Summary
Ribosomal peptide natural products are ubiquitous, yet relatively few tools exist to predict structures and clone new pathways. Cyanobactin ribosomal peptides are found in ~30% of all cyanobacteria, but the connection between gene sequence and structure was not defined, limiting the rapid identification of new compounds and pathways. Here, we report discovery of four orphan cyanobactin gene clusters by genome mining and an additional pathway by targeted cloning, which represented a tyrosine O-prenylating biosynthetic pathway. Genome mining enabled identification of five new cyanobactins, including peptide natural products from Spirulina supplements. A phylogenetic model defined four cyanobactin genotypes, which explain the synthesis of multiple cyanobactin structural classes and help direct pathway cloning and structure prediction efforts. These strategies were applied to DNA isolated from a mixed cyanobacterial bloom containing cyanobactins.
INTRODUCTION
Cyanobactin ribosomal peptides are widespread in cyanobacteria and may be produced by up to 30% of cyanobacterial strains (Figure 1) (Donia et al., 2008; Donia and Schmidt, 2010; Leikoski et al., 2009, 2010; Schmidt et al., 2005; Sivonen et al., 2010). The compounds have been isolated from cyanobacteria, marine sponges, mollusks, and ascidians, and many are potently active in bioassays. In ascidians, it has been shown that cyanobactins are synthesized by cyanobacterial symbionts, and based upon structural analysis it has been proposed that the remaining marine invertebrate cyanobactins also originate in symbiotic cyanobacteria (Donia et al., 2008; Donia and Schmidt, 2010). More than 100 cyanobactin natural products are known, exemplifying diverse structural types with diverse biological activities, and many more are apparent from genetic methods (Figure S5).
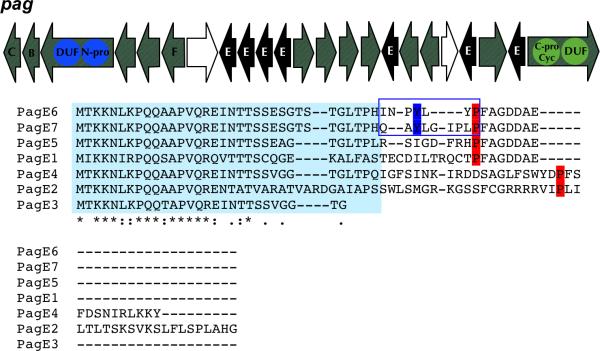
Figure 1. Prenylagaramide Gene Cluster from Planktothrix agardhii.
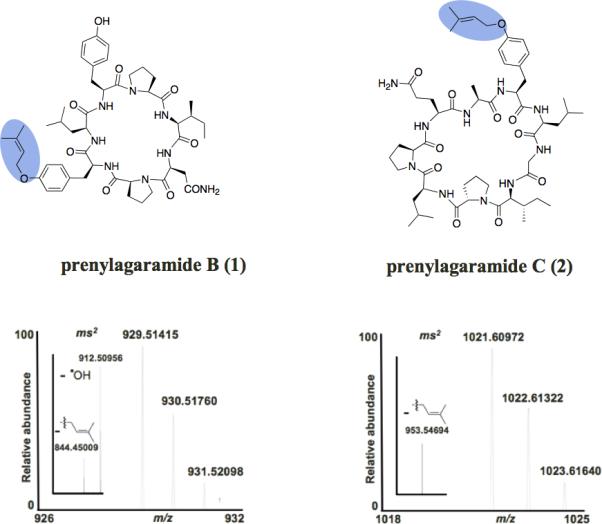
A. Genetic organization of the pag cluster and sequences of all pag precursor peptides. Black: precursor peptides. Blue: N-terminal protease. Light green: C-terminal protease / macrocyclase. Amino acid sequences of prenylagaramide B and C are boxed, prenylated Tyr residues are highlighted in blue while the actual or predicted C-terminal cleavage site Pro is highlighted in red. Asterisks indicate 100% conservation among variants. Light blue-highlighted sequences indicate the more conserved N- and C-termini of the peptides versus the hypervariable inner region. B. Prenylated tyrosine residues are highlighted in blue. Structures and HRFTMS and IRMPD-MS/MS analysis of prenylagaramide B (Murakami et al., 1999) and C showing the molecular ion and the loss of one isoprenyl group. For 2, the indicated configuration is predicted based upon biogenetic considerations and on the structures of other compounds in the series. See also Table S1 and Figure S1.
The first cyanobactin gene cluster to be analyzed was pat, leading to the patellamide group of cyanobactins (Schmidt et al., 2005). Like pat, all cyanobactin biosynthetic gene clusters share in common the presence of two proteases, which cleave precursor peptides and macrocyclize the resulting fragments from their N-to-C termini (Figures S2 and S3) (Lee et al., 2009; Leikoski et al., 2010; McIntosh et al., 2010b; Schmidt et al., 2005). These cycles are commonly 6-8 amino acids in length, but cyanobactins have been described comprising up to 20 amino acids. Many cyanobactins are simple peptide macrocycles, but most are further decorated with a plethora of other posttranslational modifications, including: regioselective Cys, Ser, and Thr heterocyclization to thiazol(in)e and oxazol(in)e; Tyr, Ser, and Thr O-prenylation; His N-methylation; and Trp C-prenylation; among others. Simple, ribosomally translated precursor peptides act as substrates for these posttranslational modifications. In addition to the posttranslational chemical diversity, cyanobactin precursor peptides are hypervariable, meaning that identical enzymes can often synthesize numerous products depending upon discrete changes in small cassettes (Donia et al., 2006). This mechanism enables bacteria to synthesize many nonribosomal peptide-like compounds with small (<20 kbp) gene clusters.
The minimal set of genes present in cyanobactin gene clusters includes the precursor peptide gene, two proteases, and two short conserved hypothetical proteins, which are named here for their similarity to the canonical patellamide (pat) gene cluster (Figure S5) (Donia and Schmidt, 2010). The first protease (PatA homolog) cleaves the N-terminus of the product encoding cassette, while the second (PatG homolog) cleaves its C-terminus in tandem with macrocyclization (Lee et al., 2009). The two conserved hypothetical proteins, PatB and PatC homologs, probably do not function as enzymes, while PatE homologs are precusor peptides. Three further protein groups are commonly encoded in cyanobactin clusters but are not universal. The first, homologs of PatD, act as heterocyclases that regioselectively install thiazoline and oxazoline residues using Cys, Ser, and Thr (McIntosh et al., 2010a; McIntosh and Schmidt, 2010). The second, homologs of PatF, are uncharacterized proteins with no close relatives of known function. Based upon genetic and biochemical studies that clearly demonstrate the function of all other proteins in these pathways, we proposed that this group is responsible for catalysis of prenylation (Donia et al., 2008; Lee et al., 2009; McIntosh et al., 2010a, 2010b.). Finally, an oxidase domain is sometimes present either in a single protein with PatG or as a freestanding protein. This domain is almost certainly responsible for oxazoline or thiazoline oxidation (McIntosh et al., 2010a; Donia et al., 2008; Sudek et al., 2006).
Cyanobactins have chemical features that are related to those in many types of natural products, including other bacterial and eukaryotic ribosomal peptides (McIntosh et al., 2009; Oman and van der Donk, 2010; Hallen et al., 2007; Ireland et al., 2010). However, cyanobactin biosynthetic sequences are very deeply branched within the ribosomal peptide family. Predictive models for cyanobactin biosynthesis have been described, (Donia et al., 2008; Leikoski et al., 2009, 2010; Sudek et al., 2006; Ziemert et al., 2008) but significant gaps remained: biosynthetic genes had not yet been described for several different cyanobactin structure types and several important posttranslational modifications. These gaps have made it difficult to predict chemical structures from gene and genome sequences, and directed cloning of divergent cyanobactin pathways, where the genes may be <90% identical to known cyanobactin pathways, was still challenging.
One of the most important of these biochemical gaps involved Tyr O-prenylation by dimethylallyl pyrophosphate (DMAPP), a rare modification that has so far only been found in severa; cyanobactins (Donia et al., 2008; Donia and Schmidt, 2010), but not gene clusters had been reported. In a previous manuscript, we predicted that a pathway identified in Lyngbya aestuarii might be Tyr O-prenylating, but this could not be confirmed through chemical analysis (Donia et al., 2008). Murakami et al. previously reported the isolation and structure elucidation of two Tyr O-prenylated cyclic peptides, prenylagaramides A and B (1) (Figure 1) (Murakami et al., 1999). Because the producing strains of Plantothrix aghardii were deposited in a culture collection, we selected P. aghardii as a model for the genetic study of Tyr O-prenylation. Prenylagaramides bear some structural resemblance to anacylamide, a cyanobactin for which a gene cluster was previously identified (Leikoski et al., 2010). We thus expected that the prenylagaramide cluster would resemble the known anacyclamide cluster, acy. A major difference is that anacyclamide is a bare N-C cycle, lacking Tyr O-prenylation or other posttranslational modifications. The acy gene cluster study led to the identification of 18 additional AcyE-like precursor peptides from different strains of cyanobacteria, four of which were shown to be prenylated using LC-MS. The resulting manuscript elegantly revealed the widespread nature of acy-like products and their hypervariability (Leikoski et al., 2010). However, position of modification, the regioselectivity of DMAPP addition (either forward or reverse prenylation), and the possibility of Tyr prenylation were not addressed. More importantly, the authors did not sequence any of the enzyme-coding genes from these prenylating pathways, leaving the Tyr O-prenylation problem unsolved.
It was expected that a thorough exploration of cyanobactin pathway types would enable both prediction of cyanobactin structures from genome sequencing and cloning of novel cyanobactin genes, especially from highly heterogeneous environmental metagenomes. Similar detailed comparisons of many related gene clusters and subsequent phylogenetic analysis has been greatly useful in the study of nonribosomal peptides and polyketides (Ansari et al., 2008; Challis et al., 2000; Chiu et al., 2001; Liu et al., 2003; Nguyen et al., 2008), but it has only rarely been applied to ribosomal peptides (Sudek et al., 2006)(Haft et al., 2010; Lee et al., 2008; Li et al., 2010). So far relatively blunt phylogenetic tools have been applied to ribosomal peptides in comparison to those available for other pathway types. For example, lantibiotics have been classified into families on the basis of known biosynthetic genes and chemicals (Chatterjee et al., 2005), but phylogenetic information has not been precise enough to apply when other information is limited, such as when a compound originates in a heterogeneous environmental sample. Here, we report discovery of new cyanobactin pathways, allowing us to create a phylogenetic model linking cyanobactin chemistry and genetics. We further demonstrate the application of this approach in cloning partial cyanobactin pathways from a metagenomic sample and by identifying four new compounds from a genome-sequenced cyanobacterium that is used widely as a food supplement.
RESULTS AND DISCUSSION
The prenylagaramide gene cluster
P. aghardii NIES-596 was cutivated, and a fragment encoding one protease with 70% identity to AcyG was obtained by PCR from its genomic DNA. In order to sequence the intact pathway, an 8000-member fosmid library was constructed and screened with exact-match primers (Figure S2). Two fosmids were used for sequencing, and the resulting pag cluster was highly similar to acy (Table 1). As expected and like acy, pag encoded homologs of the PatA and PatG proteases, responsible for N-C circularization, while it lacked a heterocyclizing PatD homolog. A PatF homolog protein was present, which previous studies of the tru pathway associate with prenylation (Donia et al., 2008). Expected homologs of the conserved hypothetical proteins PatB and PatC were also identified. In addition, multiple open reading frames encoding for hypothetical proteins were identified in the pag genomic region (Table 1).
Table 1. Analysis of coding sequences in pag.
Proteins highlighted in grey are homolgs of those known to be associated with cyanobactin biosynthesis in other systems.
| Predicted Protein | Best Cyanobactin Hit | % Identity | Predicted Function | Organism |
|---|---|---|---|---|
| PagC | AcyC | 49% | Unknown | Anabaena sp. 90 |
| PagB | AcyB | 78% | Unknown | Anabaena sp. 90 |
| PagA | AcyA and OscA | 73% and 73% | N-terminal protease | Anabaena sp. 90 and Oscillatoria sp. PCC 6506 |
| Orf1 | None | 70% | Conserved Hypothetical Protein | Oscillatoria sp. PCC 6506 |
| Orf2 | None | 89% | Conserved Hypothetical Protein | Microcystis aeruginosa NIES-843 |
| PagF | AcyF | 75% | Prenyltransferase | Anabaena sp. 90 |
| Orf3 | Acy-Orf5 | 81% | Transposase | Anabaena sp. 90 |
| PagE7 | AcyE | 56% | Precursor peptide (Prenylagaramide C) | Anabaena sp. SYKE 844B |
| PagE6 | AcyE | 57% | Precursor peptide (Prenylagaramide B) | Anabaena sp. PH262 |
| PagE5 | AcyE | 52% | Precursor peptide | Anabaena sp. PH262 |
| PagE4 | AcyE | 38% | Precursor peptide | Anabaena sp. SYKE 816 |
| Orf4 | None | 36% | Conserved Hypothetical Protein | Oscillatoria sp. PCC 6506 |
| Orf5 | None | 54% | Conserved Hypothetical Protein | Microcystis aeruginosa NIES-843 |
| Orf6 | None | 72% | Conserved Hypothetical Protein | Microcystis aeruginosa NIES-843 |
| Orf7 | None | 74% | Conserved Hypothetical Protein | Microcystis aeruginosa NIES-843 |
| PagE3 | AcyE | 71% | Precursor peptide | Anabaena sp. PH262 |
| Orf8 | None | 71% | Conserved Hypothetical Protein | Nostoc punctiforme PCC 73102 |
| Orf9 | None | 68% | Conserved Hypothetical Protein | Microcystis aeruginosa PCC 7806 |
| Orf10 | Acy-Orf5 | 77% | Transposon | Anabaena sp. 90 |
| PagE2 | AcyE | 66%-N terminus | Precursor peptide | Anabaena sp. 202A1/35 |
| Orf11 | None | 91% | Conserved Hypothetical Protein | Nostoc sp. PCC 7120 |
| PagE1 | AcyE | 47% | Precursor peptide | Anabaena sp. PH262 |
| PagG | AcyG | 70% | C-terminal protease | Anabaena sp. 90 |
The pag cluster was positively identified by the presence of a precursor peptide gene, pagE6, which directly encoded the exact peptide sequence found in prenylagaramide B (1). However, unlike any previously reported cyanobactin gene cluster, aside from the ones described in this paper, multiple (seven) precursor peptides were found in this single gene cluster. Most precursor peptides were bordered by repeating nucleotide sequences, indicating a possible mechanism for gene duplication and divergence, and additionally two copies of a transposase with ~80% identity to a transposase from the acy cluster were found around the precursor region. The precursor peptides were closely related, but identifying the N-terminal proteolysis sites was quite challenging, while the C-terminal cleavage site was readily identified. Fortunately, one of these peptides encoded prenylagaramide B (1) (INPYLYP), allowing both proteolysis sites to be defined. The C-terminal recognition sequence (P/FAGDDAE, where the slash indicates the site of proteolysis) was found in four out of the seven pag precursor peptides and was identical to the one in AcyE while the N-terminal cleavage site was quite different than that for any other known cyanobactin.
To determine which prenylagaramide variants were produced by pag, the strain was cultivated continuously over a 1-year period. This long period culture was performed to allow access to enough cell pellets for chemical extraction from small-scale cultures, since P. agardhii did not grow in large scale in our hands. The cell pellet extract was applied to a HPLC-electrospray-Fourier transform mass spectrometer, leading to identification of the known compound, prenylagaramide B (1), as the major component of the extract (Figure 1, Figure S1, Table S1). PagE6 is identical to PagE7 except that the region encoding for 1 (INPYLYP) is mutated to (QAYLGIPLP), suggesting that a similar cyanobactin could be synthesized from this peptide. Indeed LC-FT-ICR-MS led to the identification of a compound with the expected mass and a similar prenylation pattern on tyrosine. We then applied our prenylation-specific FT-IRMPD-MS/MS method to show that this new cyanobactin (termed prenylagaramide C 2) is indeed prenylated. In this method, gentle application of IRMPD specifically eliminates isoprene, providing a high-resolution molecular ion and a high-resolution product ion resulting from elimination of the prenyl group. In addition to these data, high-resolution fragmentation provided the amino acid sequences of the resulting prenylagaramides and confirmed their main-chain circularization. Both prenylagaramides B and C behaved in similar fashions in this analysis, allowing the structures to be unambiguously assigned. No peaks for any other possible predicted pag precursor product could be identified despite an exhaustive search. The configuration of 2 was tentatively assigned on the basis of the biosynthetic pathway, which encodes all-L ribosomal peptides.
Genome mining for new cyanobactin clusters
We wished to construct a phylogenetic model that would allow accurate prediction of structures in the known cyanobactin groups. In order to do so, more representative pathways of each type were required. Cyanobactin gene clusters are present in ~30% of cyanobacteria (Leikoski et al., 2009), and cyanobacterial genome mining has already led to the discovery of several new compounds (Portmann et al., 2008a; Portmann et al., 2008b; Sudek et al., 2006; Ziemert et al., 2008). Indeed, in the course of this study using BLAST searching we found new cyanobactin clusters in genome sequences of Cyanothece sp. PCC 7425, Microcystis aeruginosa NIES-843 (Kaneko et al., 2007), Arthrospira platensis NIES-39 (Fujisawa et al. 2010), and Oscillatoria sp. PCC 6506 (Mejean et al., 2010) (Figure 2). Genome regions containing the probable clusters were downloaded and analyzed manually using Vector NTI and Artemis. Only gene clusters containing an identifiable precursor peptide were considered “intact.” Many other clusters could be identified by BLAST searching in GenBank, but they appeared to be fragmented or deactivated and were disregarded. Four intact putative cyanobactin clusters are described here that substantially expand the known genotypes.
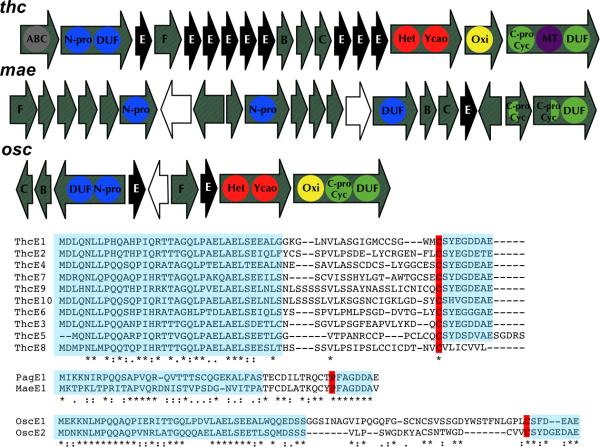
Figure 2. New Pathways by Genome Mining.
A. New gene clusters. Black: precursor peptides. Blue: N-terminal protease. Red: Heterocyclase. Light green: C-terminal protease / macrocyclase. Yellow: oxidase. White: transposon. The size of the pathways is not shown to the exact scale. B. Precursor peptide sequences from the new gene clusters. Highlighted sequences indicate the more conserved N- and C-termini of the peptides versus the hypervariable inner region. Residues highlighted in red indicate the predicted C-terminal cleavage site. Asterisks indicate 100% conservation among variants. See also Figure S3.
The Cyanothece sp. PCC 7425 genome encodes thc, which spans about 16.5 kbp and is present as one operon between the coordinates 306123 and 322611 of the circular chromosome. thc is unique in its organization and represents a new cyanobactin genotype, with new features for cyanobactins including an ABC transporter and a methyltransferase. Amazingly, thc contains a linear array of nine copies of the precursor peptide gene, each encoding for a different product. An alignment of precursor peptides clearly defines the hypervariable product regions and separates them from the more conserved leader peptide region. Although the C-terminal cleavage site can be unambiguously identified (as all products end with Cys), the N-terminal site could not be definitively assigned. Cyanothece sp. 7425 harbors three plasmids. In plasmid pP742501 (~197 kbp), a tenth precursor peptide gene was identified.
Another cluster, mae, was identified in Microcystis aeruginosa NIES-843 and spans a region of 17 kbp between the coordinates 35212 and 52235 of the circular chromosome. This cluster in particular was harder to identify as it is interrupted by multiple transposases and hypothetical proteins. There is only one precursor peptide in mae, which is similar in the primary sequence to the pag precursor peptide pagE1. Other strains of M. aeruginosa contain kawaguchipeptins and microphycin AL82, which are similar to the predicted products of mae and for which no biosynthetic genes have been identified (Gesner-Apter and Carmeli, 2008; Ishida et al., 1996; Ishida et al., 1997).
A cyanobactin gene cluster (osc) was identified in the draft genome of Oscillatoria sp. PCC 6506 (same as Oscillatoria sp. PCC 9029). osc spans ~11.5 kbp on a single contig (contig 300, coordinates 26620-37905). osc represents the first hybrid of two cyanobactin genotypes. osc contains all the modifying enzymes necessary to form oxidized heterocyle-containing cyanobactins, but in an arrangement that is closely related to pathways that do not make heterocycles. osc also encodes two precursor peptides.
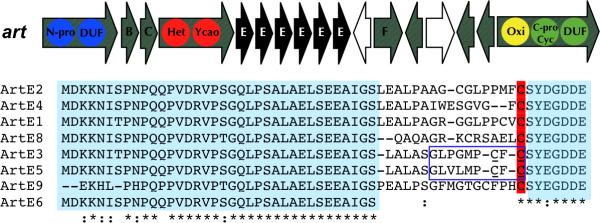
The art gene cluster was found in the genome of Arthrospira platensis NIES-39 spanning ~19 kbp on the circular chromosome between the coordinates 5,664,409 and 5,683,212. art exhibits typical features for cyanobactin pathways except that it also contains multiple precursor peptides (a total of six) encoding different products. Moreover, a transposase was identified between artF and artG (Figure 3, Figure S4, Table S2). All art genes except for the precursor peptides are also present in the draft genome of a second Arthrospira strain: A. platensis str. Paraca. Unlike NIES-39, for which the genome was largely complete, the Paraca genome was highly fragmented on small contigs, explaining the absence of the precursor peptide. More recently, a third edible Arthrospira strain was sequenced (Janssen et al., 2010) and its genome also contains a closely related cluster spanning at least three contigs of the fragmented assembly. Interestingly, this third genome harbors three new precursor peptides encoding for different products (Figure 3). A comparison between the art genes in the three strains is shown in Table 2.
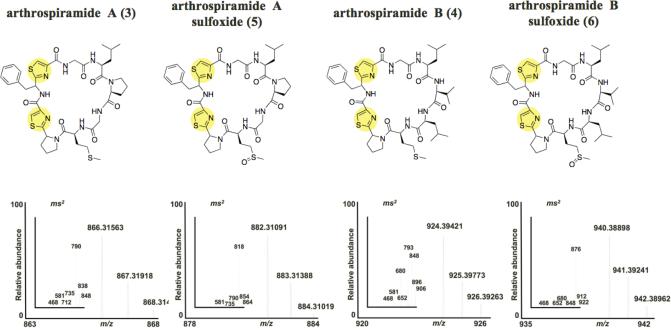
Figure 3. New cyanobactins from Spirulina supplement powders.
A. Genetic organization of the art cluster and sequences of all art precursor peptides. Color codes are the same as in Fig. 1 and Fig. 2. The exact sequences of Arthrospiramides A and B are boxed and the heterocyclized cysteine residues are underlined. B. Chemical structures of four new cyanobactins isolated from Arthrospira spirulina. Below each structure is shown the corresponding FT-MS showing the molecular ion and representative fragment ions verifying the cyclic peptide sequence. The configurational assignments are based upon biogenetic considerations. Assignments are ambiguous adjacent to thiazole residues because these stereocenters are generally chemically labile. See also Table S2 and Figure S4.
Table 2. Analysis of the open reading frames in art.
Proteins highlighted in grey are known to be associated with cyanobactin biosynthesis. Str.2 = Arthrospiraplatensis str. Paraca, Str.3 = Arthrospira sp. PCC 8005, P = partial hit due to incomplete sequencing.
| Predicted Protein | Best Cyanobactin Hit | % Identity | Predicted Function | Organism | Str. 2 | Str. 3 |
|---|---|---|---|---|---|---|
| ArtA | MicA | 73% | N-terminal protease | Microcystis aeruginosa PCC 7806 | 98% | 100% P. |
| ArtB | TruB | 63% | Unknown | Prochloron didemni | 99% | 100% |
| ArtC | TenC | 58% | Unknown | Nostoc spongiaeforme var. tenue | 98% | 100% |
| ArtD | TenD | 73% | Heterocyclase | Nostoc spongiaeforme var. tenue | 96% | 100% |
| ArtE1 | TenE | 53% | Precursor peptide | Nostoc spongiaeforme var. tenue | NO | NO |
| ArtE2 | TenE | 52% | Precursor peptide | Nostoc spongiaeforme var. tenue | NO | NO |
| ArtE3 | TenE | 54% | Precursor peptide (Arthrospiramide A) | Nostoc spongiaeforme var. tenue | NO | NO |
| ArtE4 | TenE | 69% P | Precursor peptide | Nostoc spongiaeforme var. tenue | NO | NO |
| ArtE5 | TruE1 | 55% | Precursor peptide (Arthrospiramide B) | Prochloron didemni | NO | NO |
| ArtE6 | PatE | 55% | Precursor peptide | Prochloron didemni | NO | NO |
| Orf1 | Acy-Orf5 | 71% | Transposase | Anabaena sp. 90 | 80% P | 100% P |
| ArtF | MaeF | 58% | Unknown | Microcystis aeruginosa NIES-843 | 95% | 100% |
| Orf2 | None | 85% | Conserved Hypothetical Protein | Cyanothece sp. CCY0110 | NO | NO |
| Orf3 | None | 83% | Transposase | Microcystis aeruginosa NIES-843 | NO | 100% P |
| Orf4 | None | 87% | Conserved Hypothetical Protein | Arthrospira platensis NIES-39 | NO | NO |
| Orf5 | Mic-Orf | 87% | Conserved Hypothetical Protein | Microcystis aeruginosa PCC 7806 | 90% | 100% |
| ArtG | PatG | 63% | C-terminus protease / Macrocyclase | Prochloron didemni | 85% | 100% P |
We attempted to isolate peptides from two of these genera. For the Cyanothece strain, continuous growth over a two-year period yielded insufficient material for characterization. A. platensis (“Spirulina”) is a commercial product that is widely used as a health food supplement. Two preparations of dried A. platensis were purchased from Whole Foods Market. The methanol extracts of both commercial products contained cyanobactins derived from the ArtE3 and ArtE5 precursor peptides, as judged by low-resolution LC-MS. These compounds were partially purified, and FT-ICR MS/MS confirmed the predicted amino acid sequences and high-resolution molecular ions for these compounds, which we have named arthrospiramides A (3) and B (4) (Figure 3, Table S2, Figure S4). Moreover, the sulfoxides (5) and (6) of both compounds were also identified in the extracts, further validating the structures. Configurations of 3-6 were tentatively assigned based upon biochemical considerations. Two stereocenters were not defined because they are adjacent to thiazole residues, making them chemically labile. These compounds could not easily observed in the extracts by NMR, allowing us to estimate that they are present in a concentration of <10-4% of dry weight in these products. Despite the low abundance of the compounds, their pharmacological properties should be investigated.
Phylogenetic analysis
With the growing number of sequenced cyanobactin pathways (seven previously reported and five identified here), phylogenetic relationships of the major family groups could be assessed. Maximum likelihood analysis was performed using PROMLK (Phylip) for the conserved proteins and verified using bootstrap tests and Maximum Parsimony analysis (MEGA 4.0) (Felsenstein, 1989; Tamura et al., 2007). In the resulting trees, proteins always branched based upon the cyanobactin structure and genotype, not the taxonomy of the producing strain. This observation suggests that each genotype evolved separately and was probably acquired horizontally by different species, and it also suggests a phylogeny-based strategy for further pathway cloning and structure prediction. The possibility of lateral transfer was previously noted by the Sivonen group, which described a clear phylogenetic incongruence between cyanobactin synthetase and 16S rRNA genes from different cyanobacteria (Leikoski et al., 2009). Based upon phylogenetic analysis, four genotypes were identified (Figure 4, Figure S6). Because the phylogeny was predictive of function and not of taxonomic origin, structure and sequence could be clearly linked (Figure 5). Importantly, even within these genotypes, phylogenetic analysis of individual proteins enabled prediction. For example, all of the PatF homologs from prenylating gene clusters are focused in one clade, while the homologs from non-prenylating gene clusters group in another. Regioselectivity of PatD-like activity can also be predicted using phylogenetic analysis. Therefore, the phylogenetic approach leads to a predictive model for cyanobactin synthesis.
Figure 4. Phylogeny of Cyanobactin Proteins.
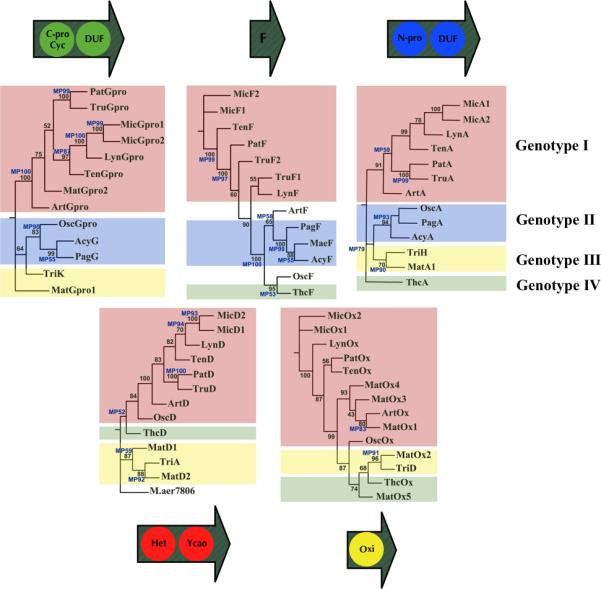
Maximum likelihood with molecular clock analysis (PROMLK, Phylip) was performed on all conserved cyanobactin proteins. Numbers indicate percentage of bootstrap replicates that support the indicated branch point. Due to the long computational times required, bootstrap numbers varied: PatG: 448; PatF: 892; PatD: 466; PatA: 602; oxidase: 442. Tree branches that were also supported by Maximum Parsimony analysis (MEGA 4.0) and verified using 1000 bootstrap replicates are indicated in blue. Representatives forming different genotypes are highlighted in different colors. “Mat” indicates the protein sequence of the metagenomic DNA fragments isolated by targeted degenerate PCR from a L. majuscula metagenomic bloom sample. See also Figure S6.
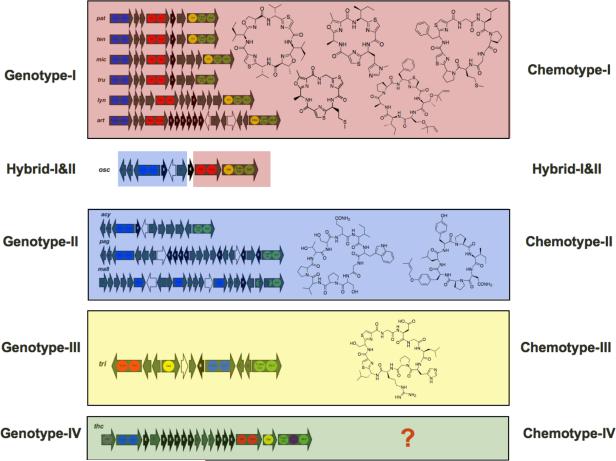
Figure 5. Cyanobactin Genotypes and Chemotypes.
Cyanobactin biosynthetic pathways can be classified into 4 genotypes that correlate to the resulting chemotypes of natural products. thc products cannot be predicted and are indicated by a question mark. Additional pathways used in this figure include: pat, patellamide; ten, tenuecyclamide; mic, microcyclamide; tru, trunkamide; lyn, lyngbya; tri, trichamide; acy, anacylamide. See also Figure S7.
Phylogenetic model of cyanobactin synthesis
A model was created that predictively links genes and chemicals in this series. Genotype I, represented by pat, is the most common in the sequenced cyanobactin gene clusters and consists of a typical single operon starting with a patA homolog and ending with a patG homolog, although sometimes additional open reading frames encoding hypothetical proteins are present between these genes. A heterocyclase (patD homolog) is always present and leads to thiazoline, and sometimes oxazoline; this selectivity is predictable from phylogenetic analysis. The presence of an oxidase domain in patG homologs is diagnostic for the oxidation to thiazole and, sometimes, oxazole, although the regiochemistry is still not predictable. Some additional clades can be observed, corresponding to the structure of the resulting cyanobactins. For example, in some cases compounds are prenylated on Ser and Thr. These modifications can be readily and accurately predicted: the TruF group prenyltransferases cluster together in the phylogenetic tree, and separately from the non-prenylating PatF-like proteins (Figure 4). Genotype II is represented by pag, acy and mae and is missing the heterocyclase and oxidase proteins. Genes usually occupy two putative operons. The mae cluster is slightly differently organized due to the extensive interruption by multiple transposition events. Genotype II leads to cyclic peptides of different sizes that lack heterocyclization, consistent with the absence of a patD homolog. Prenylation events can also occur on Tyr or, putatively, Trp, as suggested in a recent paper (Leikoski et al., 2010), and truF homologs are diagnostic of this modification. osc modifying enzymes fall in between genotypes I and II, which is consistent with the overall organization of the pathway. Although osc contains all the genes required for the biosynthesis of oxidized heterocycle-containing cyanobactins (genotype I), it is organized in a manner similar to genotype II. Therefore, it represents the first example of hybrid pathway type or an evolutionary link between two genotypes. As expected from its organization, osc proteins responsible for heterocyclization and oxidization cluster with genotype I while the ones responsible for macrocyclization cluster with genotype II (Figure S3). Based upon this result, osc is predicted to produce products that contain features of both genotypes. For example, myriastramides and haliclonamides are cyclic peptides isolated from marine sponges that contain heterocyclized Ser/Thr residues (as found in chemotype I) and prenylated Tyr residues (as found in chemotype II) (Erickson et al., 2003; Guan et al., 2001; Sera et al., 2002). Interestingly, Oscillatoria spp. are known to exist abundantly in marine sponges (Flatt et al., 2005; Ridley et al., 2005a; Ridley et al., 2005b; Thacker and Starnes, 2003).
Genotype III, exemplified by tri, contains most of the same genes as genotype I, but with some unique features. First, unlike genotype I, genotype III contains the oxidase domain as a separate open reading frame. Second, the organization of the genes in genotype III is completely different than in genotype I. Third, the precursor peptide sequence of genotype III is much different than the ones in genotype I. More sequences are required to further elucidate the resulting chemotypes of this relatively rare pathway variant. Genotype IV is exemplified by the thc pathway. Its organization is so far unique and its products are not easily predictable. It is almost certain that further genotypes await discovery, but they will be easily related to the scheme proposed here.
Sequence analysis of precursor peptides
The large number of precursor peptides identified in this and previous studies (Donia et al., 2006; Haft et al., 2010; Li et al., 2010) reinforces the notion that similar enzymes are responsible for the synthesis of diverse products, allowing relatively small gene clusters to produce a large variety of compounds. However, the point of modification of precursor peptides was not easily predictable, as shown by our results from the pag cluster. Therefore, we analyzed the precursors by sequence alignment and using jpred to predict secondary structure (Cuff et al., 1998). All of the heterocycle-containing cyanobactin leaders are similar to PatE and are uncovered by jpred. They are predicted to be helical over a large region that overlaps that identified in a recent NMR study (Houssen et al. 2010). By contrast, the non-heterocycle precursors, such as those from pag, acy, and mae, are only related to PatE in their extreme N- and C-termini. They overlap the region defined as helical by NMR, but they are missing a segment that is highly predicted to be helical in nature and that is defined by the sequence LAELSEEAL in PatE (Figure S3). Therefore, the presence of this sequence or its homolog is probably a requirement for interaction with the cyanobactin heterocyclase. Unfortunately, in the reported NMR structure construct, this sequence was too close to the C-terminus, so that it was unstructured in the experimental conditions employed (Houssen et al. 2010). Potentially helical regions of the leader sequence have been shown essential for the interaction with other ribosomal heterocyclases (Mitchell et al., 2009; Roy et al., 1998; Sinha Roy et al., 1998).
The sequence alignments enable all of the C-terminal proteolysis sites to be unambiguously predicted. Previously, it was difficult to predict N-terminal sites in new precursor peptide families, such as those encoding arthrospiramides. However, distance from the LAELSEEAL-like site is diagnostic for the N-terminus in heterocyclic cyanobactins, which allowed us to predict the N-terminus of arthrospiramides (see below). However, for new groups of peptides lacking heterocyclization, the N-terminal region remains difficult to call.
Application of phylogeny: discovery of new cyanobactins from genome sequence and new cyanonactin genes from metagenomic samples
The phylogenetic findings described above were used in proof-of-concept experiments. In the first, as described above, we were able to accurately predict the structures of arthrospiramides using the approach. Distance from the LAELSEEAL sequence defined the N-terminal cleavage site. Moreover, art lies in genotype I and contains an oxidase and a PatD-like heterocyclase, making its overall structure and posttranslational modifications easily predictable. These features allowed the resulting compounds to be found despite their low abundance.
Because many cyanobactins have been isolated from mixed environments, such as cyanobacterial blooms, sponges, ascidians and mollusks, developing strategies for identifying their biosynthetic pathways is of great importance. Therefore, we designed degenerate primers to specifically amplify all conserved cyanobactin genes (Figure S7). The primers were designed to amplify small fragments and were specifically engineered to capture all genotypes. Their utility was verified by testing them against actual samples containing multiple known pathways from different genotypes.
We selected a highly heterogenous cyanobacterial bloom of Lyngbya majuscula for metagenomic analysis. This sample was collected in Papua New Guinea and was known to contain the trichamide-like large cyanobactin, wewakazole (Nogle et al., 2003). A small sample of the original cyanobacterial mat was provided generously by W. Gerwick (UCSD). Metagenomic DNA was isolated from the sample and subjected to degenerate PCR primers designed to specifically amplify short segments of cyanobactin proteases, oxidases, and heterocyclases (Figures S7). Expected fragments were amplified, cloned, and sequenced, leading to the identification of at least five different cyanobactin gene clusters, including five different cyanobactin oxidases, two heterocyclases, and three proteases. In order to determine whether we captured the expected pieces from a trichamide-like gene cluster, we subjected them to phylogenetic analysis. Indeed, individual PatA, PatG, PatD, and oxidase homologs strongly clustered with genotype III, and translation products were closely related to predicted proteins from tri, at 75% or greater identity. Other identified fragments clustered with other genotypes (Figure 4). Therefore, this phylogenetic method led to rapid identity of the candidate genes from the mixed sample. Unfortunately, the cyanobacterial DNA was highly degraded so that these fragments could not be connected using any of multiple cloning or PCR techniques.
SIGNIFICANCE
Cyanobactins are among the major natural products isolated from cyanobacteria, which in turn are among the major groups of natural product-synthesizing organisms. We define the core genotypes of cyanobactin biosynthetic pathways, relate them to chemical structure, and use them for pathway cloning. These genotypes encompass the known chemical diversity of cyanobactin products and will be useful in cloning new pathways, in prediction of new structures, and in engineering synthesis of new cyclic peptides. Indeed, in the course of these studies using genome mining we identified peptide natural products, which despite extensive investigation were unsuspected, in the widely used supplement Spirulina. We also identified a pathway to Tyr O-prenylation in the prenylagaramides and discovered a new analog by genetic methods. There are several other related cyclic peptides containing O-prenyl Tyr originating both in other cyanobacteria and in marine sponges (Baumann et al., 2007; Erickson et al., 2003; Fujii et al., 2000; Guan et al., 2001; Sano and Kaya, 1996; Sera et al., 2002; Shin et al., 1996). We propose that these metabolites also belong to the cyanobactin group and are synthesized through a pag-like pathway of genotype II. Results of this study will be useful in accessing these clusters and in discovering cyanobactin gene clusters from other metagenomic samples, such as sponges, ascidians and mollusks, which have been the source of ~70% of the cyanobactins discovered to date (Donia and Schmidt, 2010). Here, we provide proof-of-concept for these approaches, discovering and analyzing new diverse cyanobactin genes from a highly heterogeneous sample and providing the first example of applying a phylogenetic approach to pathway identification in the diverse ribosomal peptide group of natural products. The results from this study show that cyanobactins are indeed widespread everywhere from niche microbial environments to commercial food products and provides the tools to investigate them.
EXPERIMENTAL PROCEDURES
Genome Mining
All conserved cyanobactin biosynthetic genes were compared to the NCBI published genomes database using BLAST. Hits from cyanobacterial genomes were analyzed further by downloading the genomic region of interest. Vector NTI and Artemis were used to predict open reading frames in these regions and BLAST was used to identify neighboring cyanobactin biosynthetic genes. Gene clusters containing precursor peptide genes were considered “intact” while ones lacking identifiable precursor peptide genes were disregarded for the purpose of this study. In pathways containing more than one precursor peptide gene, protein sequences of all the identified variants were aligned using CLUSTALX and then fixed manually to identify hypervariable regions versus more conserved regions.
Phylogenetic Analysis
The amino acid sequences of all the homologous proteins from the identified cyanobactin pathways were aligned using CLUSTALX. Maximum likelihood analysis with molecular clock PROMLK (PHYLIP) using the bootstrap test method was performed to assess the phylogenetic relationship between the different homologs. In most cases, the same tree branches were also supported using other phylogenetic experiments such as Maximum Parsimony (MEGA 4.0) using 1000 bootstrap replicates.
Degenerate Primer Design and Application
Homologous genes from all cyanobactin gene clusters described in this study were aligned using CLUSTALX. Conserved regions were identified manually and used to design degenerate primers by the CODEHOP algorithm (Rose et al., 1998). Primer pairs were selected to preferably amplify genomic regions of 500 bp or less. Described primer pairs were validated using multiple cyanobactin genotypes available in our lab to demonstrate broad specificity. PCR conditions are described in Supplemental Experimental Methods.
Cyanobacterial Cultivation
Planktothrix agardhii NIES-596 was obtained from the Microbial Culture Collection at the National Institute for Environmental Studies, Japan. The culture was maintained static at room temperature in a 12 / 12 light / dark cycle in 30 ml of CT medium (Shirai et al., 1989; Sun et al., 2006). The culture was grown to high density (1-2 months) then diluted two- to three-fold into fresh CT medium for further growth. Cyanothece sp. (PCC 7425) was obtained from the ATCC (ATCC number 29141) and grown in the laboratory in BG-11 medium according to the recommended supplier protocol. 12 / 12 light / dark cycle were set for 4 cultures of 500 ml. Each month repeatedly for 2 years, 400 ml of media was removed and an additional 500 ml of fresh media was added to the culture.
Chemical Analysis
Planktothrix agardhii NIES-596 was grown in small-scale cultures as mentioned above. After reaching a high density (judged by color), cultures were spun down for 10 min at 4000 RPM and pellets were combined and freeze dried overnight. The pellets extracted in this study were from combined cultures of about 1 Liter in total. Freeze-dried pellet was extracted three times with methanol (20 ml) and extracts were combined and dried. The dried methanol extract was then loaded on a small C18 column and washed with water and then 20% acetonitrile. The extract was finally eluted off the column with 100% acetonitrile, which was then diluted and used directly for FTICR-MS/MS analysis. The previously described prenylagaramide B (1) was confirmed to be produced by this strain, and a new compound prenylagaramide C (2) was found.
Earthrise Spirulina Natural Dietary Supplement (180 g dry powder) and Whole Foods Pesticide-Free Spirulina (250 tablets, 500 mg ea) were obtained from Whole Foods Market in Salt Lake City, UT. The powders and tablets were extracted 2x with 3 volume equivalents of methanol (HPLC grade). The extracts were dried to ~200 mL by rotary evaporation, then diluted with ~800 mL dH2O containing 200 mL HP20 resin. The resin was filtered and washed with 2X 300 mL each of water, then 50% methanol. Products were then eluted in 100% methanol and applied directly to a Waters Micromass ZQ HPLC-MS. Arthrospiramides A and B were predicted to be m/z = 866 and 924, respectively ([M+H]+). Indeed, these ions were clearly present in extracts of both products, although they were more enriched in the Earthrise product. In both products, the ion ratios of these peaks to the major extract components were at least ~1000:1. Therefore, this extract was further fractionated in a packed C18 column, using an aqueous methanol gradient with 10% steps. Arthrospiramides eluted in 80%-100% methanol with maximum ion sizes approximately 5% of the major components. These fractions were used directly for FTICR-MS, leading to discovery of arthrospiramides A and B and their sulfoxides, (3-6)
DNA Extraction
Cyanobacterial and metagenomic DNA were extracted using three methods: 1) A DNA extraction kit (PureGene; Qiagen); 2) A protocol related to actinomycete DNA isolation methods (Kieser et al., 2000); 3) A method derived from a cyanobacterial protocol (Cangelosi et al., 1986). For further extraction details see Supplemental Experimental Methods.
Fosmid Library Construction
Genomic DNA was cloned into the pCC1FOS or pCC2FOS fosmid vectors (Epicentre) following the manufacturer's protocol.
Discovery of pag Cluster
Exact match primers (Pren-prot-F and Pren-prot-R) were designed based on the degenerate protease amplicon from P. agardhii and used to screen 8x96 well plates containing individual fosmid clones. Plates 3 and 8 were identified as positives and screened further, yielding clones 8A2 and 3C4 as positive hits. A shotgun library was constructed from 3C4 using the TOPO® Shotgun Subcloning Kit (Invitrogen). A combination of random sequencing reactions in the shotgun library and directed walking sequencing reactions in 3C4 was used to obtain the full pag sequence, which was assembled using Sequencher (Gene Codes) and analyzed using Artemis.
Metagenomic analysis of a mixed environmental bloom sample
A frozen environmental sample of L. majuscula from Papua New Guinea was provided by W. Gerwick (UCSD). Genomic DNA from the mixed metagenome sample was screened using a PCR approach with degenerate primers for each expected gene type: two proteases, oxidase, and heterocyclase. Additionally, degenerate primers for possible precursor peptide sequences were applied. PCR products were cloned into the pCR2.1 TOPO vector and sequenced by walking in both directions, then used for phylogenetic analysis in comparison to other sequences reported here.
Prenylagramide B (1): FT-ICR MS: m/z = 929.5142 (expected for C49H68N8O10, 929.5131 [M+H]+, Δ = 1.2 ppm); ms2 (IRMPD): 912.5096 (-OH, expected for C49H67N8O9, 912.5103, Δ = -0.8 ppm); 844.4501 (-prenyl ; expected for C44H59N8O9, 844.4477, Δ = 0.7 ppm). For a list of other ions, see Table S1.
Prenylagaramide C (2): FT-ICR MS: m/z = 1021.6097 (expected for C52H80N10O11, 1021.6080 [M+H]+, Δ = 1.7 ppm); ms2 (IRMPD): 953.5469 (-prenyl ; expected for C47H72N10O11, 953.5454, Δ = 1.6 ppm). For a list of other ions, see Table S1.
Arthrospiramide A (3): FT-ICR MS: m/z = 866.3156 (expected for C40H51N9O7S, 866.3146 [M+H]+, Δ = 1.2 ppm). For ms2 (CID), see Table S2.
Arthrospiramide B (4): FT-ICR MS: m/z = 924.3944 (expected for C44H61N9O7S3, 924.3929 [M+H]+, Δ = 1.6 ppm). For ms2 (CID), see Table S2.
Arthrospiramide A sulfoxide (5): FT-ICR MS: m/z = 882.3109 (expected for C40H51N9O8S, 882.3095 [M+H] , Δ = 1.6 ppm). For ms2 (CID), see Table S2.
Arthrospiramide B sulfoxide (6): FT-ICR MS: m/z = 940.3890 (expected for C44H61N9O8S3, 940.3878 [M+H]+, Δ = 1.3 ppm). For ms2 (CID), see Table S2.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to W. Gerwick (UCSD) for providing an authentic sample of L. majuscula containing wewakazole. We thank K. Parsawar and C. Nelson for performing FT-MS runs. This work was funded by GM071425 (NIH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The pag cluster and L. majuscula fragments were deposited in GenBank with accession numbers HQ655154 and HQ655144-HQ655153, respectively.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, Tables S1 and S2, and Figures S1-S7 and can be found online at ...
REFERENCES
- Ansari MZ, Sharma J, Gokhale RS, Mohanty D. In silico analysis of methyltransferase domains involved in biosynthesis of secondary metabolites. BMC Bioinformatics. 2008;9:454. doi: 10.1186/1471-2105-9-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann HI, Keller S, Wolter FE, Nicholson GJ, Jung G, Sussmuth RD, Juttner F. Planktocyclin, a cyclooctapeptide protease inhibitor produced by the freshwater cyanobacterium Planktothrix rubescens. J. Nat. Prod. 2007;70:1611–1615. doi: 10.1021/np0700873. [DOI] [PubMed] [Google Scholar]
- Cangelosi GA, Joseph CM, Rosen JJ, Meeks JC. Cloning and expression of a Nostoc sp. leucine biosynthetic gene in Escherichia coli. Arch. Microbiol. 1986;145:315–321. [Google Scholar]
- Challis GL, Ravel J, Townsend CA. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- Chatterjee C, Paul M, Xie L, van der Donk WA. Biosynthesis and mode of action of lantibiotics. Chem. Rev. 2005;105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- Chiu HT, Hubbard BK, Shah AN, Eide J, Fredenburg RA, Walsh CT, Khosla C. Molecular cloning and sequence analysis of the complestatin biosynthetic gene cluster. Proc. Natl. Acad. Sci. U S A. 2001;98:8548–8553. doi: 10.1073/pnas.151246498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat. Chem. Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- Donia MS, Ravel J, Schmidt EW. A global assembly line for cyanobactins. Nat. Chem. Biol. 2008;4:341–343. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia MS, Schmidt EW. Comprehensive Natural Products II Chemistry and Biology. Elsevier; Oxford: 2010. Cyanobactins – ubiquitous cyanobacterial ribosomal peptide metabolites. pp. 539–558. [Google Scholar]
- Erickson KL, Gustafson KR, Milanowski DJ, Pannell LK, Klose JR, Boyd MR. Myriastramides A-C, new modified cyclic peptides from the Philippines marine sponge Myriastra clavosa. Tetrahedron. 2003;59:10231–10238. [Google Scholar]
- Felsenstein J. PHYLIP - Phylogeny Inference Package version 3.6. Cladistics. 1989;5:164–166. [Google Scholar]
- Flatt PM, Gautschi JT, Thacker RW, Musafija-Girt M, Crews P, Gerwick WH. Identification of the cellular site of polychlorinated peptide biosynthesis in the marine sponge Dysidea (Lamellodysidea) herbacea and symbiotic cyanobacterium Oscillatoria spongeliae by CARD-FISH analysis. Mar. Biol. 2005;147:761–774. [Google Scholar]
- Fujii K, Sivonen K, Naganawa E, Harada K. Non-toxic peptides from toxic cyanobacteria, Oscillatoria agardhii. Tetrahedron. 2000;56:725–733. [Google Scholar]
- Fujisawa T, Narikawa R, Okamoto S, Ehira S, Yoshimura H, Suzuki I, Masuda T, Mochimaru M, Takaichi S, Awai K, et al. Genomic structure of an economically important cyanobacterium, Arthrospira (Spirulina) platensis NIES-39. DNA Res. 2010;17:85–103. doi: 10.1093/dnares/dsq004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesner-Apter S, Carmeli S. Three novel metabolites from a bloom of the cyanobacterium Microcystis sp. Tetrahedron. 2008;64:6628–6634. [Google Scholar]
- Guan LL, Sera Y, Adachi K, Nishida F, Shizuri Y. Isolation and evaluation of nonsiderophore cyclic peptides from marine sponges. Biochem. Biophys. Res. Commun. 2001;283:976–981. doi: 10.1006/bbrc.2001.4890. [DOI] [PubMed] [Google Scholar]
- Haft DH, Basu MK, Mitchell DA. Expansion of ribosomally produced natural products: a nitrile hydratase- and Nif11-related precursor family. BMC Biol. 2010;8:70. doi: 10.1186/1741-7007-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallen HE, Luo H, Scott-Craig JS, Walton JD. Gene family encoding the major toxins of lethal Amanita mushrooms. Proc. Natl. Acad. Sci. U S A. 2007;104:19097–19101. doi: 10.1073/pnas.0707340104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houssen WE, Wright SH, Kalverda AP, Thompson GS, Kelly SM, Jaspars M. Solution structure of the leader sequence of the patellamide precursor peptide, PatE1-34. ChemBioChem. 2010;11:1867–1873. doi: 10.1002/cbic.201000305. [DOI] [PubMed] [Google Scholar]
- Ireland DC, Clark RJ, Daly NL, Craik DJ. Isolation, sequencing, and structure-activity relationships of cyclotides. J. Nat. Prod. 2010;73:1610–1622. doi: 10.1021/np1000413. [DOI] [PubMed] [Google Scholar]
- Ishida K, Matsuda H, Murakami M, Yamaguchi K. Kawaguchipeptin A, a novel cyclic undecapeptide from cyanobacterium Microcystis aeruginosa (NIES-88). Tetrahedron. 1996;52:9025–9030. doi: 10.1021/np970146k. [DOI] [PubMed] [Google Scholar]
- Ishida K, Matsuda H, Murakami M, Yamaguchi K. Kawaguchipeptin B, an antibacterial cyclic undecapeptide from the cyanobacterium Microcystis aeruginosa. J. Nat. Prod. 1997;60:724–726. doi: 10.1021/np970146k. [DOI] [PubMed] [Google Scholar]
- Janssen PJ, Morin N, Mergeay M, Leroy B, Wattiez R, Vallaeys T, Waleron K, Waleron M, Wilmotte A, Quillardet P, de Marsac NT, Talla E, Zhang CC, Leys N. Genome sequence of the edible cyanobacterium Arthrospira sp. PCC 8005. J. Bacteriol. 2010;192:2465–2466. doi: 10.1128/JB.00116-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Nakajima N, Okamoto S, Suzuki I, Tanabe Y, Tamaoki M, Nakamura Y, Kasai F, Watanabe A, Kawashima K, Kishida Y, Ono A, Shimizu Y, Takahashi C, Minami C, Fujishiro T, Kohara M, Katoh M, Nakazaki N, Nakayama S, Yamada M, Tabata S, Watanabe MM. Complete genomic structure of the bloom-forming toxic cyanobacterium Microcystis aeruginosa NIES-843. DNA Res. 2007;14:247–256. doi: 10.1093/dnares/dsm026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces genetics. John Innes Foundation; (Norwich: 2000. [Google Scholar]
- Lee J, McIntosh J, Hathaway BJ, Schmidt EW. Using marine natural products to discover a protease that catalyzes peptide macrocyclization of diverse substrates. J. Am. Chem. Soc. 2009;131:2122–2124. doi: 10.1021/ja8092168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SW, Mitchell DA, Markley AL, Hensler ME, Gonzalez D, Wohlrab A, Dorrestein PC, Nizet V, Dixon JE. Discovery of a widely distributed toxin biosynthetic gene cluster. Proc. Natl. Acad. Sci. U S A. 2008;105:5879–5884. doi: 10.1073/pnas.0801338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leikoski N, Fewer DP, Jokela J, Wahlsten M, Rouhiainen L, Sivonen K. Highly diverse cyanobactins in strains of the genus Anabaena. Appl. Environ. Microbiol. 2010;76:701–709. doi: 10.1128/AEM.01061-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leikoski N, Fewer DP, Sivonen K. Widespread occurrence and lateral transfer of the cyanobactin biosynthesis gene cluster in cyanobacteria. Appl. Environ. Microbiol. 2009;75:853–857. doi: 10.1128/AEM.02134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Sher D, Kelly L, Shi Y, Huang K, Knerr PJ, Joewono I, Rusch D, Chisholm SW, van der Donk WA. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc. Natl. Acad. Sci. U S A. 2010;107:10430–10435. doi: 10.1073/pnas.0913677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Ahlert J, Gao Q, Wendt-Pienkowski E, Shen B, Thorson JS. Rapid PCR amplification of minimal enediyne polyketide synthase cassettes leads to a predictive familial classification model. Proc. Natl. Acad. Sci. U S A. 2003;100:11959–11963. doi: 10.1073/pnas.2034291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JA, Donia MS, Schmidt EW. Ribosomal peptide natural products: bridging the ribosomal and nonribosomal worlds. Nat. Prod. Rep. 2009;26:537–559. doi: 10.1039/b714132g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JA, Donia MS, Schmidt EW. Insights into heterocyclization from two highly similar enzymes. J. Am. Chem. Soc. 2010a;132:4089–4091. doi: 10.1021/ja9107116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JA, Robertson CR, Agarwal V, Nair SK, Bulaj GW, Schmidt EW. Circular logic: nonribosomal peptide-like macrocyclization with a ribosomal peptide catalyst. J. Am. Chem. Soc. 2010b;132:15499–15501. doi: 10.1021/ja1067806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JA, Schmidt EW. Marine molecular machines: heterocyclization in cyanobactin biosynthesis. ChemBioChem. 2010;11:1413–1421. doi: 10.1002/cbic.201000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejean A, Mazmouz R, Mann S, Calteau A, Medigue C, Ploux O. The genome sequence of the cyanobacterium Oscillatoria sp. PCC 6506 reveals several gene clusters responsible for the biosynthesis of toxins and secondary metabolites. J. Bacteriol. 2010;192:5264–5265. doi: 10.1128/JB.00704-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Lee SW, Pence MA, Markley AL, Limm JD, Nizet V, Dixon JE. Structural and functional dissection of the heterocyclic peptide cytotoxin streptolysin S. J. Biol. Chem. 2009;284:13004–13012. doi: 10.1074/jbc.M900802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Itou Y, Ishida K, Shin HJ. Prenylagaramides A and B, new cyclic peptides from two strains of Oscillatoria agardhii. J. Nat. Prod. 1999;62:752–755. doi: 10.1021/np980396g. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Ishida K, Jenke-Kodama H, Dittmann E, Gurgui C, Hochmuth T, Taudien S, Platzer M, Hertweck C, Piel J. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat. Biotechnol. 2008;26:225–233. doi: 10.1038/nbt1379. [DOI] [PubMed] [Google Scholar]
- Nogle LM, Marquez BL, Gerwick WH. Wewakazole, a novel cyclic dodecapeptide from a Papua New Guinea Lyngbya majuscula. Org. Lett. 2003;5:3–6. doi: 10.1021/ol026811k. [DOI] [PubMed] [Google Scholar]
- Oman TJ, van der Donk WA. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat. Chem. Biol. 2010;6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portmann C, Blom JF, Gademann K, Juttner F. Aerucyclamides A and B: isolation and synthesis of toxic ribosomal heterocyclic peptides from the cyanobacterium Microcystis aeruginosa PCC 7806. J. Nat. Prod. 2008a;71:1193–1196. doi: 10.1021/np800118g. [DOI] [PubMed] [Google Scholar]
- Portmann C, Blom JF, Kaiser M, Brun R, Juttner F, Gademann K. Isolation of aerucyclamides C and D and structure revision of microcyclamide 7806A: Heterocyclic ribosomal peptides from Microcystis aeruginosa PCC 7806 and their antiparasite evaluation. J. Nat. Prod. 2008b;71:1891–1896. doi: 10.1021/np800409z. [DOI] [PubMed] [Google Scholar]
- Ridley CP, Bergquist PR, Harper MK, Faulkner DJ, Hooper JN, Haygood MG. Speciation and biosynthetic variation in four dictyoceratid sponges and their cyanobacterial symbiont, Oscillatoria spongeliae. Chem. Biol. 2005a;12:397–406. doi: 10.1016/j.chembiol.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Ridley CP, John Faulkner D, Haygood MG. Investigation of Oscillatoria spongeliae-dominated bacterial communities in four dictyoceratid sponges. Appl. Environ. Microbiol. 2005b;71:7366–7375. doi: 10.1128/AEM.71.11.7366-7375.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy RS, Kim S, Baleja JD, Walsh CT. Role of the microcin B17 propeptide in substrate recognition: solution structure and mutational analysis of McbA1-26. Chem. Biol. 1998;5:217–228. doi: 10.1016/s1074-5521(98)90635-4. [DOI] [PubMed] [Google Scholar]
- Sano T, Kaya K. Oscillatorin, a chymotrypsin inhibitor from toxic Oscillatoria agardhii. Tetrahedron Lett. 1996;37:6873–6876. doi: 10.1021/np9600210. [DOI] [PubMed] [Google Scholar]
- Schmidt EW, Donia MS. Cyanobactin ribosomally synthesized peptides--a case of deep metagenome mining. Methods Enzymol. 2009;458:575–596. doi: 10.1016/S0076-6879(09)04823-X. Chapter 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. U S A. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sera Y, Adachi K, Fujii K, Shizuri Y. Isolation of Haliclonamides: new peptides as antifouling substances from a marine sponge species, Haliclona. Mar. Biotechnol. 2002;4:441–446. doi: 10.1007/s10126-001-0082-6. [DOI] [PubMed] [Google Scholar]
- Shin HJ, Matsuda H, Murakami M, Yamaguchi K. Agardhipeptins A and B, two new cyclic hepta- and octapeptide, from the cyanobacterium Oscillatoria agardhii (NIES-204). Tetrahedron. 1996;52:13129–13136. [Google Scholar]
- Shirai M, Matumaru K, Ohotake A, Takamura Y, Aida T, Nakano M. Development of a solid medium for growth and isolation of axenic Microcystis strains (Cyanobacteria). Appl. Environ. Microbiol. 1989;55:2569–2571. doi: 10.1128/aem.55.10.2569-2571.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha Roy R, Belshaw PJ, Walsh CT. Mutational analysis of posttranslational heterocycle biosynthesis in the gyrase inhibitor microcin B17: distance dependence from propeptide and tolerance for substitution in a GSCG cyclizable sequence. Biochemistry. 1998;37:4125–4136. doi: 10.1021/bi9728250. [DOI] [PubMed] [Google Scholar]
- Sivonen K, Leikoski N, Fewer DP, Jokela J. Cyanobactins-ribosomal cyclic peptides produced by cyanobacteria. Appl. Microbiol. Biotechnol. 2010;86:1213–1225. doi: 10.1007/s00253-010-2482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudek S, Haygood MG, Youssef DT, Schmidt EW. Structure of trichamide, a cyclic peptide from the bloom-forming cyanobacterium Trichodesmium erythraeum, predicted from the genome sequence. Appl. Environ. Microbiol. 2006;72:4382–4387. doi: 10.1128/AEM.00380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun BK, Tanji Y, Unno H. Extinction of cells of cyanobacterium Anabaena circinalis in the presence of humic acid under illumination. Appl. Microbiol. Biotechnol. 2006;72:823–828. doi: 10.1007/s00253-006-0327-4. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thacker RW, Starnes S. Host specificity of the symbiotic cyanobacterium Oscillatoria spongeliae in marine sponges, Dysidea spp. Mar. Biol. 2003;142:643–648. [Google Scholar]
- Ziemert N, Ishida K, Quillardet P, Bouchier C, Hertweck C, de Marsac NT, Dittmann E. Microcyclamide biosynthesis in two strains of Microcystis aeruginosa: from structure to genes and vice versa. Appl. Environ. Microbiol. 2008;74:1791–1797. doi: 10.1128/AEM.02392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.