Abstract
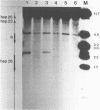
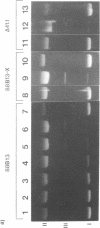
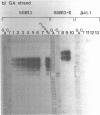
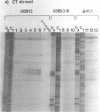
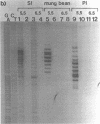
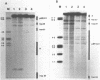
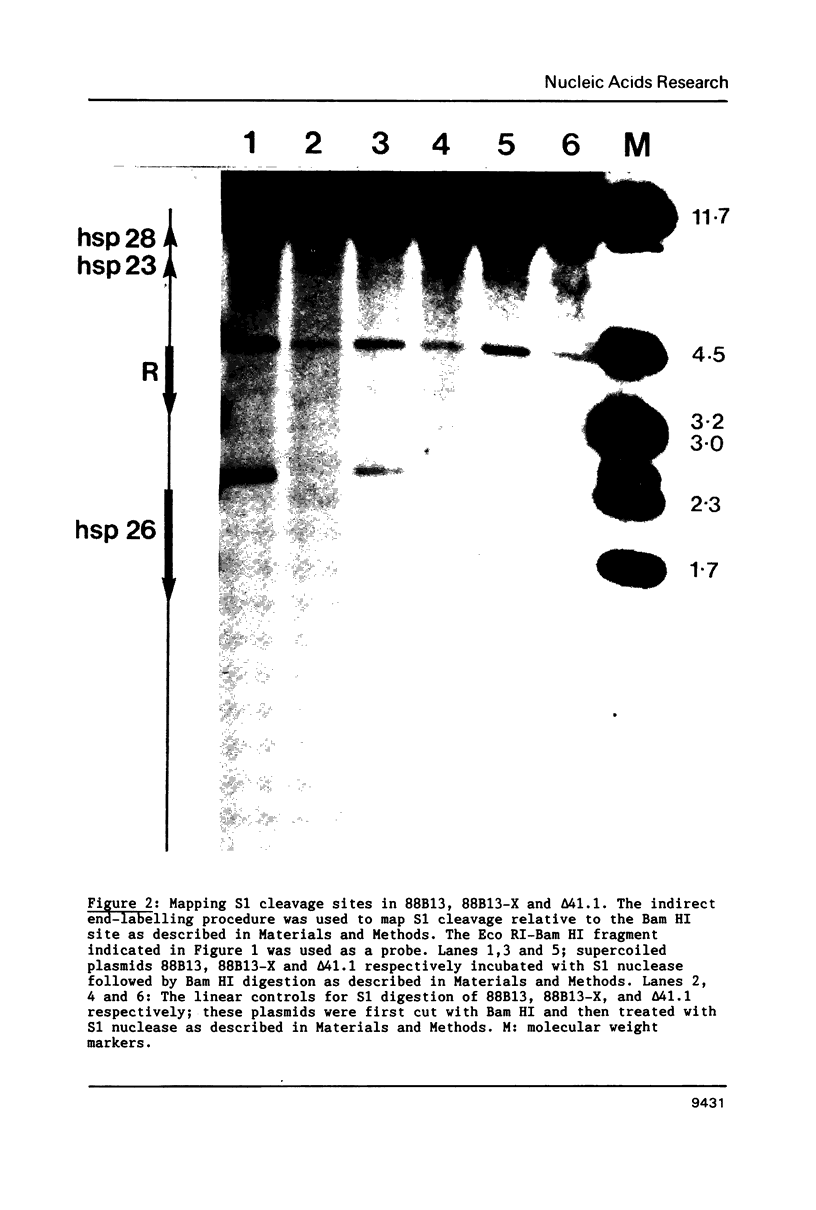
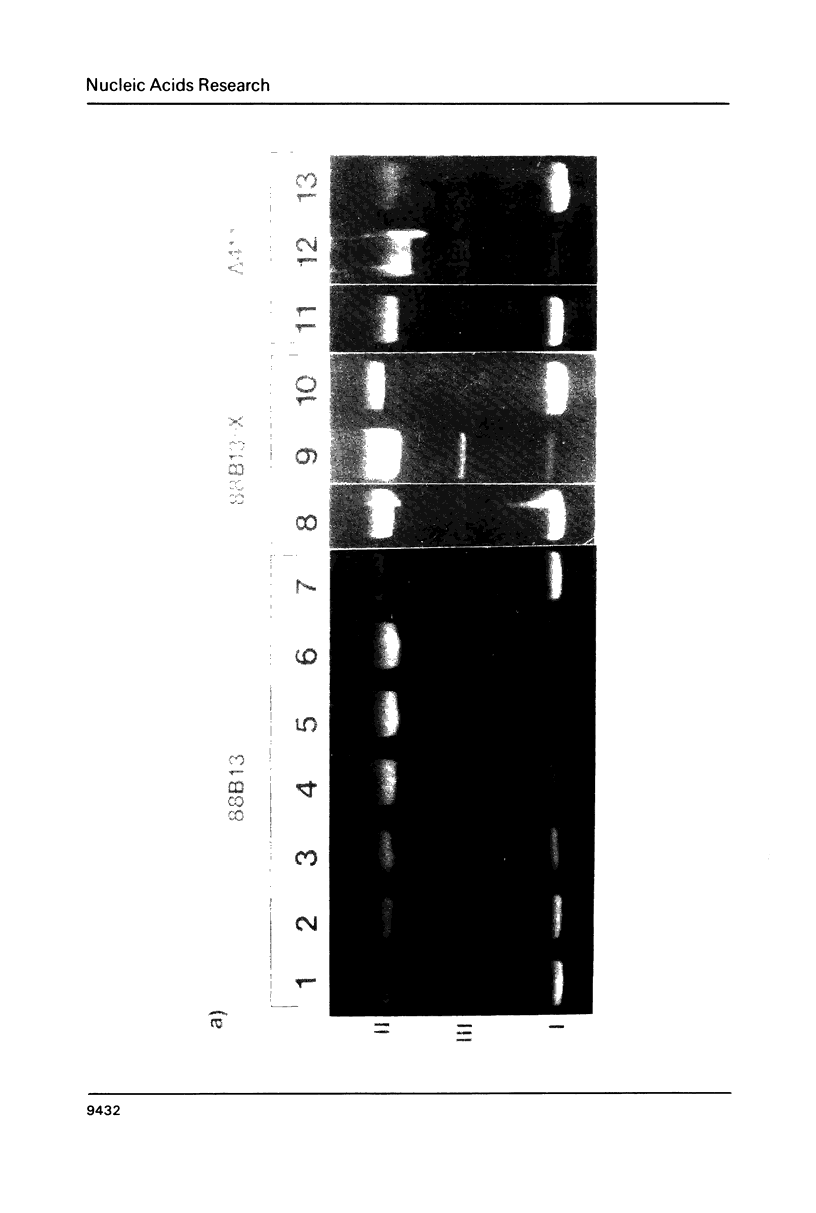
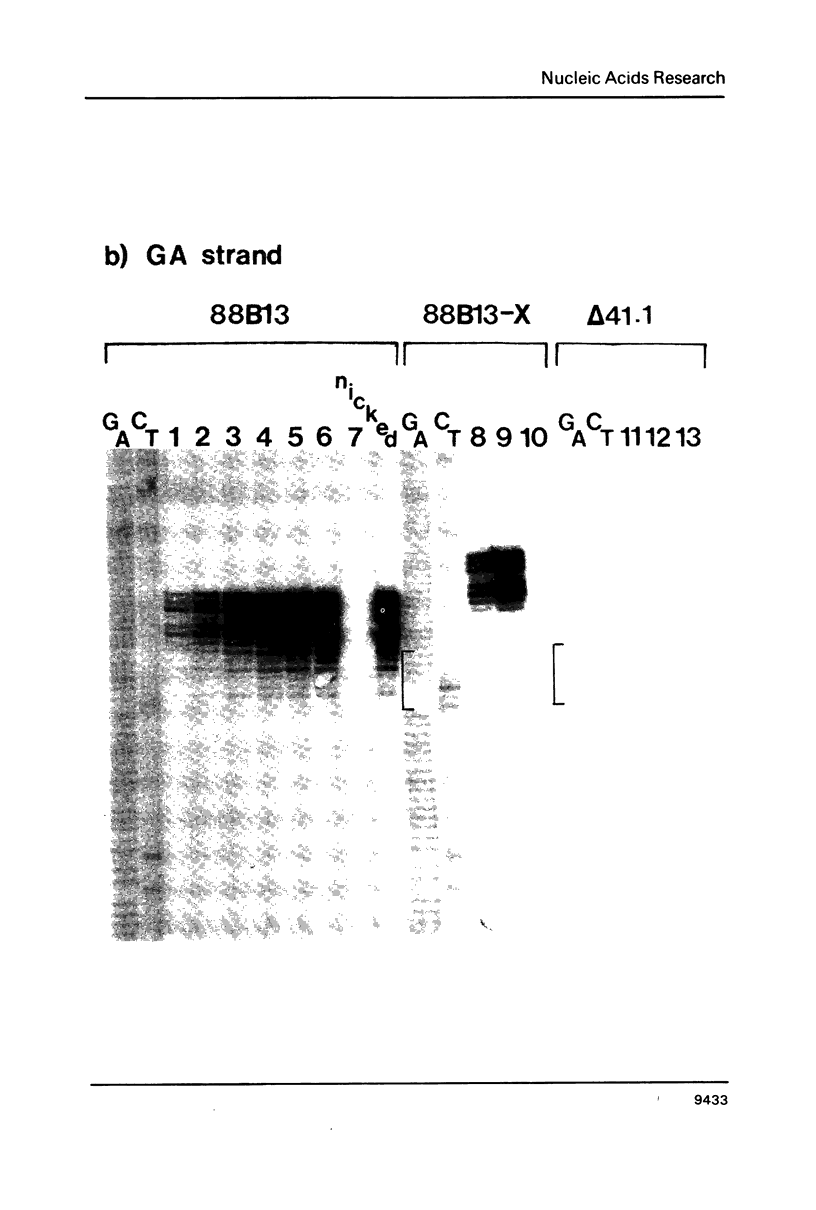
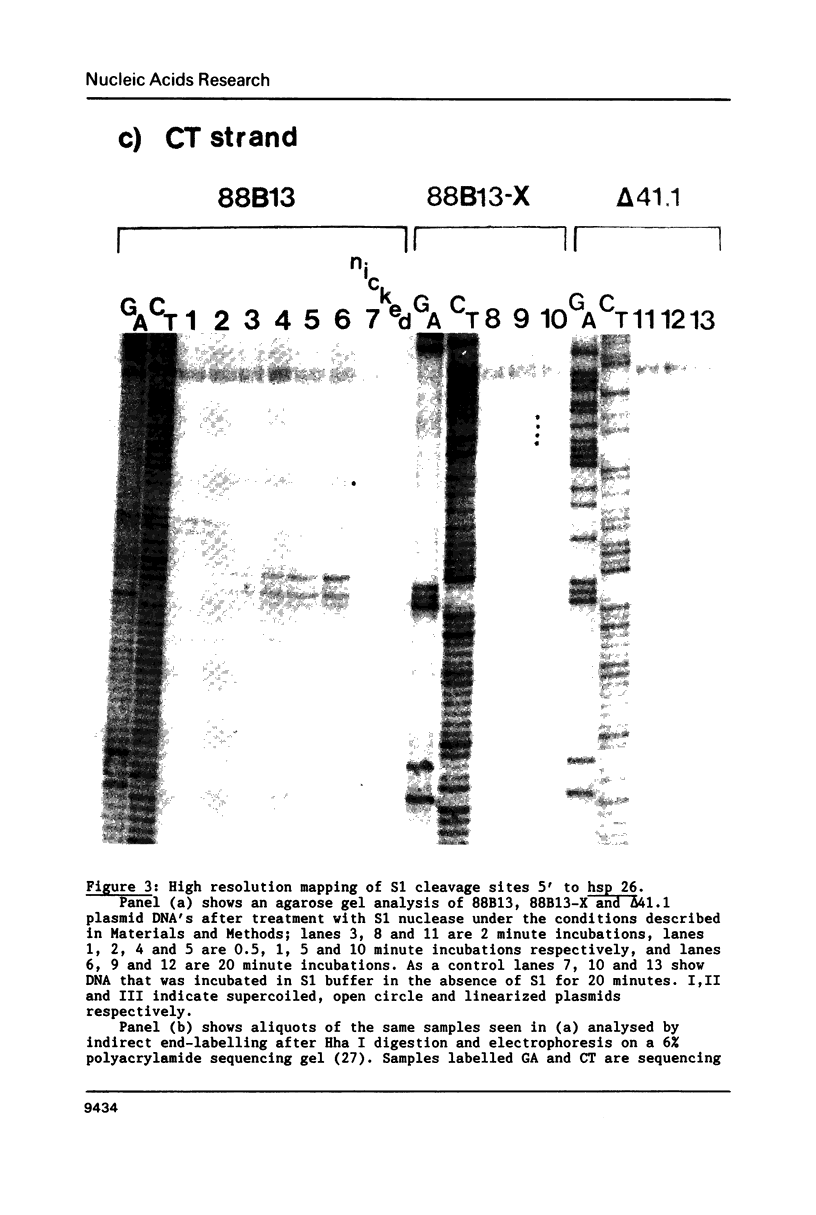
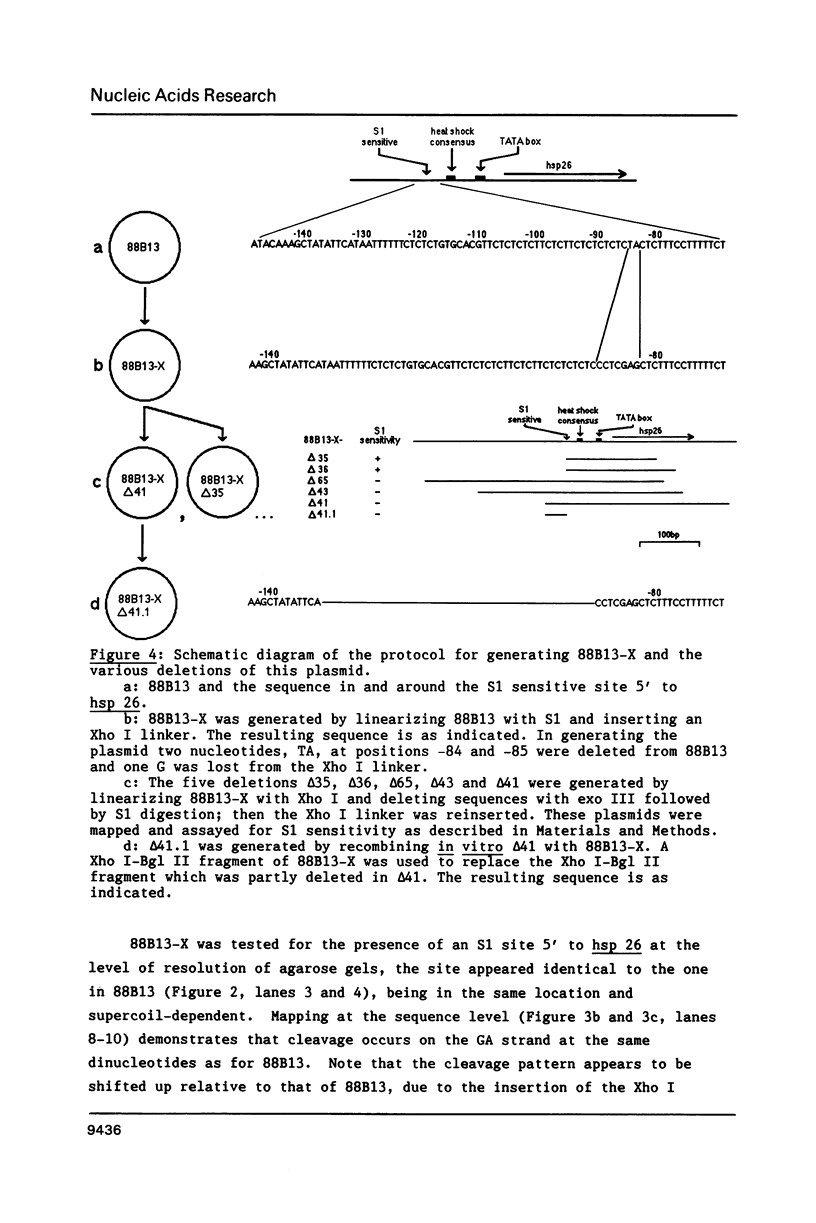
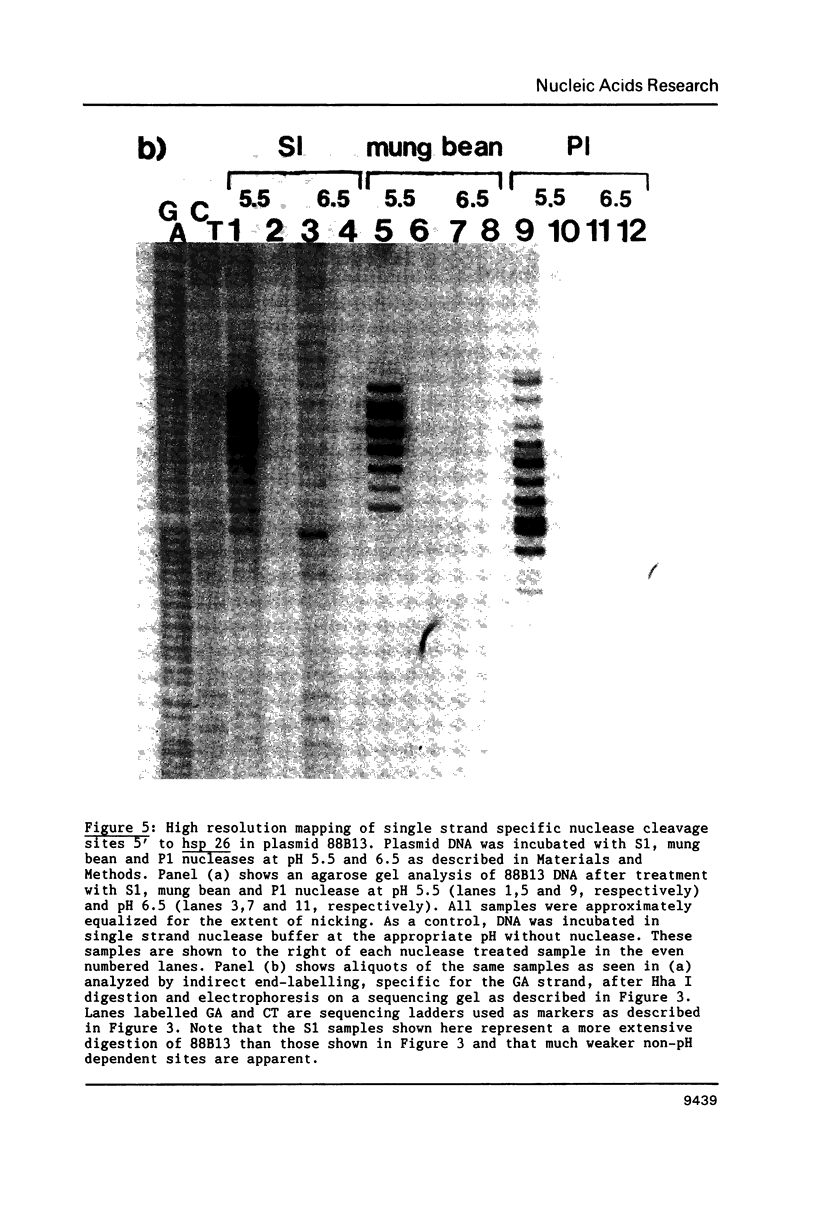
We have analyzed the prominent supercoil-dependent S1 nuclease cleavage site 5' to hsp 26 in the plasmid 88B13, which contains 11.7 kilobases from the Drosophila locus 67B1. The double stranded cleavage product is generated by initial nicking on the purine strand, six preferred sites occurring between positions -96 and -90 (relative to the start of transcription) with weaker ones extending to position -84, followed by cleavage on the pyrimidine strand at positions -86 and -84. A derivative of 88B13, 88B13-X, was generated by insertion of an Xho I linker at position -84; this does not affect the positions or strand specificity of the S1 cleavage in that region. A small deletion, delta 41.1, removes the homopurine/homopyrimidine stretch from positions -86 to -132 and is no longer sensitive to cleavage by S1 nuclease 5' to hsp 26. Mung bean and P1 nucleases recognize the same site 5' to hsp 26 and give the same general pattern of cleavage. All three nucleases show an initial cleavage of 88B13 DNA at this site at pH 5.5 but not at pH 6.5, indicating that the DNA structure there may be pH dependent in vitro.
Full text
PDF








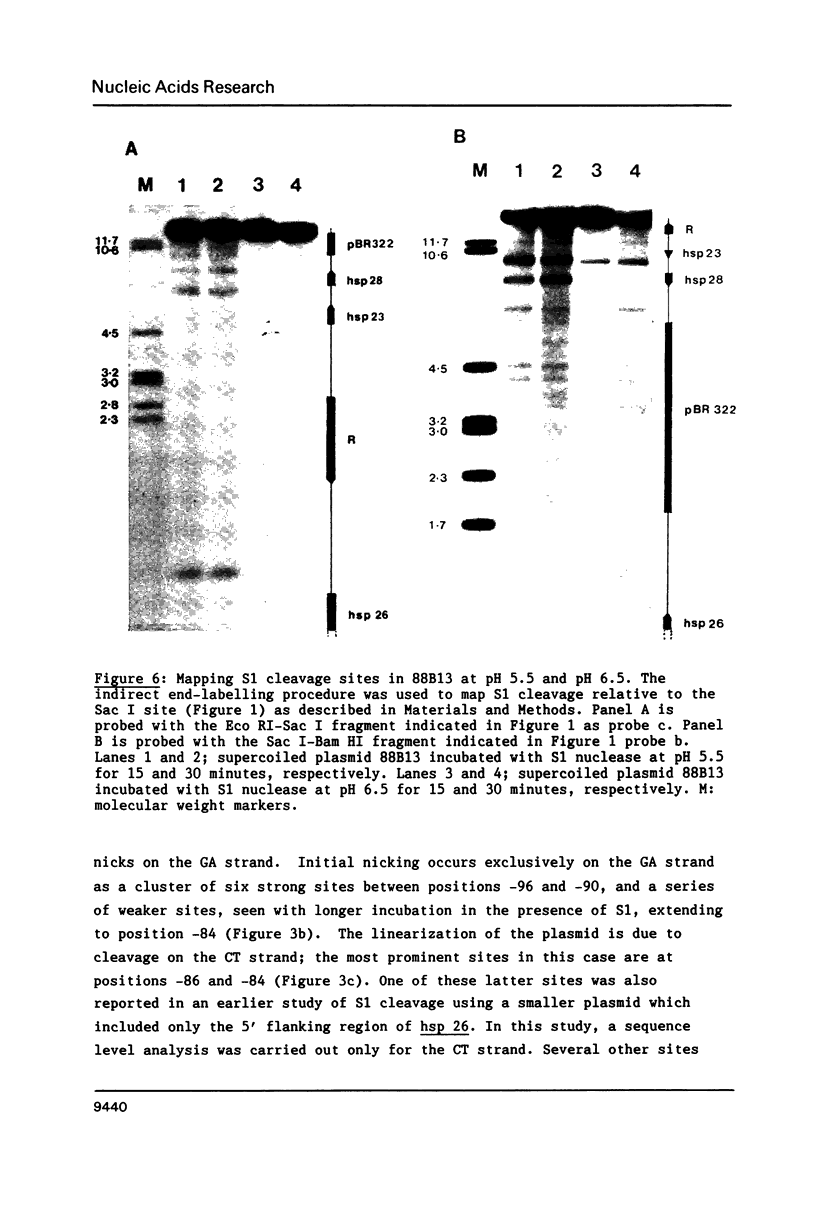




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor C. R. DNA choreography. Cell. 1981 Aug;25(2):293–295. doi: 10.1016/0092-8674(81)90045-3. [DOI] [PubMed] [Google Scholar]
- Cantor C. R., Efstratiadis A. Possible structures of homopurine-homopyrimidine S1-hypersensitive sites. Nucleic Acids Res. 1984 Nov 12;12(21):8059–8072. doi: 10.1093/nar/12.21.8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright I. L., Elgin S. C. Nucleosomal instability and induction of new upstream protein-DNA associations accompany activation of four small heat shock protein genes in Drosophila melanogaster. Mol Cell Biol. 1986 Mar;6(3):779–791. doi: 10.1128/mcb.6.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R. S., Meselson M. Separate regulatory elements for the heat-inducible and ovarian expression of the Drosophila hsp26 gene. Cell. 1985 Dec;43(3 Pt 2):737–746. doi: 10.1016/0092-8674(85)90247-8. [DOI] [PubMed] [Google Scholar]
- Corces V., Holmgren R., Freund R., Morimoto R., Meselson M. Four heat shock proteins of Drosophila melanogaster coded within a 12-kilobase region in chromosome subdivision 67B. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5390–5393. doi: 10.1073/pnas.77.9.5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J. C., Cartwright I. L., Thomas G. H., Elgin S. C. Selected topics in chromatin structure. Annu Rev Genet. 1985;19:485–536. doi: 10.1146/annurev.ge.19.120185.002413. [DOI] [PubMed] [Google Scholar]
- Evans T., Schon E., Gora-Maslak G., Patterson J., Efstratiadis A. S1-hypersensitive sites in eukaryotic promoter regions. Nucleic Acids Res. 1984 Nov 12;12(21):8043–8058. doi: 10.1093/nar/12.21.8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finer M. H., Fodor E. J., Boedtker H., Doty P. Endonuclease S1-sensitive site in chicken pro-alpha 2(I) collagen 5' flanking gene region. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1659–1663. doi: 10.1073/pnas.81.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Udvardy A., Schedl P. Transcriptionally active chromatin is sensitive to Neurospora crassa and S1 nucleases. J Mol Biol. 1984 Nov 5;179(3):469–496. doi: 10.1016/0022-2836(84)90076-7. [DOI] [PubMed] [Google Scholar]
- Harland R. M., Weintraub H., McKnight S. L. Transcription of DNA injected into Xenopus oocytes is influenced by template topology. Nature. 1983 Mar 3;302(5903):38–43. doi: 10.1038/302038a0. [DOI] [PubMed] [Google Scholar]
- Hentschel C. C. Homocopolymer sequences in the spacer of a sea urchin histone gene repeat are sensitive to S1 nuclease. Nature. 1982 Feb 25;295(5851):714–716. doi: 10.1038/295714a0. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Htun H., Lund E., Dahlberg J. E. Human U1 RNA genes contain an unusually sensitive nuclease S1 cleavage site within the conserved 3' flanking region. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7288–7292. doi: 10.1073/pnas.81.23.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchnik A. N., Bakayev V. V., Zbarsky I. B., Georgiev G. P. Elastic torsional strain in DNA within a fraction of SV40 minichromosomes: relation to transcriptionally active chromatin. EMBO J. 1982;1(11):1353–1358. doi: 10.1002/j.1460-2075.1982.tb01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D. A pH-dependent structural transition in the homopurine-homopyrimidine tract in superhelical DNA. J Biomol Struct Dyn. 1985 Oct;3(2):327–338. doi: 10.1080/07391102.1985.10508420. [DOI] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D. Structures of homopurine-homopyrimidine tract in superhelical DNA. J Biomol Struct Dyn. 1986 Feb;3(4):667–669. doi: 10.1080/07391102.1986.10508454. [DOI] [PubMed] [Google Scholar]
- Mace H. A., Pelham H. R., Travers A. A. Association of an S1 nuclease-sensitive structure with short direct repeats 5' of Drosophila heat shock genes. Nature. 1983 Aug 11;304(5926):555–557. doi: 10.1038/304555a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedospasov S. A., Georgiev G. P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980 Jan 29;92(2):532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- Nickol J. M., Felsenfeld G. DNA conformation at the 5' end of the chicken adult beta-globin gene. Cell. 1983 Dec;35(2 Pt 1):467–477. doi: 10.1016/0092-8674(83)90180-0. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Haniford D. B., Morgan A. R. A structural basis for S1 nuclease sensitivity of double-stranded DNA. Cell. 1985 Aug;42(1):271–280. doi: 10.1016/s0092-8674(85)80122-7. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Ryoji M., Worcel A. Chromatin assembly in Xenopus oocytes: in vivo studies. Cell. 1984 May;37(1):21–32. doi: 10.1016/0092-8674(84)90297-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon E., Evans T., Welsh J., Efstratiadis A. Conformation of promoter DNA: fine mapping of S1-hypersensitive sites. Cell. 1983 Dec;35(3 Pt 2):837–848. doi: 10.1016/0092-8674(83)90116-2. [DOI] [PubMed] [Google Scholar]
- Selleck S. B., Elgin S. C., Cartwright I. L. Supercoil-dependent features of DNA structure at Drosophila locus 67B1. J Mol Biol. 1984 Sep 5;178(1):17–33. doi: 10.1016/0022-2836(84)90228-6. [DOI] [PubMed] [Google Scholar]
- Shen C. K. Superhelicity induces hypersensitivity of a human polypyrimidine . polypurine DNA sequence in the human alpha 2-alpha 1 globin intergenic region to S1 nuclease digestion--high resolution mapping of the clustered cleavage sites. Nucleic Acids Res. 1983 Nov 25;11(22):7899–7910. doi: 10.1093/nar/11.22.7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K., Klysik J., Wells R. D. Conformational flexibility of junctions between contiguous B- and Z-DNAs in supercoiled plasmids. Proc Natl Acad Sci U S A. 1983 May;80(9):2447–2451. doi: 10.1073/pnas.80.9.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R. DNA supercoiling: another level for regulating gene expression. Cell. 1981 Jun;24(3):599–600. doi: 10.1016/0092-8674(81)90085-4. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Weintraub H. A dominant role for DNA secondary structure in forming hypersensitive structures in chromatin. Cell. 1983 Apr;32(4):1191–1203. doi: 10.1016/0092-8674(83)90302-1. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Cheng P. F., Conrad K. Expression of transfected DNA depends on DNA topology. Cell. 1986 Jul 4;46(1):115–122. doi: 10.1016/0092-8674(86)90865-2. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Goodman T. C., Hillen W., Horn G. T., Klein R. D., Larson J. E., Müller U. R., Neuendorf S. K., Panayotatos N., Stirdivant S. M. DNA structure and gene regulation. Prog Nucleic Acid Res Mol Biol. 1980;24:167–267. doi: 10.1016/s0079-6603(08)60674-1. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Yu Y. T., Manley J. L. Structure and function of the S1 nuclease-sensitive site in the adenovirus late promoter. Cell. 1986 Jun 6;45(5):743–751. doi: 10.1016/0092-8674(86)90788-9. [DOI] [PubMed] [Google Scholar]