Abstract
Chinese hamster ovary (CHO) cells are the predominant cell factory for the production of recombinant therapeutic proteins. Nevertheless, the lack in publicly available sequence information is severely limiting advances in CHO cell biology, including the exploration of microRNAs (miRNA) as tools for CHO cell characterization and engineering. In an effort to identify and annotate both conserved and novel CHO miRNAs in the absence of a Chinese hamster genome, we deep-sequenced small RNA fractions of 6 biotechnologically relevant cell lines and mapped the resulting reads to an artificial reference sequence consisting of all known miRNA hairpins. Read alignment patterns and read count ratios of 5′ and 3′ mature miRNAs were obtained and used for an independent classification into miR/miR* and 5p/3p miRNA pairs and discrimination of miRNAs from other non-coding RNAs, resulting in the annotation of 387 mature CHO miRNAs. The quantitative content of next-generation sequencing data was analyzed and confirmed using qPCR, to find that miRNAs are markers of cell status. Finally, cDNA sequencing of 26 validated targets of miR-17-92 suggests conserved functions for miRNAs in CHO cells, which together with the now publicly available sequence information sets the stage for developing novel RNAi tools for CHO cell engineering.
Keywords: microRNA, Chinese hamster ovary cells, Next-generation sequencing
1. Introduction
The Chinese hamster, Cricetulus griseus, has come a long way from being an important model organism for cytogenetic research to becoming the origin of a cell line (Tjio and Puck, 1958) that is now the most frequently used cell factory for the production of recombinant protein therapeutics with an annual market value exceeding 70 billion dollars (Jayapal et al., 2007). The continuous improvement of CHO-based bioprocesses, which is essential to meet the increasing demand for complex glycosylated protein therapeutics, is based on various strategies (Wurm, 2004), including their targeted genetic engineering (Kramer et al., 2010). In the striking absence of public Chinese hamster DNA sequence information, functional genomic and proteomic tools have been developed in several labs to identify promising cellular pathways (Kantardjieff et al., 2009, 2010) as well as specific genes (Doolan et al., 2010) that are significantly deregulated under conditions of high productivity or fast growth and which could therefore serve as targets for cell engineering approaches. In this respect, the miRNA dependent post-transcriptional regulation of gene expression in CHO cells was only recently proposed as a potential tool to characterize and engineer CHO cell lines (Barron et al., 2010; Müller et al., 2008), as they are well recognized to regulate many physiological processes like cell cycle (Carleton et al., 2007), metabolism (Gao et al., 2009), and cell death (Subramanian and Steer, 2010).
Being small, non-coding RNAs, miRNAs are transcribed within the nucleus, processed by RNaseIII Drosha (Lee et al., 2003) and exported as ∼70 nucleotide long hairpins to the cytoplasm, where they are enzymatically cleaved by Dicer (Hutvagner et al., 2001) to give rise to two ∼22 nucleotide long mature miRNA sequences in the form of a complementary duplex structure (Carthew and Sontheimer, 2009). Depending on the thermodynamic properties of this duplex, one strand is preferably incorporated into the RNA-induced-silencing complex (RISC), to become the guide miRNA. By binding partially complementary regions in the 3′ untranslated regions (UTR) of target mRNAs, the guide miRNA enables RISC to either degrade or repress translation of the target mRNA (Bartel, 2009). As individual miRNAs have the potential to bind numerous different mRNAs, and since the 3′UTR of a single mRNA can contain binding sites for several different miRNAs, the resulting multiplicity of potential interactions allows miRNAs to modulate complex regulatory pathways (Baek et al., 2008; Selbach et al., 2008). Consequently, it has been proposed that specific miRNA transcription signatures might not only be linked to undifferentiated, differentiated or cancerous cellular phenotypes, but could also facilitate the emergence of entirely new cell types (Kosik, 2010). From a bioprocessing point of view, this opens a wide area for the use of miRNAs as tools for characterizing and engineering industrially relevant CHO cell lines (Müller et al., 2008).
MicroRNA transcription was first described in CHO cells in 2007, when Gammell et al. used a cross-species microarray platform to profile changes in miRNA expression patterns upon temperature shifts to 31 °C (Gammell et al., 2007), a condition commonly observed to increase specific protein productivity (Rössler et al., 1996; Sunley et al., 2008; Trummer et al., 2006). Results of this study indicated that miRNA sequences are likely to be highly conserved between mouse and CHO cells, but experimental verification of this assumption could only be given for one miRNA, cgr-miR-21. In contrast to hybridization based strategies such as microarray technology or quantitative real-time PCR, next-generation sequencing (NGS) provides a valid alternative for miRNA expression profiling, especially if no or little sequence information is available (Morozova and Marra, 2008). Using this technology the existence of several conserved mature miRNAs was recently reported in CHO cells (Johnson et al., 2010) using BLASTn alignment of Illumina sequencing reads to known mature and star miRNA sequences taken from the miRNA sequence repository miRBase (Griffiths-Jones et al., 2008). However, no precise annotations were introduced for these conserved CHO miRNAs, most likely since BLASTn alignment does not allow for an accurate mismatch control and therefore cannot reliably differentiate members of closely related miRNA species as they occur in many miRNA families such as the let-7 family or miR-17 family. Besides, such an approach also fails to provide reliable information on the miR/miR* identity of processed miRNA transcripts, which describes whether the 5′ or 3′ arm of the miRNA precursor hairpin gives rise to the predominant mature miRNA species. Especially in the light of absent genomic sequence information for the Chinese hamster, finding the best annotation for each individual conserved CHO miRNA is, however, crucial in establishing their functionality, as this often implies the use of “cross-species” target prediction algorithms for the alleged orthologous miRNA in human, mouse or rat.
In an effort to identify, annotate and profile miRNA expression in CHO cell lines for the identification of promising targets for cell engineering (“engimiRs”), we sequenced the small RNA transcriptome of 6 CHO cell lines, developed a novel method for miRNA identification and annotation in the absence of genomic sequence information and provide insights in the regulation of miRNA transcription under biotechnologically relevant conditions. By submitting sequence information of all conserved and novel CHO miRNAs to the miRBase repository (www.mirbase.org) we further provide the basis for the CHO research community to establish the necessary tools to improve miRNA research in the Chinese hamster.
2. Materials and methods
2.1. Cell lines and culture conditions
Chinese hamster ovary cell lines were cultivated at 37 °C and 7% atmospheric CO2. Serum-dependent CHO-K1 cell lines (ECACC CCL-61) were grown in 1:1 DMEM/Ham's F12 media (Biochrom, Germany) in the presence of 5% fetal calf serum (PAA, Austria) and 4 mM l-Glutamine (l-Gln). Serum-dependent CHO-DUXB11 cells (ATCC CRL-9096) were cultivated in the same medium plus 1× HT (hypoxanthine/thymidine) supplement. CHO-K1 cells were in-house adapted to serum-free growth in chemically defined CD CHO media (Gibco, Carlsbad, CA) supplemented with 8 mM l-Gln. Recombinant antibody producing CHO-K1 cells (ECACC 85051005) were serum-free adapted and cultivated in 1:1 DMEM/Ham's F12 supplemented with 2 mM methionine-sulfoximine (MSX), 0.25% soy peptone, 0.1% Pluronic F68 (BASF, Germany), PF supplement (Polymun Scientific, Austria) and GS supplement (SAFC, St. Louis, MO). Serum-free adapted CHO-DUXB11 cells were cultivated in 1:1 DMEM/Ham's F12 media supplemented with 4 mM l-Gln, 0.25% soy peptone, 0.1% Pluronic F68 and 1x PF and HT supplement. The recombinant DUXB11 cells were transfected with an Erythropoietin-Fc fusion protein (Lattenmayer et al., 2007) and cultivated in the same medium with the addition of 0.19 μM methotrexate (MTX).
2.2. RNA Isolation and Illumina small RNA library preparation
For RNA isolation, CHO cells were harvested during exponentially growth 48 h after seeding. Additionally an RNA pool was prepared comprising equal amounts of total RNA from the following conditions: (I) stationary growth phase after 120 h of batch cultivation (K1 fcs, DXB11 sf, and DXB11 rec); (II) heat shock treatment at 42 °C for 30 min (K1 sf and DXB11 rec); III) cold shock at 33 °C for 48 h (DXB11 fcs and K1 rec); and IV) sodium butyrate (NaBu, 0.3 M) treatment for 48 h at 33 °C (DXB11 sf and DXB11 rec). Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad CA) according to the manufacturer's recommendations. Quality of total RNA was controlled using Nanodrop (Thermo Scientific) and 21000 Bioanalyzer (Agilent Technologies, Germany) analyses, where RNA integrity numbers were required to be >9 for subsequent library preparation: therefore, small RNA fragments of 18–36 nucleotides were purified from 10 μg of total RNA on a 15% TBE Urea RNA Gel (Invitrogen, Carlsbad, CA). Apart from this intital purification of small RNA fractions, Illumina sequencing libraries were prepared according to the Illumina v1.5 preparation kit protocol.
2.3. Library quantity and quality assessment, cluster amplification and sequencing
Quantities of all libraries were analyzed using the Quant-iT PicoGreen dsDNA kit (Invitrogen) and the Tecan Infinite 200 Microplate Reader (Tecan, Austria) according to the manufacturer's instructions. The average fragment size of each library was measured by a DNA 1000 LabChip using the 2100 Bioanalyzer (Agilent Technologies, Germany). The molar concentration of each library was calculated from the average fragment size and the corresponding quantity. Subsequently, the libraries were diluted to 1 nM stock solutions with elution buffer EB (Qiagen GmbH, Hilden, Germany). Consequently, 120 μl of a 6 pM dilution of each library were used for cluster generation with the Single-Read Cluster Generation Kit v2 on the Cluster Station (Illumina Inc., San Diego, USA) according to the manual provided by the manufacturer (Part # 1006080 Rev A) applying the Single-Read Multi-Primer One-Step protocol. Thereby, each library was amplified in a separate lane of the flow cell including the PhiX control in lane no. 5. After cluster generation, the flow cell was sequenced on the Genome Analyzer IIx using one SBS Sequencing Kit v3 generating 36 bp single-reads. All reads were submitted to the Sequence Read Archive (SRA; www.ncbi.nlm.nih.gov/sra) at NCBI (Shumway et al., 2009), and are accessible under the accession number SRA024456.1.
2.4. Conserved miRNA identification
Sequencing reads together with quality scores were generated for all 7 libraries using Illumina's GA pipeline 1.5. Trimming of 5′ and 3′ adaptors was performed using an in-house developed Perl script and low quality reads containing adenosine stretches longer than 7 (polyAs) or other low complexity features were discarded. Unique sequence reads were derived for each library and stored in FASTA format, where the total read count for each unique sequence was added to the end of the respective sequence header after a hash symbol. The entire set of miRNA precursor sequences as available in miRBase v14.0 was used to generate an artificial genome by concatenating these sequences leaving stretches of 50 Ns in between into a 1.6 Mb sequence (supplemental data 1). The respective positions of miRNA precursors within the artificial genome were stored in a Genbank database (supplemental data 1). The SARUMAN software (Blom et al., 2011) was used to map all unique reads to the artificial reference genome by allowing up to 3 mismatches or insertions/deletions. In order to be annotated as conserved miRNA, a unique sequence read had to have a minimum abundance of 5 reads. Multiple unique reads mapping the same position of a hairpin sequence (isomiRs) were further represented by the sequence of the most abundant read. For each hairpin the total read counts found at the 5′ or 3′ arms were retrieved, and if both arms were mapped a ratio 5p/3p was calculated. The final denotation given to a conserved hamster sequence read consisted of “cgr” as the species prefix, “miR-xy” as the miRNA identifier and a final suffix of either “-5p”, “-3p” depending on the alignment position of the read to the respective hairpin.
2.5. Novel miRNA predictions
Novel miRNAs were predicted using the following procedure: reads that could not be matched to known small RNAs were mapped to the mouse genome using segemehl (Hoffmann et al., 2009) with two allowed mismatches or insertions/deletions in the seed region and a minimum accuracy of 80%. This led to a mapping of 960,000 unique reads. The matched reads were combined into 317,000 block-clusters using Blockbuster (Langenberger et al., 2009a). By applying published (Langenberger et al., 2009a) and two additional descriptors defining the sharpness of blocks, a support vector machine (SVM) was trained to identify miRNA candidates among these 317,000 clusters. The SVM classified 131,000 potential miRNA clusters, which were filtered according to their length (with a minimum length of 40 and a maximum length of 170), resulting in 14,378 candidates. The mouse genomic sequences of these candidates (plus 15 nt up and downstream) were retrieved from UCSC genome browser, and the sequences were folded in silico using RNAfold (Hofacker and Stadler, 2006). Only perfect hairpins without multi-loops and stretches of unpaired bases longer than 50 were kept, resulting in 1435 candidate novel miRNAs. Of these, 122 that were located in mouse intergenic regions, were subject to manual inspection of (1) overall secondary structure predicted by RNAfold; (2) duplex complementarity using a support vector machine trained to distinguish Dicer cleaved duplexes from other duplexes; and (3) of short read alignment patterns.
2.6. Statistical analysis of miRNA expression data
MicroRNA read counts were normalized to the individual lane size by dividing each read count by the total number of reads in million per lane. Log10 transformation of the resulting normalized values was performed to approximate a Gaussian distribution of expression values. Statistical data analysis was generally performed in R 2.9.1: hierarchical unsupervised clustering of cell lines was calculated using the hclust function and complete linkage distance calculation. For principal component analysis of the miRNA expression matrix consisting of 6 samples (cell lines) and 365 variables (miRNAs) values were centered and single value decomposition was calculated using the prcomp function. For biplot illustration, principal components were retrieved (x <-pca$x) multiplied by 10 and rounded (round(x*10)). Differential expression analysis for the contrasts serum-free (n = 4) versus serum-dependent (n = 2) as well as recombinant (n = 2) versus host (n = 2), was calculated using normalized and log10 transformed read counts and one-way ANOVA statistics as available in Genesis (Sturn et al., 2002). Low abundant miRNAs with read counts below 500 were not included in the analysis, and the null hypotheses of no difference in mean values were tested on a significance level of p = 0.05.
2.7. Quantitative real-time PCR
Quantitative real-time PCR was performed on 200 ng of total RNA extracts that had been poly-adenylated and reverse-transcribed into cDNA using an anchored oligo(dT) primer (Invitrogen, Carlsbad CA). PCRs were run using the Platinum SYBR Green kit system, an universal poly(A) primer and gene specific primers that were designed based on sequence data acquired in this study (Supplementary Table 3). Chinese hamster Glycerinaldehyd-3-phosphat-Dehydrogenase (GAPDH) was used as internal control. qRT PCRs were run on the Corbett Rotorgene rotorcycler (Qiagen, Germany) including 4 technical replicates per sample. Data was analyzed using the delta–delta–Ct method (Livak and Schmittgen, 2001). The resulting log2 fold changes were used for correlation of qPCR and sequencing expression data. The Pearson correlation coefficient was calculated in R 2.9.1 using the cor(x,y) function, where x and y are vectors of log2 fold differences of 10 miRNAs as determined by next generation sequencing and by qRT PCR.
3. Results
3.1. Illumina sequencing of CHO small RNA libraries
Two different CHO cell subtypes, CHO-K1 (K1) and the dihydrofolate reductase negative mutant CHO-DUXB11 (Urlaub and Chasin, 1980) (DXB11) were used for preparation of small RNA libraries (Table 1). From both subtypes, 3 distinct cell lines were chosen, which represent three biotechnologically relevant stages during cell line development: (i) adherent cells with serum containing media (fcs), (ii) serum-free, non-adherent host cells (sf), and (iii) recombinant protein producing cells under serum-free conditions (rec). In addition, RNA was isolated from CHO cells undergoing cold shock, heat shock, or sodium butyrate treatment and from cells in stationary growth phase (Table 1) and pooled. The resulting seven RNA libraries were loaded into separate lanes of the flow cell for cluster generation and subsequent sequencing on the Illumina Genome Analyzer IIx in a 36 nt single-read run. By this means, more than 129 million clusters were sequenced corresponding to an average of about 16 million high quality sequence reads per lane and sample. These reads were further filtered for polyA sequences, as well as reads with 3′ adaptors before position 18 and reads with 5′ adaptor contaminations. This approach generated about 14 million reads (18–36 nt) per library, which were collapsed into sets of about 0.6 to 1 million unique reads per library (Supp. Table 1).
Table 1.
Chinese hamster ovary cell lines and culture conditions.
| # | Library ID | Cell line ID | Description | Culture condition at total RNA harvest | Cell line collection | References |
|---|---|---|---|---|---|---|
| 1 | K1 fcs | CHO-K1 fcs | Host/5% serum/adherent | Exponential phase | ECACC CCL 61 | Tjio and Puck (1958) |
| 2 | K1 sf | CHO-K1 sf | Host/serum-free/suspension | Exponential phase | ECACC CCL 61 | Hernandez-Bort et al. (2010) |
| 3 | K1 rec | CHO-K1 (GS) | Recombinant/serum-free/suspension | Exponential phase | ECACC 85051005 | Jeffs et al. (2006) |
| 4 | DXB11 fcs | DUXB11 fcs | Host/5% serum/adherent | Exponential phase | ATCC CRL-9096 | Urlaub and Chasin (1980) |
| 5 | DXB11 sf | DUXB11 sf | Host/serum-free/suspension | Exponential phase | ATCC CRL-9096 | Lattenmayer et al. (2007) |
| 6 | DXB11 rec | EpoFc 14F2 | Recombinant/serum-free/suspension | Exponential phase | ATCC CRL-9096 | Lattenmayer et al. (2007) |
| 7.1 | Pool | CHO-K1 sf | Host/serum-free/suspension | heat shock (42 °C) | ECACC CCL 61 | See above |
| 7.2 | Pool | DUKXB11 fcs | Host/5% serum/adherent | Cold shock (33 °C) | ATCC CRL-9096 | See above |
| 7.3 | Pool | EpoFc 14F2 | Recombinant/serum-free/suspension | Heat shock (42 °C) | ATCC CRL-9096 | See above |
| 7.4 | Pool | CHO-K1 (GS) | Recombinant/serum-free/suspension | cold shock (33 °C) | ECACC 85051005 | See above |
| 7.5 | Pool | CHO-K1 fcs | Host/5% serum/adherent | Late stationary phase | ECACC CCL 61 | See above |
| 7.6 | Pool | DUKXB11 sf | Host/serum-free/suspension | late stationary phase | ATCC CRL-9096 | See above |
| 7.7 | Pool | EpoFc 14F2 | Recombinant/serum-free/suspension | Late stationary phase | ATCC CRL-9096 | See above |
| 7.8 | Pool | DUKXB11 sf | Host/serum-free/suspension | NaBu (2 mM 48 h) | ATCC CRL-9096 | See above |
| 7.9 | Pool | EpoFc 14F2 | Recombinant/serum-free/suspension | NaBu (2 mM 48 h) | ATCC CRL-9096 | See above |
sf, serum free; fcs, fetal calf serum; rec, recombinant; NaBu, sodium butyrate; GS, glutamine synthase selection system.
3.2. Conserved CHO microRNA discovery and annotation
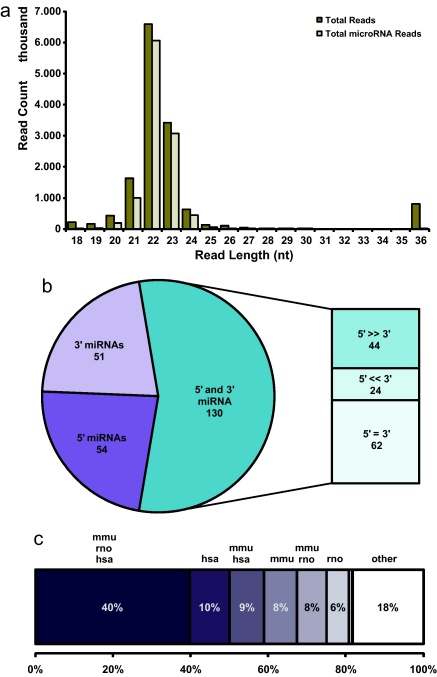
The common strategy for the discovery of mature miRNA sequences within a set of small RNA reads derived from a deep sequencing experiment, is based on read alignment to a reference genome followed by filtering of alignments according to several criteria (Berezikov et al., 2006; Friedlander et al., 2008). Since in the case of the Chinese hamster no genomic sequences are publicly available, an alternative strategy for the discovery and correct annotation of conserved miRNAs was developed (Fig. 1a): first, as a substitute for a hamster genome, an “artificial” reference sequence was generated by concatenating the entire set of miRNA hairpin sequences available in miRBase (Griffiths-Jones et al., 2008) into a 1.6 Mb sequence (termed comprehensive miRNA hairpin reference, CMR) and creating a corresponding GenBank file (available as supplemental data 1). The CMR then served as a reference for the alignment of unique sequencing reads using the SARUMAN software, which was developed as a GPU-supported short-read mapping approach that guarantees to find all possible alignments under a given error tolerance of 3 mismatches or insertions/deletions (Blom et al., 2011). Alignments for all hairpins were visualized using VAMP (developed at the Center for Biotechnology in Bielefeld, Germany), resulting in short read alignment patterns harboring the known characteristics of mature miRNAs: reads corresponding to the mature ∼22 nt long form of miRNAs, align in non-overlapping blocks to either the 5′ or 3′ arm of a hairpin reference or adjacent regions (Fig. 1b and c), for which Langenberger et al. recently introduced the name microRNA-offset RNAs (Langenberger et al., 2009a). Another typical feature of miRNAs is the occurrence of numerous miRNA isoforms, which are characterized by uniform 5′ termini and variations at the 3′ termini. Kuchenbauer et al. have introduced the term “isomiR” for these sequences and reasoned their existence as a consequence of variable enzymatic cleavage sites (Kuchenbauer et al., 2008). The presence of isomiRs, and the average miRNA read length of ∼22 nucleotides together with a characteristic distribution of read frequency over read length (Fig. 2a), suggested a successful enrichment of mature miRNAs in all libraries.
Fig. 1.
Identification and annotation of conserved CHO miRNAs. (a) Small RNA reads were mapped to the entire set of known miRNA hairpin sequences, in the form of a concatenated sequence leaving spacers of 50 bases (N50) between each hairpin sequence (1). In the second step, miRNA isoforms (isomiRs) were grouped and further represented by the most abundant isomiR sequence (2). For annotation of miRNA reads, three scenarios were differentiated: mapping of both arms of the hairpin duplex (A); mapping of only one arm of the hairpin duplex (B) and mapping of regions adjacent to the duplex (C). For the visualization of short read alignments to the miRNA hairpin reference sequence, VAMP, a software developed at the Center for Biotechnology in Bielefeld, Germany was used: orange bars in the upper section represent annotated hairpin sequences while the lower section shows the single-basepair coverage computed from read alignments; green color indicates perfect coverage with no mismatches, yellow color best-match coverage (containing 1–3 mismatches), and red color represents the complete coverage (reads with 1–3 mismatches that were found to align to a different hairpin at lower mismatch rate). (b) The coverage pattern for hsa-miR-18b at single-basepair level is shown in: both hairpin arms are mapped at high perfect coverage, with more reads mapping to the 5′ arm of the hairpin. (c) A locus in the hairpin genome containing 9 miRNA hairpin sequences from Rattus norvegicus is shown at lower zoom: high perfect coverage is generally observed at the 5′ and 3′ duplex positions within a hairpin. In most cases a predominant hairpin-arm exists (high coverage), while in some cases (mir-106b) both hairpins-arms show equal coverage. In a few cases, antisense alignments (mir-96, mir-98) are observed, indicated by coverage facing downwards. (For interpretation of the references to color in text, the reader is referred to the web version of the article.)
Fig. 2.
Hairpin classification and Chinese hamster ovary miRNA conservation. (a) Bar chart showing total read counts over read length for the complete read set (dark) compared to reads that had mapped the comprehensive miRNA genome and can therefore be considered as conserved miRNA reads (bright). (b) Of 235 canoncial miRNA hairpins that were discovered in CHO cells, 105 miRNA had been mapped at either the 5′ (54) or 3′ (51) position, while 130 hairpins had been mapped at both hairpin arms. The ratio of 5′ and 3′ read abundances was calculated for these 130 hairpins, resulting in 44 instances where the 5p/3p ratio exceeded an arbitrary ratio cut-off of 20:1, while in 24 instances it was below 1:20. (c) Out of 224 miRNAs that showed perfect identity to miRBase miRNA sequences, 82% had a human, mouse, or rat ortholog. Among the remaining 18% that did not have a perfect human, or a rodent ortholog, cow, platypus, and chicken were the most frequently found species.
For miRNA annotation, all isomiRs mapping to the same position within a hairpin were grouped and subsequently represented by the most abundant sequence read (Fig. 1a), which conforms to the current understanding that a heterogenous 3′ terminus should not affect miRNA target recognition (Bartel, 2009). Names were then given following the established workflow (Griffiths-Jones et al., 2006) by using the prefix cgr for Cricetulus griseus, the species name of the Chinese hamster, the miRNA name and suffixes of “-5p”, “-3p” according to the exact alignment position relative to the hairpin (Ambros et al., 2003; Griffiths-Jones et al., 2006). In total, 235 canonical miRNA hairpin sequences were mapped by at least 5 small RNA reads with no more than 3 mismatches. Of these 235 hairpins, (i) 130 were mapped at both the 5′ and 3′ duplex position while (ii) 105 hairpins were either mapped at the 5′ or 3′ duplex position (Fig. 2b), thus, adding up to a total of 365 highly conserved mature miRNA sequences (Table 2).
Table 2.
Numbers of conserved Chinese hamster ovary miRNAs.
| Pool | K1 fcs | DXB11 fcs | K1 sf | DXB11 sf | K1 rec | DXB11 rec | Total | |
|---|---|---|---|---|---|---|---|---|
| Total number of conserved miRNA hairpins | 195 | 197 | 194 | 195 | 184 | 208 | 188 | 235 |
| (i) Both hairpin-arms mapped | 119 | 123 | 122 | 119 | 118 | 121 | 119 | 130 |
| (ii) Single hairpin-arm mapped | 76 | 74 | 72 | 76 | 66 | 87 | 69 | 105 |
| Total number of conserved mature miRNAs | 311 | 317 | 312 | 311 | 299 | 327 | 304 | 365 |
| Conserved mature miRNAs with perfect match to miRBase | 178 | 178 | 176 | 171 | 166 | 183 | 170 | 224 |
| Cell line/culture condition specific microRNAs | 2 | 5 | 5 | 0 | 2 | 10 | 1 | 25 |
We refrained from introducing annotations as “mature” and “star” miRNAs for conserved Chinese hamster miRNAs, as this nomenclature would be arbitrary at this stage where only the epithelial ovary cells of this organism have been sequenced. Nevertheless, the ratio of miRNA read counts showed that for 68 out of 130 hairpins with both duplex positions mapped, a strong bias to either the 5′ mature miRNA or 3′ mature miRNA exists by using an arbitrary ratio cut-off of 20:1 (Fig. 2b). Assuming an annotation as miR/miR* for miRNA pairs with high ratios, and of “5p/3p” for pairs with equal abundances, 16 pairs would have been annotated differently than their conserved mouse orthologs in miRBase. This shows that a mere BLAST alignment of sequence reads to mature or star sequences stored in miRBase for the identification of conserved miRNAs is likely to result in imprecise annotations. In addition, the finding that 4 hairpins were mapped at a hairpin-arm (either 5′ or 3′), where no mature miRNA had yet been observed according to miRBase, suggests the presence of 4 so far unknown conserved mature miRNAs in CHO cells (Table 3), and underlines the effectiveness of the presented strategy.
Table 3.
Conserved hairpins give rise to previously unknown mature miRNAs.
| Hairpin ID | miRBase accession | Hairpin length | Pos. of annotated mature miRNA | Alignment pos. of CHO miRNA read | CHO mature miRNA sequence | CHO mature miRNA ID |
|---|---|---|---|---|---|---|
| mmu-mir-1903 | MI0008317 | 80 | 11–32 | 51–68 | CUGGAAGAGGAACAAGUG | cgr-miR-1903-3p |
| mmu-mir-1935 | MI0009924 | 60 | 8–29 | 34–54 | UCGAGGCCAGCCUGGACUACAC | cgr-miR-1935-3p |
| mmu-mir-1944 | MI0009933 | 74 | 40–66 | 5–27 | CACAAAUGAUGAACCUUCUGACG | cgr-miR-1944-5p |
| mmu-mir-702 | MI0004686 | 109 | 88–108 | 10–30 | GUGAGUGGGGUGGUUGGCAUG | cgr-miR-702-5p |
In terms of sequence identity, 224 out of the entire 365 CHO miRNAs aligned perfectly to homologous hairpin sequences in miRBase, with most perfect matches (82%) occurring to human, rat or mouse miRNAs (Fig. 2c). Of the remaining 18% (41 CHO miRNAs) that did not match miRNAs in these three species, the majority mapped to cow, platypus, or chicken miRNAs.
3.3. Identification of non-coding RNAs and prediction of novel CHO microRNAs
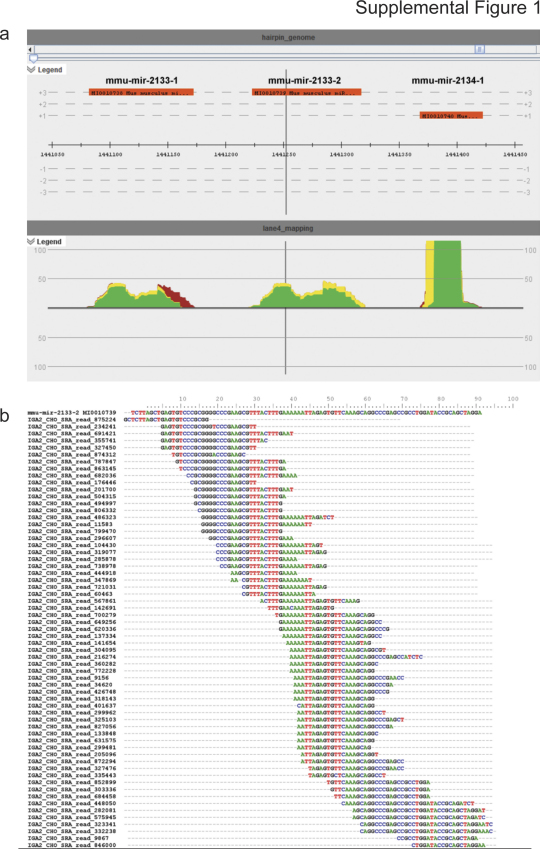
The alignment patterns obtained from mapping short RNA reads to the comprehensive miRNA hairpin reference were further used for the discrimination between several classes of small non-coding RNAs (ncRNAs) (Langenberger et al., 2009b) by filtering for hairpins exhibiting alignment patterns clearly deviating from the typical miRNA alignment pattern (Langenberger et al., 2009a, 2009b). This way, 17 miRNA hairpin sequences were identified in miRBase version 14.0 that, at least for CHO cells, are likely to be of a non-miRNA origin (Supp. Fig. 1) and of which 7 still represent valid entries in miRBase v16.0 (ClustalW alignments of these reads to the respective hairpin sequences are available in supplemental data 2) while 10 have been experimentally verified as ncRNAs and were consequently removed in miRBase version 16 (Table 4).
Table 4.
miRNA hairpins with short read alignment patterns that resemble non-coding RNAs.
| Hairpin ID | miRBase Accession | miRBase Status |
|---|---|---|
| mmu-mir-685 | MI0004649 | removed in miRBase v15 |
| mmu-mir-1935 | MI0009924 | still present* |
| mmu-mir-1957 | MI0009954 | still present* |
| hsa-mir-1973 | MI0009983 | still present* |
| mmu-mir-2133-1 | MI0010738 | removed in miRBase v16 |
| mmu-mir-2133-2 | MI0010739 | removed in miRBase v16 |
| mmu-mir-2134-1 | MI0010740 | removed in miRBase v16 |
| mmu-mir-2134-2 | MI0010741 | removed in miRBase v16 |
| mmu-mir-2134-3 | MI0010742 | removed in miRBase v16 |
| mmu-mir-2134-4 | MI0010743 | removed in miRBase v16 |
| mmu-mir-2134-5 | MI0013182 | removed in miRBase v16 |
| mmu-mir-2134-6 | MI0013183 | removed in miRBase v16 |
| mmu-mir-2135-1 | MI0010744 | removed in miRBase v16 |
| mmu-mir-2135-4 | MI0010745 | removed in miRBase v16 |
| mmu-mir-2135-5 | MI0010746 | removed in miRBase v16 |
| mmu-mir-2135-2 | MI0010747 | removed in miRBase v16 |
| mmu-mir-2135-3 | MI0010748 | removed in miRBase v16 |
| mmu-mir-2140 | MI0010753 | removed in miRBase v16 |
| mmu-mir-2141 | MI0010754 | removed in miRBase v16 |
| mmu-mir-2142 | MI0010755 | removed in miRBase v15 |
| mmu-mir-2143-1 | MI0010756 | removed in miRBase v15 |
| mmu-mir-2143-2 | MI0010757 | removed in miRBase v15 |
| mmu-mir-2143-3 | MI0010758 | removed in miRBase v15 |
| mmu-mir-2144 | MI0010759 | removed in miRBase v15 |
| mmu-mir-2145-1 | MI0010760 | still present* |
| mmu-mir-2145-2 | MI0010761 | still present* |
| mmu-mir-2146 | MI0010762 | removed in miRBase v16 |
| mmu-mir-690 | MI0004658 | still present* |
| mmu-mir-709 | MI0004693 | still present* |
| mmu-mir-712 | MI0004696 | still present* |
In miRBase v16.
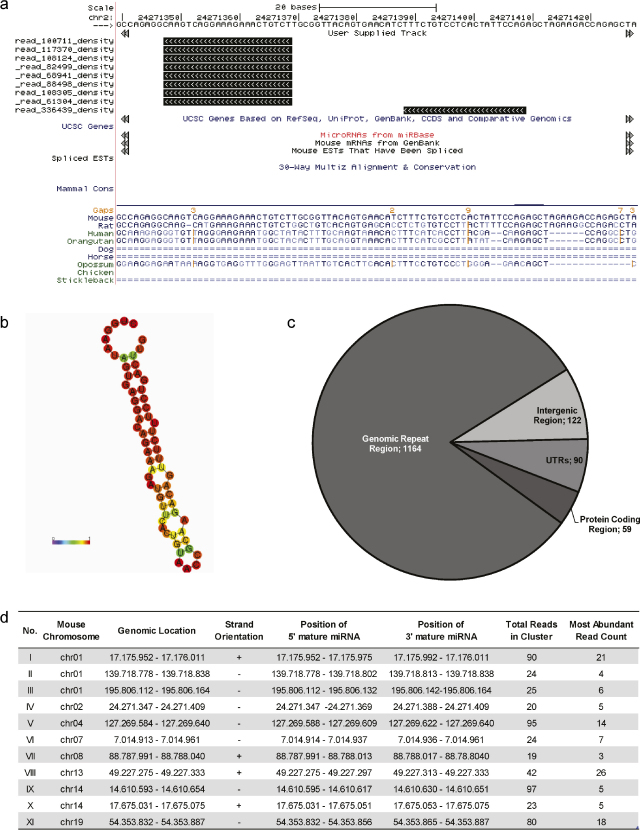
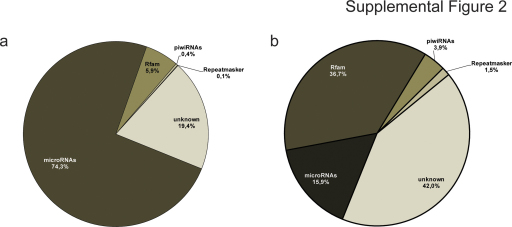
For the prediction of novel miRNAs from reads not mapping to the comprehensive hairpin genome, an initial BLAST alignment to ncRNAs in Rfam (Gardner et al., 2009), RNAdb (Pang et al., 2007) and rodent repetitive elements in Repbase v15 repository (Jurka et al., 2005) was performed (Supp. Fig. 2). In the absence of a hamster genome sequence, all unique reads that failed to map either known miRNAs or non-coding RNAs (referred to as “unknown” reads) were aligned to the mouse genome using segemehl (Hoffmann et al., 2009). In order to unmask putative novel miRNAs within a total of 1 million unique aligned reads, several important characteristics of canonical miRNAs had to be fulfilled (Berezikov et al., 2006). First, read alignments were combined into clusters of adjacent blocks using blockbuster (Langenberger et al., 2009a). These clusters were then filtered for clusters consisting of non-overlapping blocks with a uniform 5′ terminus using a support vector machine (Fig. 3a). Second, mouse genomic sequences of these clusters were retrieved from UCSC genome browser (Rhead et al., 2010) and filtered for lengths between 40 and 170 basepairs. Third, sequences of all 14,000 clusters that fulfilled criteria (1) and (2) were folded in silico using RNAfold (Hofacker and Stadler, 2006), to check whether RNA transcripts from these genomic locations are likely to exhibit hairpin-like structures (Fig. 3b). This was true for 1435 clusters of which 1164 were located in genomic repeat regions, 149 in protein coding regions and 122 clusters in intergenic regions that were chosen for further analysis (Fig. 3c) to check whether the short reads aligning to these regions resembled features characteristic to Dicer cleavage. Therefore a support vector machine was trained on known miR/miR* pairs using published descriptors (van der Burgt et al., 2009) to identify double strand Dicer cleavage products at a 90% recall rate. When subjected to this SVM, putative miR/miR* reads of 11 out of 122 intergenic clusters were found to form duplexes that had all features of known Dicer cleaved duplexes and are consequently proposed as novel miRNAs (Fig. 3d).
Fig. 3.
Prediction of novel miRNAs. Several criteria were defined for the identification of novel miRNA genes and are exemplarily shown for novel miRNA candidate IV: (a) previously reported descriptors were used in blockbuster (Langenberger et al., 2009b; van der Burgt et al., 2009) to identify genomic loci with miRNA-like alignment patterns such as “sharp” blocks with uniform 5′ termini and coverage of both hairpin-arms. (b) RNAfold was used for prediction of RNA secondary structures of these genomic regions. Sequences that did not fold in silico into miRNA hairpin-like structures were filtered and discarded. The remaining sequences between 40 and 170 nucleotides in length were sorted according to their genomic location (c). Short read sequences located in intergenic regions were subjected to a support vector machine that was trained to identify Dicer cleaved duplexes at a 90% recall rate. These were manually screened to identify 11 putative novel miRNAs, which are listed in table-format (d) giving the mouse genomic location of the cluster as well as locations of the most abundant 5′ and 3′ reads.
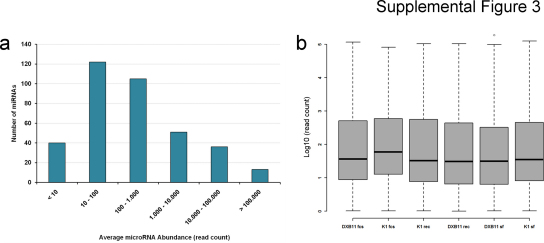
3.4. Quantitative analysis of miRNA transcription in CHO cell lines
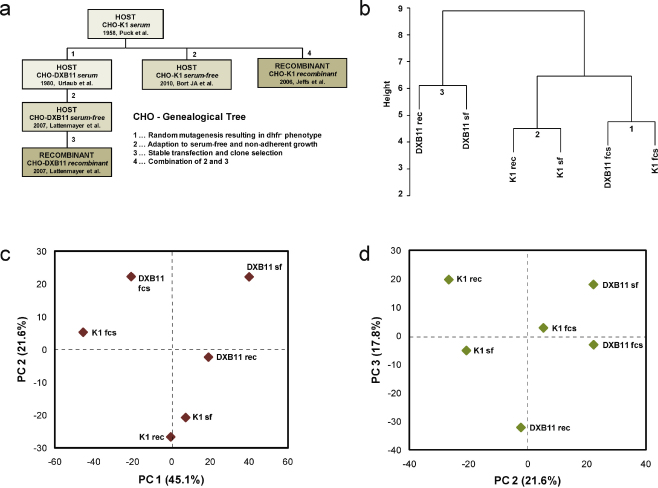
For a quantitative analysis of conserved miRNA expression in CHO cell lines, miRNA read counts that ranged from <10 to >100,000 (Supp. Fig. 3a) were normalized and log10 transformed according to previous reports (Glazov et al., 2008), resulting in a uniform distribution of miRNA read counts throughout all cell lines (Supporting Fig. 3b). In order to visualize similarities in miRNA transcription levels between all 6 sequenced CHO cell lines, which can be linked in a genealogical tree (Fig. 4a), the normalized and log10-transformed read counts were of all miRNAs were used for unsupervised hierarchical clustering analysis. The results clearly show that CHO cells grown in the presence of serum (node 1, Fig. 4b) cluster together, as well as serum-free adapted cell lines of the K1 and DXB11 subtype (nodes 2 and 3, Fig. 4b) indicating pronounced changes in miRNA transcription upon removal of serum from the cultivation media. The very similar transcription patterns in K1 fcs and DXB11 fcs are remarkable, since the dihydrofolate reductase (DHFR) negative DXB11 cells were established from K1 cells by strong mutagenesis, suggesting that the inclusion of fetal calf serum in the cultivation media strongly determines miRNA transcription. To further explore the variance of miRNA transcription in CHO cell lines, we applied principal component analysis (PCA) to the miRNA expression matrix consisting of 6 cell lines and 365 canonical conserved miRNAs. The uncorrelated principal components 1, 2, and 3 were sufficient to explain 84% of the observed variability, and were visualized as 2D-biplots (Fig. 4c and d). The relative positions of CHO cell lines in these 2D-biplots indicate again a considerable distance between serum-dependent and serum-free cell lines, but also significant variation between host and recombinant cell lines.
Fig. 4.
miRNA transcription provides information on the cellular state of CHO cell lines. (a) Cartoon depicting the biological relationship of sequenced CHO cell lines. (b) Unsupervised hierarchical clustering of CHO cell lines according to their miRNA transcription profiles identified 3 nodes, corresponding to serum-dependent K1 and DXB11 cell lines (1), serum-free adapted host and recombinant K1 cell lines (2), and serum-free host and recombinant DUXB11 cell lines (3). Principal component analysis of a miRNA expression matrix consisting of 6 samples (CHO cell lines) and 365 variables (conserved miRNAs) was centered and used for singular value decomposition using R. Principal components were retrieved, and biplot graphs were chosen for their illustration as PC1 versus PC2 (c) and PC2 versus PC3 (d).
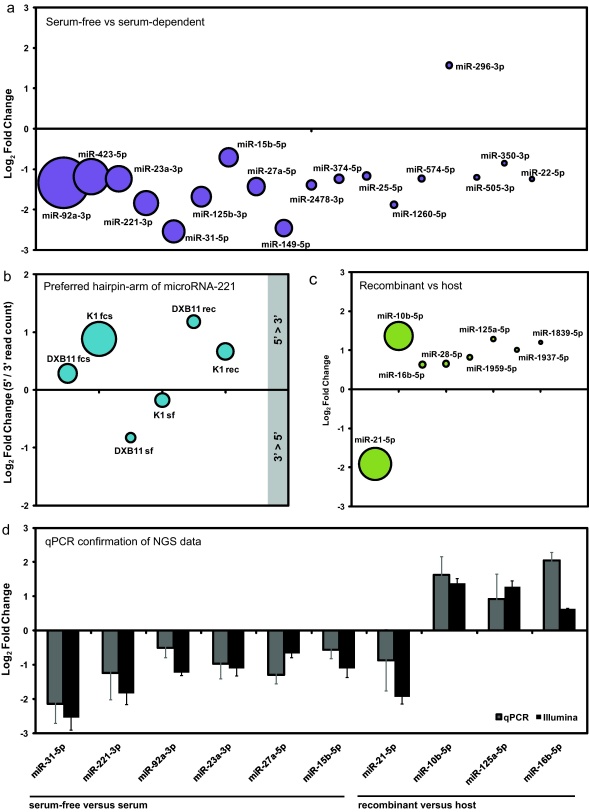
Consequently, we first tested for differentially transcribed miRNAs (one-way ANOVA, p < 0.05) between serum-dependent and serum-free adapted cells, and found that 17 miRNAs were repressed in serum-free adapted cell lines, while only one miRNA was found overexpressed (Fig. 5a). Among the repressed miRNAs, cgr-miR-31-5p exhibited the strongest repression with log2 fold reduction of −2.54 (83% repression), followed by cgr-miR-149-5p and miR-221-3p with a −2.45 (82%) and −1.88 (73%) log2 fold reduction, respectively (Supp. Table 2). In the case of mir-221, the strong repression under serum-free growth was accompanied by a switch in the preferred hairpin-arm from 3′ to 5′, which, however, was restored in the recombinant serum-free cell lines (Fig. 5b). Secondly, miRNA transcription was compared between recombinant and serum-free cell lines using one-way ANOVA statistics, which revealed that cgr-miR-21-5p is strongly repressed in recombinant cell lines (Fig. 5c), while 7 other miRNAs are overexpressed in both recombinant CHO cell lines (Supp. Table 2). Quantitative PCR analysis of 10 significantly regulated miRNAs taken from both contrasts showed good correlation with sequencing data (Pearson = 0.89), and supports that biotechnologically relevant cell variations can be differentiated by transcriptional profiling of a small set of marker miRNAs (Fig. 5d).
Fig. 5.
Analysis of differential miRNA transcription in CHO cell lines. (a) Differential expression analysis for the contrast serum-free versus serum-dependent (one-way ANOVA, p ≤ 0.05) was performed considering only miRNAs with read counts > 500. Log2 fold changes of 18 significantly regulated miRNAs are depicted in a bubble plot, where miRNAs are sorted according to mean expression levels, represented by the bubble size. (b) The significant reduction of miR-221-3p in serum-free adapted cells was accompanied by an overall switch of the ratio of 5′ and 3′ mature miRNA levels originating from mir-221 from positive to negative, wich was restored again in recombinant cell lines. (c) Differential expression analysis of miRNAs between recombinant and serum-free CHO host-cells (one-way ANOVA, p < 0.05, read count > 500) identified 8 significantly regulated miRNAs. (d) Six out of 18 miRNAs that were found regulated between serum-free and serum-dependent growth, and 4 miRNAs that were found regulated in recombinant versus host cells were chosen for qPCR validation. Log2 transformed fold changes for both contrasts are given as bar chart, where black bars represent log2 fold changes as determined by sequencing and grey bars as determined by quantitative PCR.
The degree of conservation of miRNA target sites in CHO messenger RNAs (mRNAs) was evaluated by sequencing the CHO homologs of 26 validated targets of miR-17-92, and aligning the resulting CHO contigs (supplied in supplemental data 3) to the homologous mouse cDNA sequences. For 19 out of 26 mRNA targets, the TargetScan (www.targetscan.org) predicted binding sites of miR-17-92 (Friedman et al., 2009) were identified in our CHO cDNA sequences and found to be highly conserved, with 8mer and 7mer-m8 seed regions being perfectly conserved throughout (Table 5).
Table 5.
miR-17-92 target regions are commonly conserved in Chinese hamster ovary cells.
| No. | Gene symbol | RefSeq accession | miR-17-92 seed family | Seed pos. in mouse 3’ UTR | Seed pairing Type | pCT score | Alignment | Percentage identity |
|---|---|---|---|---|---|---|---|---|
| 1 | APP | NM_007471.2 | miR-17 family | 726–732 | 7mer-m8 | 0.60 | 91.0 | |
| 2 | BCL2L11 (Bim) | NM_207680.2 | miR-17 family | 2107–2113 | 8mer | 0.93 | n/a | |
| 3 | CCND1 | NM_007631.2 | miR-17 family | 925–931 | 7mer-m8 | 0.87 | 100.0 | |
| 4 | CDKN1A (p21) | NM_007669.3 | miR-17 family | 436–442 | 7mer-m8 | 0.85 | n/a | |
| 5 | CTGF | NM_010217.1 | miR-18 family | 1023–1029 | 7mer-m8 | 0.39 | 100.0 | |
| 6 | E2F1 | NM_007891.2 | miR-17 family | 469–475 | 7mer-m8 | 0.59 | 91.7 | |
| E2F1 | NM_007891.2 | miR-17 family | 984–990 | 7mer-m8 | 0.77 | n/a | ||
| 7 | GAB1 | NM_021356.2 | miR-17 family | 263–269 | 7mer-m8 | 0.68 | n/a | |
| 8 | HIF-1α | NM_010431.1 | miR-17 family | 975–981 | 7mer-m8 | 0.36 | 95.0 | |
| HIF-1α | NM_010431.1 | miR-18 family | 304–310 | 7mer-m8 | 0.51 | 100.0 | ||
| 9 | HIPK3 | NM_005734.3 | miR-25 family | 118–124 | 7mer-m8 | 0.73 | 100.0 | |
| HIPK3 | NM_005734.3 | miR-19 family | 165–171 | 8mer | 0.79 | n/a | ||
| 10 | IRF1 | NM_008390.1 | miR-17 family | 584–590 | 7mer-m8 | 0.44 | 100.0 | |
| 11 | ITCH | NM_008395.2 | miR-17 family | 1102–1108 | 7mer-m8 | 0.74 | n/a | |
| 12 | MAPK9 | NM_016961.2 | miR-17 family | 361–367 | 7mer-m8 | < 0.1 | 95.0 | |
| 13 | MAPK14 | NM_011951.2 | miR-19 family | 1819–1825 | 8mer | 0.39 | n/a | |
| 14 | MYLIP | NM_153789.3 | miR-25 family | 1200–1206 | 8mer | 0.96 | 95.0 | |
| MYLIP | NM_153789.3 | miR-19 family | 1314–1320 | 8mer | 0.90 | 100.0 | ||
| 15 | NCOA3 | NM_008679.2 | miR-17 family | 588–594 | 8mer | 0.95 | 95.0 | |
| 16 | PKD1. PKD2 | NM_013630.2 | miR-17 family | 192–198 | 8mer | 0.90 | 82.6 | |
| 17 | PTEN | NM_008960.2 | miR-19 family | 1236–1242 | 8mer | 0.58 | 100.0 | |
| 18 | RB1 | NM_009029.1 | miR-17 family | 844–850 | 7mer-m8 | 0.31 | 95.0 | |
| 19 | RB2/p130 | NM_011250.2 | miR-17 family | 598–604 | 8mer | 0.83 | 100.0 | |
| 20 | RUNX1 | NM_009821.1 | miR-17 family | 1748–1756 | 7mer-m8 | 0.88 | n/a | |
| 21 | SOCS-1 | NM_009896.2 | miR-19 family | 293–299 | 8mer | 0.9 | 100.0 | |
| 22 | STAT3 | NM_213659.2 | miR-17 family | 156–162 | 7mer-m8 | 0.56 | 96.0 | |
| 23 | TGFBR2 | NM_009371.2 | miR-17 family | 298–304 | 8mer | 0.96 | 95.0 | |
| 24 | THBS1 | NM_011580.3 | miR-19 family | 1840–1846 | 7mer-1A | 0.36 | n/a | |
| 25 | TSG101 | NM_021884.3 | miR-17 family | 170–176 | 7mer-m8 | < 0.1 | 100.0 | |
| 26 | VEGFA | NM_001025250.2 | miR-17 family | 109–115 | 7mer-m8 | 0.87 | 100.0 | |
4. Discussion
In order to follow up our hypothesis that miRNAs play a crucial role in the regulation of biological processes in CHO cells (Müller et al., 2008), we have identified 235 conserved as well as 11 novel miRNA genes, provided proof-of-principle that CHO miRNAs are subject to regulation in biotechnologically relevant cellular states and provided experimental evidence that conserved miRNAs are likely to have a conserved function, by sequencing miRNA binding sites in CHO orthologs of 26 validated target mRNAs of miR-17-92.
The presented strategy of conserved miRNA identification can be universally applied to any organism without published genome sequence data. Compared to BLAST alignments to mature and star miRNA sequences (Johnson et al., 2010), the use of hairpin sequences as reference allows for a more precise annotation of conserved miRNAs, since the calculation of a 5p/3p read count ratio prevents from inheriting potentially erroneous denotations as “mature” and “star” from homologous miRNAs in related species. Moreover, short read alignment patterns to the hairpin references contain information on the nature of non-coding RNAs so that the chances of misinterpretations of non-coding RNAs as mature miRNAs can be reduced. This, together with the newly available option of including deep sequencing data in miRBase (Kozomara and Griffiths-Jones, 2010) will improve the identification and annotation of process of miRNAs in species with incomplete genomic sequence information.
The question how many miRNAs remain to be identified in epithelial derived Chinese hamster ovary cells, is difficult to answer. In the light of the well-known tissue-specificity of miRNA expression, however, we expect the number of miRNAs in CHO cells will be below those identified in closely related species such as mouse or rat where a variety of tissues and cell lines have been sequenced. Therefore, taken into account that a recent study reported 312 conserved miRNA genes in mouse (Chiang et al., 2010), the 235 confidently identified conserved miRNA genes are likely to represent the majority of functionally relevant miRNAs in CHO cells. The number of additional CHO specific miRNAs is even harder to estimate as long as the genomic sequence is missing. Nevertheless, by using the mouse genome assembly as reference, our presented strategy of novel miRNA prediction resulted in 11 candidates that resemble all currently expected miRNA characteristics (Ambros et al., 2003; Berezikov et al., 2006), and might represent a fraction of novel rodent specific miRNAs. While the functional relevance of these low abundant, novel and species specific miRNAs remains to be elucidated, we could show that the transcription of conserved miRNAs in CHO cells is differentially regulated in biotechnologically relevant stages of CHO cell line development. Statistical analysis identified 18 miRNAs to be consistently regulated upon adaption to serum-free and non-adherent growth, which included several hamster orthologs of well characterized miRNAs, such as miR-31, miR-221-3p, or miR-92a that have been linked to the regulation of cell proliferation (Creighton et al., 2010), to apoptosis (Dai et al., 2010), tumor development (Ivanov et al., 2010), and to aging (Grillari et al., 2010). The switch in the preferred hairpin-arm of mir-221, a phenomenon so far only observed across different tissues (Chiang et al., 2010), shows that miRNA expression in CHO cells is highly responsive to culture conditions. From a biotechnological perspective this is of interest, since serum-free growth was shown to result in decreased proliferation capacities and apoptosis resistance (Zanghi et al., 1999) and might negatively impact the production and quality of recombinant proteins (Lefloch et al., 2006). Hence, our data indicate that a fast and good adaption to serum-free growth might in part be influenced by miRNA expression, especially since the overexpression of two prominent miRNA targets, BCL-2 and CDKN1A, has been shown to shorten the duration of this process (Astley and Al-Rubeai, 2008). The experimental verification, whether overexpression of miRNAs that are repressed in serum-free adapted cell lines can restore some of the growth characteristics observed for CHO cells grown in the presence of serum is currently ongoing. Of further interest from a biotechnological perspective are miRNA transcription signatures that are specific to recombinant protein producing CHO cell lines, as these clonal cell lines are the result of gene amplification (Lattenmayer et al., 2007) and selection of clones with high specific recombinant protein production. Hence, the differential regulation of cgr-miR-21 in recombinant CHO cells is of high interest, not least, since human miR-21 is known to play an important role the regulation of cell growth and apoptosis (Krichevsky and Gabriely, 2009). The 4-fold (75%) repression of cgr-miR-21 in optimized recombinant cells as identified in this study, together with the upregulation observed in batch cultivations upon temperature shift from 37 °C to 31–33 °C (Gammell et al., 2007), which is accompanied by growth arrest and increased specific productivity, leads us to conclude that miR-21 could be an attractive target for engineering in CHO cells (“engimiR”).
The specific genes and pathways, which are controlled by these miRNAs in CHO cells can currently only be predicted based on their preferential conservation in other mammalian species (Friedman et al., 2009). By sequencing the cDNA of 26 validated mRNA targets of miR-17-92 in CHO cells we were able to identify the conserved target sites in 19 of these cDNAs, which supports that the targets, and therefore also the functions, of miRNAs are conserved in Chinese hamster. However, for 7 validated targets of miR-17-92 the predicted miRNA binding sites could not be detected. This absence can be of technical (incomplete sequencing coverage) or biological nature, since it is known that certain genes, for example in human cancer cell lines, have evaded miRNA control by altering their 3′UTR structures using alternative polyadenylation sites or alternative cleavage (Mayr and Bartel, 2009).
This study has now provided the basis for establishing miRNAs as relevant tools in CHO cell line development by identifying and giving precise annotations to conserved and novel CHO miRNAs, so that conservation based approaches for their target prediction can be used reliably in the absence of genomic sequence information of the Chinese hamster. Nevertheless, the public availability of CHO sequence information is of utmost importance in order to improve these tools and consequently miRNA research in Chinese hamster.
Funding
This work was supported by the GEN-AU project “Non-coding RNAs” [grant number 820982] to JG and IH; the BMBF GenoMik-Transfer program [grant number 0315599B] to JB; and the BOKU DOC grant to MH.
Acknowledgements
JG and RK would like to acknowledge support by the FWF Doctoral Programme “Biotop”; DD the support by the International Program “Bioinformatics of Signaling Networks”; MW is supported by the Austrian Center for Industrial Biotechnology (ACIB); and TJ receives a scholarship from the CLIB Graduate Cluster Industrial Biotechnology.
Footnotes
All relevant sequence data was submitted to the Sequence Read Archive at http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi? and can be accessed under the accession SRA024456.1.
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jbiotec.2011.02.011.
Appendix A. Supplementary data
Contains the comprehensive miRNA hairpin reference as fasta file and the respective annotations as Genbank file.
Multiple alignments of Illumina reads to the respective hairpin reference sequence are shown for all miRNA hairpins that were unmasked as non-coding RNAs and still represent valid entries in miRBase.
Excel sheet containing the entire list of cDNA contigs used for identification of conserved miR-17-92 binding sites in 26 Chinese hamster ovary mRNA transcripts.
Excel sheet containing sequences, read counts and normalized log10 transformed read counts for the entire set of conserved CHO miRNAs for each sequenced CHO cell line.
Supplemental Figure 1.
Discrimination of other non-coding RNAs from miRNAs by read alignment patterns. (a) VAMP visualization of read alignments to three murine miRNA hairpins, which have been recently experimentally validated as non-coding RNAs. In contrast to canonical miRNAs, reads do not align in non-overlapping blocks with uniform 5′ termini, as expected for mature miRNA reads. (b) ClustalW multiple read alignment of sequencing reads to mmu-mir-2133-2.
Supplemental Figure 2.
Comparison of total and unique read annotation as observed for Pool small RNAs. Pie charts were used for illustrating the distribution of read annotation as miRNA, Rfam ncRNA, piwiRNA (piRNA), repetitive rodent RNA, or unknown. Pie charts are given for the total read set (a) and the unique read set (b).
Supplemental Figure 3.
Quantitative analysis of miRNA transcription. The histogram plot in (a) shows that the majority of miRNAs exhibit read counts between 10 and 1.000. (b) Read counts were then normalized to the individual size of each library (in million read counts) and log10-transformed to achieved a Gaussian distribution of miRNA expression across all 6 CHO cell lines, visualized using a boxplot diagram.
References
- Ambros V., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astley K., Al-Rubeai M. The role of Bcl-2 and its combined effect with p21CIP1 in adaptation of CHO cells to suspension and protein-free culture. Appl. Microbiol. Biotechnol. 2008;78:391–399. doi: 10.1007/s00253-007-1320-2. [DOI] [PubMed] [Google Scholar]
- Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron N., Sanchez N., Kelly P., Clynes M. MicroRNAs: tiny targets for engineering CHO cell phenotypes? Biotechnol. Lett. 2010;2010:25. doi: 10.1007/s10529-010-0415-5. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E., Cuppen E., Plasterk R.H. Approaches to microRNA discovery. Nat. Genet. 2006;38(Suppl):S2–S7. doi: 10.1038/ng1794. [DOI] [PubMed] [Google Scholar]
- Blom, J., Jakobi, T., Doppmeier, D., Jaenicke, S., Kalinowski, J., Stoye, J., and Goesmann, A., 2011. Exact and complete short read alignment to microbial genomes using GPU programming. Bioinformatics, in press. [DOI] [PubMed]
- Carleton M., Cleary M.A., Linsley P.S. MicroRNAs and cell cycle regulation. Cell Cycle. 2007;6:2127–2132. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- Carthew R.W., Sontheimer E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang H.R., Schoenfeld L.W., Ruby J.G., Auyeung V.C., Spies N., Baek D., Johnston W.K., Russ C., Luo S., Babiarz J.E. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton C.J., Fountain M.D., Yu Z., Nagaraja A.K., Zhu H., Khan M., Olokpa E., Zariff A., Gunaratne P.H., Matzuk M.M. Molecular profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. 2010;70:1906–1915. doi: 10.1158/0008-5472.CAN-09-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai R., Li J., Liu Y., Yan D., Chen S., Duan C., Liu X., He T., Li H. miR-221/222 suppression protects against endoplasmic reticulum stress-induced apoptosis via p27(Kip1)- and MEK/ERK-mediated cell cycle regulation. Biol. Chem. 2010;391:791–801. doi: 10.1515/BC.2010.072. [DOI] [PubMed] [Google Scholar]
- Doolan P., Meleady P., Barron N., Henry M., Gallagher R., Gammell P., Melville M., Sinacore M., McCarthy K., Leonard M. Microarray and proteomics expression profiling identifies several candidates, including the Valosin-Containing Protein (VCP), involved in regulating high cellular growth rate in production CHO cell lines. Biotechnol. Bioeng. 2010;106:42–56. doi: 10.1002/bit.22670. [DOI] [PubMed] [Google Scholar]
- Friedlander M.R., Chen W., Adamidi C., Maaskola J., Einspanier R., Knespel S., Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammell P., Barron N., Kumar N., Clynes M. Initial identification of low temperature and culture stage induction of miRNA expression in suspension CHO-K1 cells. J. Biotechnol. 2007;130:213–218. doi: 10.1016/j.jbiotec.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Gao P., Tchernyshyov I., Chang T.C., Lee Y.S., Kita K., Ochi T., Zeller K.I., De Marzo A.M., Van Eyk J.E., Mendell J.T., Dang C.V. C-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner P.P., Daub J., Tate J.G., Nawrocki E.P., Kolbe D.L., Lindgreen S., Wilkinson A.C., Finn R.D., Griffiths-Jones S., Eddy S.R., Bateman A. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazov E.A., Cottee P.A., Barris W.C., Moore R.J., Dalrymple B.P., Tizard M.L. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res. 2008;18:957–964. doi: 10.1101/gr.074740.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H.K., Van Dongen S., Enright A.J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillari J., Hackl M., Grillari-Voglauer R. miR-17-92 cluster: ups and downs in cancer and aging. Biogerontology. 2010;11:501–506. doi: 10.1007/s10522-010-9272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Bort CHO-K1 host cells adapted to growth in glutamine free medium by FACS-assisted evolution. Biotechnol. J. 2010;5(October (10)):1090–1097. doi: 10.1002/biot.201000095. [DOI] [PubMed] [Google Scholar]
- Hofacker I.L., Stadler P.F. Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics. 2006;22:1172–1176. doi: 10.1093/bioinformatics/btl023. [DOI] [PubMed] [Google Scholar]
- Hoffmann S., Otto C., Kurtz S., Sharma C.M., Khaitovich P., Vogel J., Stadler P.F., Hackermuller J. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput. Biol. 2009;5:e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G., McLachlan J., Pasquinelli A.E., Balint E., Tuschl T., Zamore P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Ivanov S.V., Goparaju C.M., Lopez P., Zavadil J., Toren-Haritan G., Rosenwald S., Hoshen M., Chajut A., Cohen D., Pass H.I. Pro-tumorigenic effects of miR-31 loss in mesothelioma. J. Biol. Chem. 2010;285:22809–22817. doi: 10.1074/jbc.M110.100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayapal K.P., Wlaschin K.F., Hu W.S., Yap M.G.S. Recombinant protein therapeutics from CHO Cells – 20 years and counting. Chem. Eng. Prog. 2007;103:40–47. [Google Scholar]
- Jeffs S.A., Goriup S. Comparative analysis of HIV-1 recombinant envelope glycoproteins from different culture systems. Appl. Microbiol. Biotechnol. 2006;72(2):279–290. doi: 10.1007/s00253-005-0256-7. [DOI] [PubMed] [Google Scholar]
- Johnson K.C., Jacob N.M., Nissom P.M., Hackl M., Lee L.H., Yap M., Hu W.S. Conserved MicroRNAs in Chinese hamster ovary cell lines. Biotechnol. Bioeng. 2010;2010:9. doi: 10.1002/bit.22940. [DOI] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kantardjieff A., Jacob N.M., Yee J.C., Epstein E., Kok Y.J., Philp R., Betenbaugh M., Hu W.S. Transcriptome and proteome analysis of Chinese hamster ovary cells under low temperature and butyrate treatment. J. Biotechnol. 2010;145:143–159. doi: 10.1016/j.jbiotec.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Kantardjieff A., Nissom P.M., Chuah S.H., Yusufi F., Jacob N.M., Mulukutla B.C., Yap M., Hu W.S. Developing genomic platforms for Chinese hamster ovary cells. Biotechnol. Adv. 2009;27:1028–1035. doi: 10.1016/j.biotechadv.2009.05.023. [DOI] [PubMed] [Google Scholar]
- Kosik K.S. MicroRNAs and cellular phenotype. Cell. 2010;143:21–26. doi: 10.1016/j.cell.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2010;2010:30. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer O., Klausing S., Noll T. Methods in mammalian cell line engineering: from random mutagenesis to sequence-specific approaches. Appl. Microbiol. Biotechnol. 2010;88:425–436. doi: 10.1007/s00253-010-2798-6. [DOI] [PubMed] [Google Scholar]
- Krichevsky A.M., Gabriely G. miR-21: a small multi-faceted RNA. J. Cell Mol. Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchenbauer F., Morin R.D., Argiropoulos B., Petriv O.I., Griffith M., Heuser M., Yung E., Piper J., Delaney A., Prabhu A.L. In-depth characterization of the microRNA transcriptome in a leukemia progression model. Genome Res. 2008;18:1787–1797. doi: 10.1101/gr.077578.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenberger D., Bermudez-Santana C., Hertel J., Hoffmann S., Khaitovich P., Stadler P.F. Evidence for human microRNA-offset RNAs in small RNA sequencing data. Bioinformatics. 2009;25:2298–2301. doi: 10.1093/bioinformatics/btp419. [DOI] [PubMed] [Google Scholar]
- Langenberger D., Bermudez-Santana C.I., Stadler P.F., Hoffmann S., Langenberger D., Bermudez-Santana C., Hertel J., Hoffmann S., Khaitovich P., Stadler P.F. Identification and classification of small RNAs in transcriptome sequence data. Pac. Symp. Biocomput. 2009;2010:80–87. doi: 10.1142/9789814295291_0010. [DOI] [PubMed] [Google Scholar]
- Lattenmayer C., Loeschel M., Schriebl K., Steinfellner W., Sterovsky T., Trummer E., Vorauer-Uhl K., Muller D., Katinger H., Kunert R. Protein-free transfection of CHO host cells with an IgG-fusion protein: selection and characterization of stable high producers and comparison to conventionally transfected clones. Biotechnol. Bioeng. 2007;96:1118–1126. doi: 10.1002/bit.21183. [DOI] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Kim V.N. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lefloch F., Tessier B., Chenuet S., Guillaume J.M., Cans P., Goergen J.L., Marc A. Related effects of cell adaptation to serum-free conditions on murine EPO production and glycosylation by CHO cells. Cytotechnology. 2006;52:39–53. doi: 10.1007/s10616-006-9039-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mayr C., Bartel D.P. Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova O., Marra M.A. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Müller D., Katinger H., Grillari J. MicroRNAs as targets for engineering of CHO cell factories. Trends Biotechnol. 2008;26:359–365. doi: 10.1016/j.tibtech.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Pang K.C., Stephen S., Dinger M.E., Engstrom P.G., Lenhard B., Mattick J.S. RNAdb 2.0—an expanded database of mammalian non-coding RNAs. Nucleic Acids Res. 2007;35:D178–182. doi: 10.1093/nar/gkl926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B., Karolchik D., Kuhn R.M., Hinrichs A.S., Zweig A.S., Fujita P.A., Diekhans M., Smith K.E., Rosenbloom K.R., Raney B.J. The UCSC genome browser database: update 2010. Nucleic Acids Res. 2010;38:D613–619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rössler B., Lübben H., Kretzmer G. Temperature: a simple parameter for process optimization in fed-batch cultures of recombinant Chinese hamster ovary cells. Enzyme Microbial. Technol. 1996;18:423–427. [Google Scholar]
- Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Shumway M., Cochrane G., Sugawara H. Archiving next generation sequencing data. Nucleic Acids Res. 2009;38:D870–871. doi: 10.1093/nar/gkp1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturn A., Quackenbush J., Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- Subramanian S., Steer C.J. MicroRNAs as gatekeepers of apoptosis. J. Cell Physiol. 2010;223:289–298. doi: 10.1002/jcp.22066. [DOI] [PubMed] [Google Scholar]
- Sunley K., Tharmalingam T., Butler M. CHO cells adapted to hypothermic growth produce high yields of recombinant gamma-interferon. Biotechnol. Prog. 2008;24:898–906. doi: 10.1002/btpr.9. [DOI] [PubMed] [Google Scholar]
- Tjio J.H., Puck T.T. Genetics of somatic mammalian cells. II. chromosomal constitution of cells in tissue culture. J. Exp. Med. 1958;108:259–268. doi: 10.1084/jem.108.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trummer E., Fauland K., Seidinger S., Schriebl K., Lattenmayer C., Kunert R., Vorauer-Uhl K., Weik R., Borth N., Katinger H., Müller D. Process parameter shifting: Part II. Biphasic cultivation - A tool for enhancing the volumetric productivity of batch processes using Epo-Fc expressing CHO cells. Biotechnol. Bioeng. 2006;94:1045–1052. doi: 10.1002/bit.20958. [DOI] [PubMed] [Google Scholar]
- Urlaub G., Chasin L.A. Isolation of Chinese hamster cell mutants deficient in dihydrofolate reductase activity. Proc. Natl. Acad. Sci. U. S. A. 1980;77:4216–4220. doi: 10.1073/pnas.77.7.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burgt A., Fiers M.W., Nap J.P., van Ham R.C. In silico miRNA prediction in metazoan genomes: balancing between sensitivity and specificity. BMC Genomics. 2009;10:204. doi: 10.1186/1471-2164-10-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm F.M. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat. Biotechnol. 2004;22:1393–1398. doi: 10.1038/nbt1026. [DOI] [PubMed] [Google Scholar]
- Zanghi J.A., Fussenegger M., Bailey J.E. Serum protects protein-free competent Chinese hamster ovary cells against apoptosis induced by nutrient deprivation in batch culture. Biotechnol. Bioeng. 1999;64:108–119. doi: 10.1002/(sici)1097-0290(19990705)64:1<108::aid-bit12>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Contains the comprehensive miRNA hairpin reference as fasta file and the respective annotations as Genbank file.
Multiple alignments of Illumina reads to the respective hairpin reference sequence are shown for all miRNA hairpins that were unmasked as non-coding RNAs and still represent valid entries in miRBase.
Excel sheet containing the entire list of cDNA contigs used for identification of conserved miR-17-92 binding sites in 26 Chinese hamster ovary mRNA transcripts.
Excel sheet containing sequences, read counts and normalized log10 transformed read counts for the entire set of conserved CHO miRNAs for each sequenced CHO cell line.