature. 1999;398:686–690. doi: 10.1038/19491. [DOI] [PubMed] [Google Scholar]
- (2).Cheley S, Gu L-Q, Bayley H. Stochastic Sensing of Nanomolar Inositol 1,4,5-Trisphosphate with an Engineered Pore. Chem. Biol. 2002;9:829–838. doi: 10.1016/s1074-5521(02)00172-2. [DOI] [PubMed] [Google Scholar]
- (3).Guan X, Gu L-Q, Cheley S, Braha O, Bayley H. Stochastic Sensing of TNT with a Genetically Engineered Pore. ChemBioChem. 2005;6:1875–1881. doi: 10.1002/cbic.200500064. [DOI] [PubMed] [Google Scholar]
- (4).Jayawardhana DA, Crank JA, Zhao Q, Armstrong DW, Guan X. Nanopore Stochastic Detection of a Liquid Explosive Component and Sensitizers using Boromycin and an Ionic Liquid Supporting Electrolyte. Anal. Chem. 2009;81:460–464. doi: 10.1021/ac801877g. [DOI] [PubMed] [Google Scholar]
- (5).Wang D, Zhao Q, de Zoysa RSS, Guan X. Detection Of Nerve Agent Hydrolytes in an Engineered Nanopore. Sens. Actuators B. 2009;139:440–446. [Google Scholar]
- (6).Braha O, Walker B, Cheley S, Kasianowicz JJ, Song L, Gouaux JE, Bayley H. Designed Protein Pores as Components for Biosensors. Chem. Biol. 1997;4:497–505. doi: 10.1016/s1074-5521(97)90321-5. [DOI] [PubMed] [Google Scholar]
- (7).Braha O, Gu L-Q, Zhou L, Lu X, Cheley S, Bayley H. Simultaneous Stochastic Sensing of Divalent Metal Ions. Nat. Biotechnol. 2000;18:1005–1007. doi: 10.1038/79275. [DOI] [PubMed] [Google Scholar]
- (8).Kang X, Cheley S, Guan X, Bayley H. Stochastic Detection of Enantiomers. J. Am. Chem. Soc. 2006;128:10684–10685. doi: 10.1021/ja063485l. [DOI] [PubMed] [Google Scholar]
- (9).Movileanu L, Howorka S, Braha O, Bayley H. Detecting Protein Analytes that Modulate Transmembrane Movement of a Polymer Chain within a Single Protein Pore. Nat. Biotechnol. 2000;18:1091–1095. doi: 10.1038/80295. [DOI] [PubMed] [Google Scholar]
- (10).Howorka S, Nam J, Bayley H, Kahne D. Stochastic Detection of Monovalent and Bivalent Protein-Ligand Interactions. Angew. Chem. Int. Ed. 2004;43:842–846. doi: 10.1002/anie.200352614. [DOI] [PubMed] [Google Scholar]
- (11).Shim JW, Tan Q, Gu L-Q. Single-Molecule Detection of Folding and Unfolding of the G-Quadruplex Aptamer in a Nanopore Nanocavity. Nucleic Acids Res. 2009;37:972–982. doi: 10.1093/nar/gkn968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Luchian T, Shin S-H, Bayley H. Single-Molecule Covalent Chemistry with Spatially Separated Reactants. Angew. Chem. Int. Ed. 2003;42:3766–3771. doi: 10.1002/anie.200351313. [DOI] [PubMed] [Google Scholar]
- (13).Shin S-H, Luchian T, Cheley S, Braha O, Bayley H. Kinetics of a Reversible Covalent-Bond-Forming Reaction Observed at the Single-Molecule Level. Angew. Chem. Int. Ed. 2002;41:3707–3709. doi: 10.1002/1521-3773(20021004)41:19<3707::AID-ANIE3707>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- (14).Zhao Q, Jayawardhana DA, Guan X. Stochastic Study of the Effect of Ionic Strength on Non-Covalent Interactions in Protein Pores. Biophys. J. 2008;94:1267–1275. doi: 10.1529/biophysj.107.117598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zhao Q, de Zoysa RSS, Wang D, Jayawardhana DA, Guan X. Real-Time Monitoring of Peptide Cleavage using a Nanopore Probe. J. Am. Chem. Soc. 2009;131:6324–6325. doi: 10.1021/ja9004893. [DOI] [PubMed] [Google Scholar]
- (16).Kasianowicz JJ, Brandin E, Branton D, Deamer D. Characterization of Individual Polynucleotide Molecules using a Membrane Channel. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid Nanopore Discrimination between Single Polynucleotide Molecules. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Howorka S, Cheley S, Bayley H. Sequence-Specific Detection of Individual DNA Strands using Engineered Nanopores. Nat. Biotechnol. 2001;19:636–639. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- (19).Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Single-Nucleotide Discrimination in Immobilized DNA Oligonucleotides with a Biological Nanopore. Proc. Natl. Acad. Sci. U.S.A. 2009;106:7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Clarke J, Wu H, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous Base Identification for Single-Molecule Nanopore DNA Sequencing. Nat. Nanotechnol. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- (21).Butler TZ, Gundlach JH, Troll M. Ionic Current Blockades from DNA and RNA Molecules in the Alpha-Hemolysin Nanopore. Biophys. J. 2007;93:3229–3240. doi: 10.1529/biophysj.107.107003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure Of Staphylococcal Alpha-Hemolysin, A Heptameric Transmembrane Pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- (23).Li J, Stein D, McMullan C, Branton D, Aziz MJ, Golovchenko JA. Ion-Beam Sculpting at Nanometre Length Scales. Nature. 2001;412:166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- (24).Storm AJ, Storm C, Chen J, Zandbergen H, Joanny JF, Dekker C. Fast DNA Translocation through a Solid-State Nanopore. Nano Lett. 2005;5:1193–1197. doi: 10.1021/nl048030d. [DOI] [PubMed] [Google Scholar]
- (25).van den Hout M, Krudde V, Janssen XJ, Dekker NH. Distinguishable Populations Report on the Interactions of Single DNA Molecules with Solid-State Nanopores. Biophys J. 2010;99:3840–3848. doi: 10.1016/j.bpj.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Chen Z, Jiang Y, Dunphy DR, Adams DP, Hodges C, Liu N, Zhang N, Xomeritakis G, Jin X, Aluru NR, et al. DNA Translocation through an Array of Kinked Nanopores. Nat Mater. 2010;9:667–675. doi: 10.1038/nmat2805. [DOI] [PubMed] [Google Scholar]
- (27).Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Single-Molecule DNA Detection with an Engineered Mspa Protein Nanopore. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20647–20652. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Wendell D, Jing P, Geng J, Subramaniam V, Lee TJ, Montemagno C, Guo P. Translocation of Double-Stranded DNA through Membrane-Adapted phi29 Motor Protein Nanopores. Nat. Nanotechnol. 2009;4:765–772. doi: 10.1038/nnano.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Maglia G, Henricus M, Wyss R, Li Q, Cheley S, Bayley H. DNA Strands from Denatured Duplexes are Translocated through Engineered Protein Nanopores at Alkaline pH. Nano Lett. 2009;9:3831–6. doi: 10.1021/nl9020232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.