Abstract
Chromatin is important for the regulation of transcription and other functions, yet the diversity of chromatin composition and the distribution along chromosomes is still poorly characterized. By integrative analysis of genome-wide binding maps of 53 broadly selected chromatin components in Drosophila cells, we show that the genome is segmented into five principal chromatin types that are defined by unique, yet overlapping combinations of proteins, and form domains that can extend over >100 kb. We identify a repressive chromatin type that covers about half of the genome and lacks classic heterochromatin markers. Furthermore, transcriptionally active euchromatin consists of two types that differ in molecular organization and H3K36 methylation, and regulate distinct classes of genes. Finally, we provide evidence that the different chromatin types help to target DNA-binding factors to specific genomic regions. These results provide a global view of chromatin diversity and domain organization in a metazoan cell.
INTRODUCTION
Chromatin consists of DNA and all associated proteins. The scaffold of chromatin is formed by nucleosomes, which are histone octamers in a tight complex with DNA. This scaffold serves as the docking platform for hundreds of structural and regulatory proteins. Furthermore, histones carry a variety of post-translational modifications that form recognition sites for specific proteins (Berger, 2007; Rando and Chang, 2009). The local composition of chromatin is a major determinant of the transcriptional activity of a gene; some chromatin proteins enhance transcription, while others have repressive effects.
Traditionally, chromatin was divided into heterochromatin and euchromatin. There is now ample evidence that a finer classification is required. For example, in Drosophila at least two types of heterochromatin exist that have distinct regulatory functions and consist of different proteins. The first type is marked by Polycomb Group (PcG) proteins and methylation of lysine 27 of histone H3 (H3K27). PcG chromatin forms large continuous domains; it is a repressive type of chromatin that primarily regulates genes with developmental functions (Sparmann and van Lohuizen, 2006). The second type is marked by Heterochromatin Protein 1 (HP1) and several associated proteins, combined with methylation of H3K9. This type of heterochromatin can also cover large genomic segments, particularly around centromeres. Reporter genes integrated in or near HP1 heterochromatin tend to be repressed, but paradoxically many genes that are naturally bound by HP1 are transcriptionally active (Hediger and Gasser, 2006). Direct comparison of genome-wide binding maps indicates that PcG and HP1 heterochromatin are non-overlapping (de Wit et al., 2007).
HP1 and PcG chromatin illustrate two important principles of chromatin organization: each type is marked by unique combinations of proteins, and can cover long stretches of DNA. But are there other major types of chromatin that follow these same principles? For example, is euchromatin also organized into domains with distinct protein compositions? Are there additional types of repressive chromatin that have remained unnoticed?
In order to address these questions we generated genome-wide location maps of 53 broadly selected chromatin proteins and four key histone modifications in Drosophila cells, providing a rich description of chromatin composition along the genome. By integrative computational analysis we identified, besides PcG and HP1 chromatin, three additional principal chromatin types, which are defined by unique combinations of proteins. One of these is a type of repressive chromatin that covers ~50% of the genome. In addition, we identified two types of transcriptionally active euchromatin that are bound by different proteins and harbor distinct classes of genes.
RESULTS
Genome-wide location maps of 53 chromatin proteins
We constructed a database of high-resolution binding profiles of 53 chromatin proteins in the embryonic Drosophila melanogaster cell line Kc167 (Figure 1A and Supplementary Figure 1A). In order to obtain a representative cross-section of the chromatin proteome, we selected proteins from most known chromatin protein complexes, including a variety of histone-modifying enzymes, proteins that bind specific histone modifications, general transcription machinery components, nucleosome remodelers, insulator proteins, heterochromatin proteins, structural components of chromatin, and a selection of DNA binding factors (DBFs) (Supplementary Table 1). For ~40 of these proteins, full-genome high-resolution binding maps have not previously been reported in any Drosophila cell type or tissue. While chromatin immunoprecipitation (ChIP) is widely used to map protein-genome interactions (Collas, 2009), large-scale application of this method is hampered by the limited availability of highly specific antibodies. Moreover, at least for some chromatin proteins, ChIP results can greatly depend on the choice of crosslinking reagents (Wang et al., 2009) and can be unreliable for proteins with short residence times (Gelbart et al., 2005; Schmiedeberg et al., 2009). We therefore used the DamID technology, which does not require crosslinking or antibodies. With DamID, DNA adenine methyltransferase (Dam) fused to a chromatin protein of interest deposits a stable adenine-methylation ‘footprint’ in vivo at the interaction sites of the chromatin protein, so that even transient interactions may be detected (van Steensel et al., 2001). Note that the fusion protein is expressed at very low levels, averting overexpression artifacts. The DamID profiles of all 53 proteins were generated in duplicate under standardized conditions and detected using oligonucleotide microarrays that query the entire fly genome at ~300 bp intervals. Comparisons to published and new ChIP data confirm the overall reliability of the DamID data (Supplementary Figure 1B), which was also reported in previous comparative studies (Moorman et al., 2006; Negre et al., 2006). For reference purposes, we also generated ChIP maps of histone H3 and the histone marks H3K4me2, H3K9me2, H3K27me3 and H3K79me3 on the same array platform.
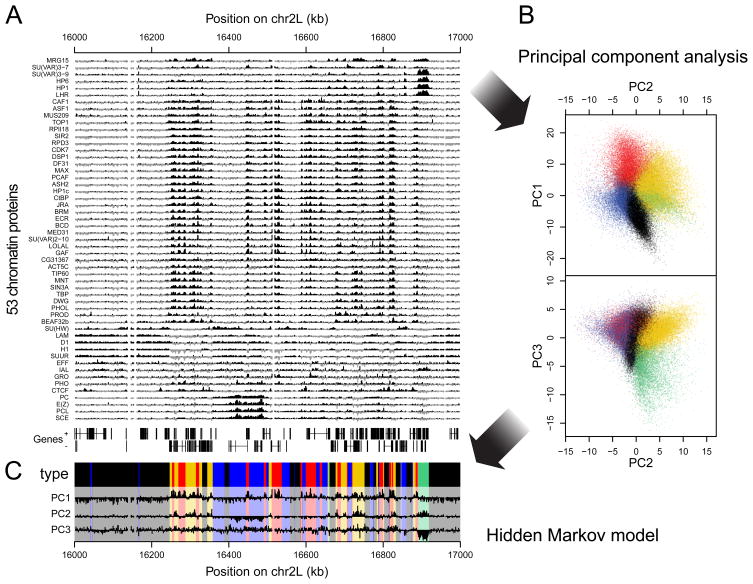
Figure 1.
Overview of protein binding profiles and derivation of the 5-type chromatin segmentation. (A) Sample plot of all 53 DamID profiles (log2 enrichment over Dam-only control). Positive values are plotted in black, negative values in grey for contrast. Below the profiles, genes on both strands are depicted as lines with blocks indicating exons. (B) Two-dimensional projections of the data onto the first three principal components. Colored dots indicate the chromatin type of probed loci as inferred by a 5-state HMM. (C) Values of the first three principal components along the region shown in (A), with domains of the different chromatin types after segmentation by the 5-state HMM highlighted by the same colors as in (B). See also Supplementary Figure S1 and Table S1.
Most of the fly genome interacts with non-histone chromatin proteins
Comparison of the DamID profiles for all 53 proteins shows a variety of binding patterns (Figure 1A). Nevertheless, several sets of proteins exhibit profiles that are similar. Some similarities were anticipated, such as for PC, PCL, SCE and E(Z), which are all PcG proteins (Sparmann and van Lohuizen, 2006); and for HP1, SU(VAR)3-9, LHR and HP6, which are part of classic HP1-type heterochromatin (Greil et al., 2007). We also observe extensive colocalization of Lamin (LAM), histone H1 (H1), Effete (EFF), Suppressor of Underreplication (SUUR) and the AT-hook protein D1, which have not been linked previously except for LAM and SUUR (Pindyurin et al., 2007). There is a prominent overlap in the binding patterns of a large set of approximately 30 proteins including histone modifying enzymes (e.g. RPD3 and SIR2), components of the basal transcription machinery (e.g., CDK7, TBP), and others detailed below.
In order to identify target and non-target loci for each protein, we applied a 2-state Hidden Markov Model (HMM) to each individual binding map (Supplementary Methods). This method identifies the most likely segmentation into “bound” and “unbound” probed loci. According to the resulting binary classifications, the genome-wide occupancy by individual proteins varies broadly, ranging from about 2% (GRO) to 79% (IAL). Interestingly, 99.99% of the probed loci are bound by at least one protein, and 99.6% by at least three proteins. This indicates that, at least at the resolution of our maps, essentially no part of the fly genome is permanently in a configuration that consists of nucleosomes only. Approximately 1% of the genome shows extremely high protein occupancy, being bound by 36 to 44 of the 53 mapped proteins.
Principal chromatin types defined by combinations of proteins
Next, we used a computational classification strategy to identify the major types of chromatin, defined as distinct combinations of proteins that are recurrent throughout the genome. To identify such combinations, we initially performed Principal Component Analysis on the 53 quantitative DamID profiles to reduce the dimensionality of the data. We then focused on the first three principal components, which together account for 57.7% of the total variance. By projecting the genomic sites on the principal components, we could distinguish five distinct lobes in the three-dimensional scatter plot (Figure 1B). No additional distinct lobes could be observed upon further inspection of higher-level principal components. Importantly, the five groups were also clearly separated when using the previously defined binary target definitions (Supplementary Figure 1C), showing that this result is robust to different quantification methods.
Having established that classification into five types properly summarizes the data, we fitted a 5-state HMM onto the first three principal components. Thus, every probed sequence in the genome was assigned one of five exclusive chromatin types (Supplementary Methods). To avoid semantic confusion, and in line with the Greek word chroma (color), we labeled each of the five protein signatures with a color (BLUE, GREEN, BLACK, RED and YELLOW). The HMM classification produced a mosaic pattern of chromosomal domains that vary widely in length (Figure 1C). We emphasize that this segmentation is purely data-driven, without using any other knowledge besides the 53 DamID profiles. The segmentation is generally robust: removal of any of the proteins except for PC still yields a 5-state classification that is on average 96.7% identical to the model obtained with all 53 proteins. A detailed analysis of the robustness is summarized in Supplementary Figure 1D.
Domain organization of chromatin types
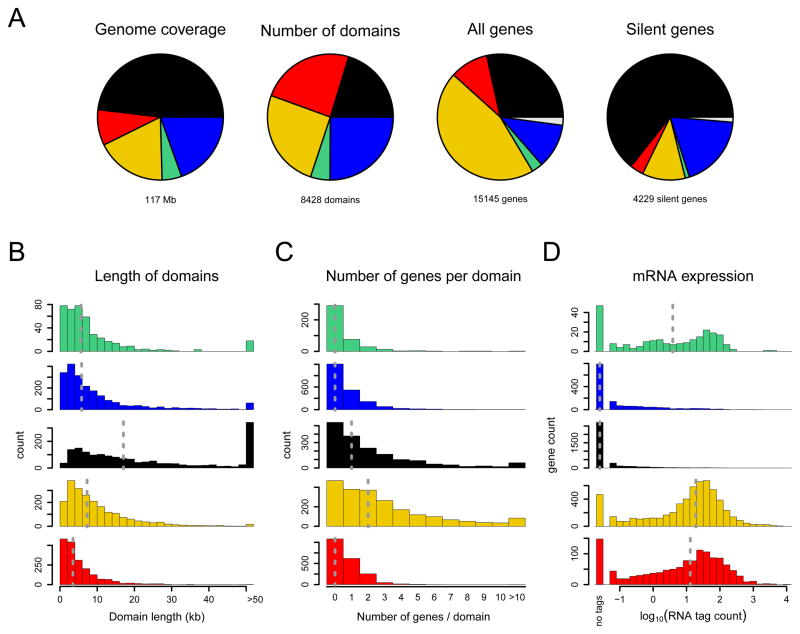
The five types of chromatin differ substantially in their genome coverage, numbers of domains, and numbers of genes (Figure 2A). We identified a total of 8,428 domains that typically range from ~1 to 52 kb (5th-95th percentiles) with a median length of 6.5 kb, although the size distribution depends on the chromatin type (Figure 2B). 441 domains are larger than 50 kb, and 155 are larger than 100 kb, with the largest domain being 737 kb. Many individual domains include multiple neighboring genes (Figure 2C); the largest number of which within a single domain is 139 (for a centromere-proximal GREEN domain). Taken together, these data indicate that the fly genome is generally organized into large regions that are covered by specific combinations of proteins.
Figure 2.
Characteristics of the five chromatin types. (A) Coverage and gene content of chromatin domains of each type. The chromatin type of a gene is defined as the chromatin type at its transcription start site (TSS). Grey sectors correspond to genes whose TSS maps at the transition between two chromatin types. Silent genes have an average RNA tag count below 1 per million total tags (see (D)). (B) Length distribution of chromatin domains, i.e. genomic segments covered contiguously by one chromatin type. (C) Distribution of the number of genes per chromatin domain. Because some genes overlap with more than one domain, genes are assigned to a chromatin type based on the type at the transcription start site. (D) Histogram of mRNA expression determined by RNA tag profiling. Data are represented as log10 (tags per million total tags). Dashed vertical lines in (B)-(D) indicate medians.
BLUE and GREEN chromatin correspond to known heterochromatin types
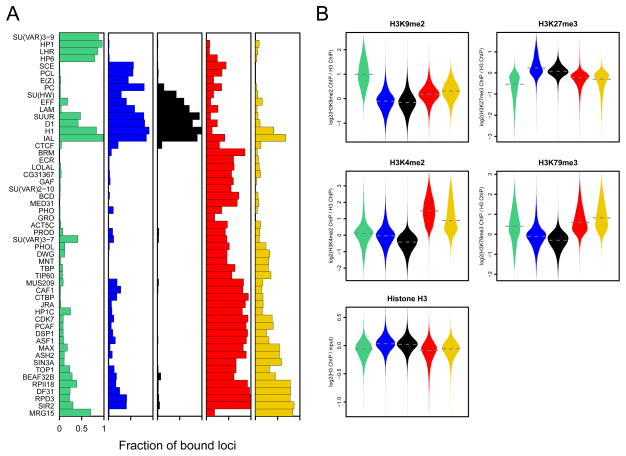
Visualization of the protein occupancy in each of the five chromatin types (Figure 3A) shows that most proteins are not confined to a single chromatin type. Rather, the five chromatin types are defined by unique combinations of proteins. Importantly, BLUE and GREEN chromatin closely resemble previously identified chromatin types. GREEN chromatin corresponds to classic heterochromatin that is marked by SU(VAR)3-9, HP1, and the HP1-interacting proteins LHR and HP6. As described previously (Ebert et al., 2006; Greil et al., 2007), this type of chromatin is prominent in pericentric regions and on chromosome 4 (Supplementary Figure 2A). To further validate this classification, we conducted genome-wide ChIP of H3K9me2, a histone mark that is predominantly generated by SU(VAR)3-9 and bound by HP1 (Hediger and Gasser, 2006) . Indeed, H3K9me2 is highly and specifically enriched in GREEN chromatin (Figure 3B).
Figure 3.
Chromatin types are characterized by distinctive protein combinations and histone modifications. (A) Fraction of all probed genomic loci within each chromatin type that is bound by each protein. Bound loci were determined separately for each protein as described in the text. (B) Levels of histone H3 and four histone modifications as determined by genome-wide ChIP. The distribution of values is shown as “violin plots”, which are symmetrized density plots of binding values per chromatin type: the wider the violin, the more data points are associated to that value. Dashed horizontal lines indicate the median binding value for each chromatin type. Histone modification ChIP data were normalized to H3 occupancy. See also Supplementary Figure S2.
BLUE chromatin corresponds to PcG chromatin as shown by the extensive binding by the PcG proteins PC, E(Z), PCL and SCE. Indeed, well-known PcG target loci such as the Hox gene clusters are localized in BLUE domains (Supplementary Figure 2B). Furthermore, genome-wide ChIP of H3K27me3, the histone mark that is generated by E(Z) and recognized by PC (Sparmann and van Lohuizen, 2006) is highly enriched in BLUE chromatin (Figure 3B). We emphasize that these histone modification profiles serve as independent validation because they were not used in the 5-state HMM classification. The fact that two major well-known chromatin types were faithfully recovered indicates that our chromatin classification strategy is biologically meaningful.
Interestingly, we identified several additional proteins that mark BLUE or GREEN chromatin, or both. For example, moderate degrees of occupancy of the histone deacetylase (HDAC) RPD3 occur in both BLUE and GREEN chromatin, in accordance with known biochemical and genetic interactions of RPD3 with PcG proteins as well as SU(VAR)3-9 (Czermin et al., 2001; Tie et al., 2003). The presence of EFF in BLUE chromatin is consistent with a reported role of this protein in PcG-mediated silencing (Fauvarque et al., 2001).
BLACK chromatin is the prevalent type of repressive chromatin
BLACK chromatin covers 48% of the probed genome and is thus by far the most abundant type (Figure 2A). With a median size of 17 kb and with 134 domains larger than 100 kb, BLACK chromatin domains tend to be longer than domains of the four other types (Figure 2B). BLACK chromatin is overall relatively gene-poor (Figure 2A; compare genome coverage and number of genes), but it nevertheless harbors 4,162 genes.
By mRNA high-throughput sequencing we detected no transcriptional activity (< 1 mRNA molecule per 10 million) for 66% of the genes in BLACK chromatin, while the remaining 34% have very low activity (Figure 2D). This is in agreement with the low coverage of BLACK chromatin by RPII18, a subunit shared by all three RNA polymerases (Figure 3A) and a lack of the active histone marks H3K4me2 and H3K79me3 as detected by ChIP (Figure 3B). We note that the majority of silent genes in the genome are located in BLACK chromatin (Figure 2A). Thus, BLACK chromatin is a distinctively silent type of chromatin that covers a large part of the genome.
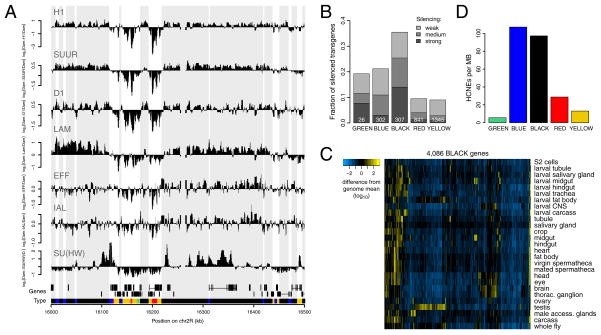
BLACK chromatin is almost universally marked by four of the 53 mapped proteins: histone H1, D1, IAL and SUUR, while SU(HW), LAM and EFF are also frequently present (Figure 3A). Close-up views show that H1, D1, IAL, SUUR and LAM have a broad distribution within BLACK domains, while SU(HW) exhibits a distinct, more focal pattern (Figure 4A).
Figure 4.
Properties of BLACK chromatin. (A) Sample plots of binding profiles of the six proteins that are the most prevalent in BLACK chromatin. Genes on both strands as well as chromatin types are depicted below the profiles. Grey blocks in the background correspond to BLACK chromatin domains. (B) Silencing of a white reporter gene in 2,852 P-element insertions in adult eyes (Babenko et al., 2010) separated by chromatin type in Kc cells. The fraction of silenced insertions is higher among those overlapping with BLACK regions than in the rest of the genome (p<2.2*10−16, Chi-squared test). (C) Relative expression levels (log10 scale, normalized to genome-wide average) of BLACK genes in various tissues (Chintapalli et al., 2007). (D) Density of highly conserved non-coding elements (HCNEs) per chromatin type.
Given that genes in BLACK chromatin are expressed at very low levels, we asked whether BLACK chromatin actively represses transcription, or merely forms secondary to a lack of transcription. In the former model, transgenes inserted into BLACK chromatin may exhibit reduced transcription, while in the latter model transgenes should be unaffected. To test this, we examined a dataset of 2,852 random P-element insertions that carry a mini-white eye color reporter gene. For each of these insertions the expression level was previously scored and the integration site mapped (Babenko et al., 2010). Strikingly, of 307 insertions located in BLACK regions 36% exhibited various degrees of w silencing, compared to 13% genome-wide (Figure 4B). Moreover, repression of transgene insertions in BLACK chromatin is more pronounced than in BLUE and GREEN chromatin. This result strongly indicates that BLACK chromatin has an active role in transcriptional silencing.
Developmental regulation of genes in BLACK chromatin
Not all genes in BLACK regions are expected to remain silenced in various tissues. Indeed, a survey of tissue expression profiling data (Chintapalli et al., 2007) indicates that genes in BLACK chromatin can become active, although their expression tends to be restricted to a few tissues only (Figure 4C). This suggests that BLACK chromatin domains as defined in Kc167 cells can be remodeled into a different chromatin type in some cell types. Consistent with this dynamic regulation, BLACK chromatin is particularly rich in highly conserved non-coding elements (HCNEs) (Figure 4D), which are thought to mediate gene regulation (Engstrom et al., 2007). The density of HCNEs in BLACK chromatin is comparable to that in BLUE chromatin, which harbors many developmentally regulated genes (Tolhuis et al., 2006), and is much higher than in the other three chromatin types. Together, these data suggest that BLACK chromatin is at least in part under developmental control.
YELLOW and RED chromatin are two distinct types of euchromatin
In contrast to BLACK and BLUE chromatin, RED and YELLOW chromatin have hallmarks of transcriptionally active euchromatin: Most genes in these two chromatin types produce substantial amounts of mRNA (Figure 2D), and levels of RNA polymerase (Figure 3A), H3K4me2 and H3K79me3 are typically high, whereas levels of H3K9me2 and H3K27me3 are low (Figure 3B).
RED and YELLOW chromatin share various chromatin proteins (Figure 3A). Among these are the HDACs RPD3 and SIR2, as well as the RPD3-interacting protein SIN3A. HDACs have recently also been found in transcriptionally active chromatin in human cells (Wang et al., 2009). Other proteins that are highly abundant in both RED and YELLOW chromatin include DF31, a little-studied protein that drives chromatin decondensation in vitro (Crevel et al., 2001); ASH2, a homolog of a subunit of a H3K4 methyltransferase complex in yeast and vertebrate cells (Nagy et al., 2002); and MAX, a DBF that is part of the MYC network of regulators of growth and proliferation (Orian et al., 2003).
Besides these similarities, RED and YELLOW chromatin display striking differences. RED chromatin is abundantly marked by several proteins that are mostly absent from the four other chromatin types (Figure 3A). Among these are the nucleosome remodeling ATPase Brahma (BRM); the regulator of chromosome structure SU(VAR)2-10; the Mediator subunit MED31; the 55 kDa subunit of CAF1, present in various histone-modifying complexes (Martinez-Balbas et al., 1998; Tie et al., 2001); and several DBFs including the ecdysone receptor (ECR), GAGA factor (GAF), and Jun-related antigen (JRA).
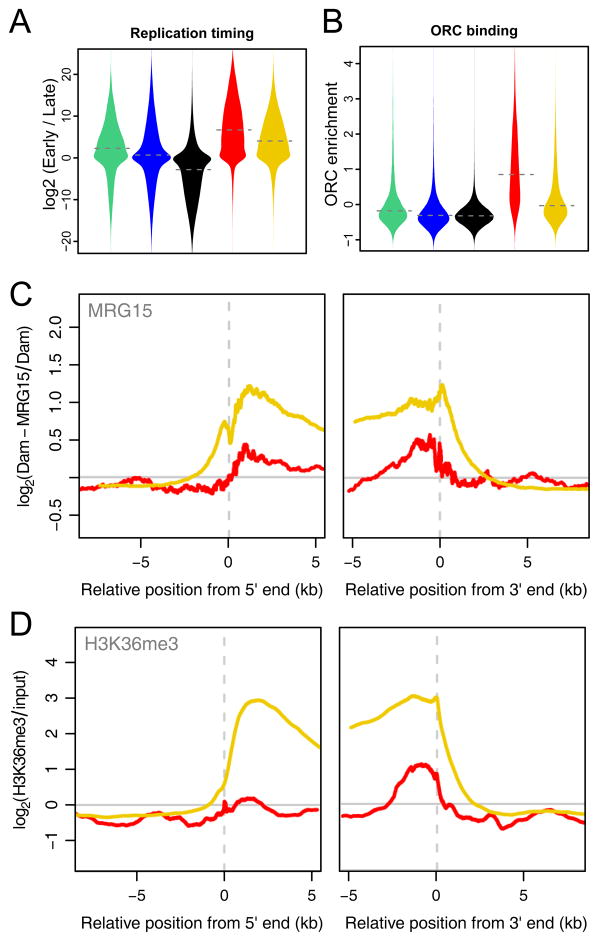
These differences in protein composition prompted us to investigate the timing of DNA replication during S-phase, which is known to differ in relation with chromatin marks (Gilbert, 2002). Analysis of a genome-wide replication timing map from Kc167 cells (Schwaiger et al., 2009) shows that DNA in RED and YELLOW chromatin is generally replicated early in S-phase, as may be expected for euchromatin. However, RED chromatin tends to be replicated even earlier than YELLOW chromatin (Figure 5A). This coincides with a strong enrichment of origin recognition complex (ORC) binding in RED chromatin as mapped by ChIP (MacAlpine et al., 2010) (Figure 5B), suggesting that DNA replication is often initiated in RED chromatin. These observations further underscore that RED and YELLOW chromatin are distinct types of euchromatin.
Figure 5.
RED and YELLOW are two distinct types of euchromatin. (A) Violin plots of replication timing (Schwaiger et al., 2009) per chromatin type. (B) Violin plots of origin of replication complex 2 (ORC2) binding (MacAlpine et al., 2010) per chromatin type. (C) Average binding of MRG15 around 5′ and 3′ ends of genes in RED and YELLOW chromatin. Left panel, alignment to transcript 5′ ends; right panel, alignment to 3′ ends. Only genes that are entirely within one chromatin type are depicted. (D) Average enrichment of H3K36me3 (Bell et al., 2010), plotted as in (C).
Active genes in YELLOW but not RED chromatin carry H3K36me3
Only one protein of the dataset is abundant in YELLOW but not in RED chromatin: MRG15, which is a chromodomain-containing protein. Because human MRG15 has previously been reported to bind H3K36me3 (Zhang et al., 2006), we compared the fine distribution of MRG15 and H3K36me3 along genes within the two chromatin types (Bell et al., 2010). Indeed, both are highly enriched along genes in YELLOW chromatin, but nearly absent from RED chromatin (Figure 5C, D). These data are consistent with binding of MRG15 to H3K36me3 in vivo. Interestingly, H3K36me3 was previously thought to be a universal marker of elongating transcription units (Lee and Shilatifard, 2007; Rando and Chang, 2009). Our analysis reveals that, at least in Drosophila Kc167 cells, this histone mark is mostly absent from genes lying in RED chromatin, even though these genes are expressed at similar levels as genes in YELLOW chromatin (Figure 2D).
RED and YELLOW chromatin mark different types of genes
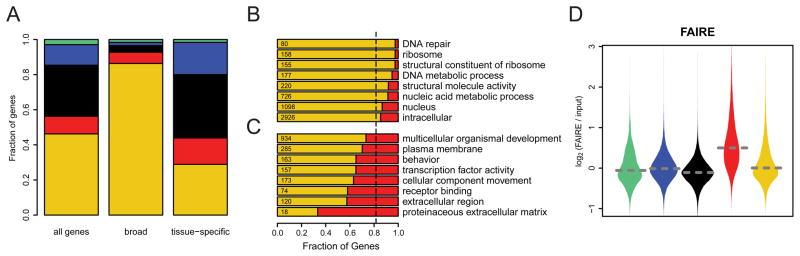
The substantial differences between RED and YELLOW chromatin suggested that the genes they harbor may be regulated by two globally distinct pathways. We therefore investigated whether genes located in RED and YELLOW chromatin have different characteristics. We began by comparing the embryonic tissue expression patterns of genes in the two chromatin types. Strikingly, genes with a broad expression pattern over many embryonic stages and tissues (Tomancak et al., 2007) are highly enriched in YELLOW chromatin, while genes with more restricted expression patterns are depleted (Figure 6A). Consistent with this, Gene Ontology (GO) analysis revealed that universal cellular functions such as “ribosome”, “DNA repair” and “nucleic acid metabolic process” are almost exclusively found in YELLOW chromatin (Figure 6B), while genes in RED chromatin are linked to more specific processes such as “receptor binding”, “defense response”, “transcription factor activity” and “signal transduction” (Figure 6C). Such specific functions and expression patterns require complex mechanisms of gene regulation. Indeed, intergenic regions in RED domains contain about twofold more HCNEs than YELLOW chromatin (Figure 4D), although not as much as BLACK and BLUE chromatin. Furthermore, genome wide formaldehyde-assisted identification of regulatory elements (FAIRE) (Braunschweig et al., 2009; Giresi et al., 2007) points to a high density of regulatory chromatin complexes in RED chromatin (Figure 6D).
Figure 6.
Genes in RED and YELLOW differ in regulation and function. (A) Distribution of genes having “broad” and “tissue-specific” expression patterns (defined in (Tomancak et al., 2007)) over the five chromatin types. Left bar shows distribution of all genes for comparison. (B)-(C) GO slim categories that are significantly enriched (B) or depleted (C) in RED compared to YELLOW genes. Bars indicate the fraction of RED and YELLOW genes for the given category (BLACK, GREEN and BLUE are not considered here). Vertical dotted line represents the distribution expected by random chance. The total number of RED and YELLOW genes within each category are indicated on the left. (D) Violin plots of the log2 FAIRE signal per chromatin type (Braunschweig et al., 2009). See also Supplementary Figure S3.
Motif binding by DBFs is guided by chromatin types
Chromatin can affect the ability of DBFs to bind to their cognate binding sequences, which is thought to explain why in vivo most DBFs bind to only a small subset of their recognition motifs in the genome (Beato and Eisfeld, 1997). We investigated how the five chromatin types might modulate DBF-DNA interactions. We focused on five DBFs in our dataset (JRA, MNT, GAF, CTCF and SU(HW)) for which the sequence-specificity is well-characterized. We first calculated the expected genomic binding pattern of each DBF, based on the occurrence of sequence motifs that match the known DBF recognition motif. The exactness of these matches is taken into account, yielding for each DamID-probed locus a predicted relative affinity for the DBF (Foat et al., 2006). Genome-wide comparison of this sequence-based predicted affinity and actual protein occupancy indicated only weak to moderate correlations (Spearman’s rho ranging from 0.04 to 0.35; dashed grey curves in Figure 7A; Supplementary Figure 4). This suggests that chromatin indeed has substantial modulating effects on DBF-motif interactions.
Figure 7.
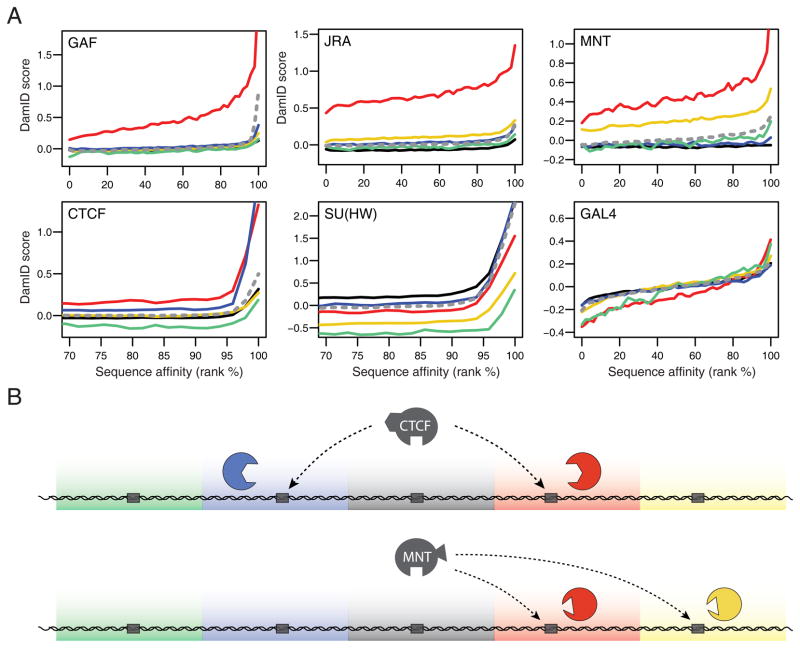
Binding of DBFs to their cognate motifs is differentially guided by chromatin types. (A) Correlations between predicted DNA affinity and actual binding detected by DamID, genome-wide (grey dashed lines) or for each chromatin type (solid lines), for six DBFs as indicated. Curves are loess-fitted lines; raw data is shown in Supplementary Figure S4. (B) Cartoon model depicting the specific guidance of DBFs to their cognate motifs in only certain chromatin types, illustrated for CTCF and MNT. DBF binding to its cognate motif (grey box) is guided by protein-protein interactions. The presence of specific interactors (colored shapes) only in some chromatin types may account for targeting. See also Supplementary Figure S4.
We then repeated this correlation analysis by chromatin type. Surprisingly, this revealed that each DBF has its own dependence on chromatin context (Figure 7A and Supplementary Figure 4): GAF and JRA both bind to their respective motif variants over a range of affinities in RED chromatin, but not in the other chromatin types; MNT binds to its motifs only in RED and YELLOW; CTCF preferentially binds its motifs in RED and BLUE chromatin; SU(HW) recognizes its motifs most efficiently in BLACK, BLUE and RED chromatin. Thus, each of the five chromatin types is conducive to DNA binding by specific subsets of DBFs. Some chromatin types may also weakly bind certain DBFs independently of DNA interactions, as suggested by the varying DamID baseline levels in loci that lack high-affinity motifs (e.g. for SU(HW) and CTCF; Figure 7A).
Four out of five DBFs exhibit a preference for their motif in RED chromatin. We wondered whether RED chromatin might have an intrinsic property such as ‘openness’ or nucleosome remodeling activity that would generally facilitate DBF access. To test this, we generated a DamID profile for the DNA-binding domain (DBD) of yeast Gal4. This foreign DBD is not expected to have specific protein-protein interactions with Drosophila chromatin, and its recognition motif occurs randomly throughout the fly genome. We observed similar interactions of Gal4-DBD with its cognate motifs in all five chromatin types (Figure 7A, bottom right panel). This indicates that RED chromatin does not have a general positive effect on protein-DNA interactions, and that high DBF occupancy in this chromatin type is more likely due to specific targeting mechanisms for each DBF. In summary, these results indicate that the five chromatin types together act as guides that help to target DBFs to specific regions of the genome, even though the cognate binding motifs are broadly distributed (Figure 7B).
DISCUSSION
By systematic integration of 53 protein location maps we found that the Drosophila genome is packaged into a mosaic of five principal chromatin types, each defined by a unique combination of proteins. Extensive evidence demonstrates that the five types differ in a wide range of characteristics besides protein composition, such as biochemical properties, transcriptional activity, histone modifications, replication timing, DBF targeting, as well as sequence properties and functions of the embedded genes. This validates our classification by independent means and provides important insights into the functional properties of the five chromatin types.
The number of chromatin states
Identifying five chromatin states out of the binding profiles of 53 proteins comes out as a surprisingly low number (one can form approximately 1016 subsets of 53 elements). We emphasize that the five chromatin types should be regarded as the major types. Some may be further divided into sub-types, depending on how fine-grained one wishes the classification to be. For example, within each of the transcriptionally active chromatin types, promoters and 3′ ends of genes exhibit (mostly quantitative) differences in their protein composition (data not shown) and thus could be regarded as distinct sub-types. However, these local differences are minor relative to the differences between the five principal types that we describe here. We cannot exclude that the accumulation of binding profiles of additional proteins would reveal other novel chromatin types. We also anticipate that the pattern of chromatin types along the genome will vary between cell types. For example, many genes that are embedded in BLACK chromatin (defined in Kc167 cells) are activated in some other cell types (Figure 4C). Thus, the chromatin of these genes is likely to switch to an active type.
While the integration of data for 53 proteins provides substantial robustness to the classification of chromatin along the genome, a subset of only five marker proteins (histone H1, PC, HP1, MRG15 and BRM), which together occupy 97.6% of the genome, can recapitulate this classification with 85.5% agreement (Supplementary Figure 1E). Assuming that no unknown additional principal chromatin types exist in some cell types, DamID or ChIP of this small set of markers may thus provide an efficient means to examine the distribution of the five chromatin types in various cells and tissues, with acceptable accuracy.
BLACK chromatin: a distinct type of repressive chromatin
Previous work on the expression of integrated reporter genes (Handler and Harrell, 1999; Kelley and Kuroda, 2003; Markstein et al., 2008) had suggested that most of the fly genome is transcriptionally repressed, contrasting with the low coverage of PcG and HP1-marked chromatin. BLACK chromatin, which consists of a previously unknown combination of proteins and covers about half of the genome, may account for these observations. Essentially all genes in BLACK chromatin exhibit extremely low expression levels, and transgenes inserted in BLACK chromatin are frequently silenced, indicating that BLACK chromatin constitutes a strongly repressive environment. Importantly, BLACK chromatin is depleted of PcG proteins, HP1, SU(VAR)3-9 and associated proteins, and is also the latest to replicate, underscoring that it is different from previously characterized types of heterochromatin (here identified as BLUE and GREEN chromatin).
The proteins that mark BLACK domains provide important clues to the molecular biology of this type of chromatin. Loss of LAM, EFF or histone H1 causes lethality during Drosophila development (Cenci et al., 1997; Lenz-Bohme et al., 1997; Lu et al., 2009). Extensive in vitro and in vivo evidence has suggested a role for H1 in gene repression, most likely through stabilization of nucleosome positions (Laybourn and Kadonaga, 1991; Wolffe and Hayes, 1999; Woodcock et al., 2006). The enrichment of LAM points to a role of the nuclear lamina in gene regulation in BLACK chromatin (Pickersgill et al., 2006), consistent with the long-standing notion that peripheral chromatin is silent (Towbin et al., 2009). Depletion of LAM causes derepression of several LAM-associated genes (Shevelyov et al., 2009), while artificial targeting of genes to the nuclear lamina can reduce their expression (Finlan et al., 2008; Reddy et al., 2008), suggesting a direct repressive contribution of the nuclear lamina in BLACK chromatin. D1 is a little-studied protein with 11 AT-hook domains. Overexpression of D1 causes ectopic pairing of intercalary heterochromatin (Smith and Weiler, 2010), suggesting a role in the regulation of higher-order chromatin structure. SUUR specifically regulates late replication on polytene chromosomes (Zhimulev et al., 2003), which is of interest because BLACK chromatin is particularly late-replicating. EFF is highly similar to the yeast and mammalian ubiquitin ligase Ubc4 that mediates ubiquitination of histone H3 (Liu et al., 2005; Singh et al., 2009), raising the possibility that nucleosomes in BLACK chromatin may carry specific ubiquitin marks. These insights suggest that BLACK chromatin is important for chromosome architecture as well as gene repression and provide important leads for further study of this previously unknown yet prevalent type of chromatin.
RED and YELLOW: distinct types of euchromatin
In RED and YELLOW chromatin most genes are active, and the overall expression levels are similar between these two chromatin types. However, RED and YELLOW chromatin differ in many respects. One of the conspicuous distinctions is the disparate levels of H3K36me3 at active transcription units. This histone mark is thought to be laid down in the course of transcription elongation and may block the activity of cryptic promoters inside the transcription unit (Li et al., 2007). Why active genes in RED chromatin lack H3K36me3 remains to be elucidated.
The remarkably high protein occupancy in RED chromatin suggests that RED domains are “hubs” of regulatory activity. This may be related to the predominantly tissue-specific expression of genes in RED chromatin, which presumably requires many regulatory proteins. We note that our DamID assay integrates protein binding events over nearly 24 hours, so it is likely that not all proteins bind simultaneously; some proteins may bind only during a specific stage of the cell cycle. It is highly unlikely that the high protein occupancy in RED chromatin originates from an artifact of DamID, e.g. caused by a high accessibility of RED chromatin. First, all DamID data are corrected for accessibility using parallel Dam-only measurements. Second, several proteins, such as EFF, SU(VAR)3-9 and histone H1 exhibit lower occupancies in RED than in any other chromatin type. Third, ORC also shows a specific enrichment in RED chromatin, even though it was mapped by ChIP, by another laboratory and on another detection platform (MacAlpine et al., 2010). Fourth, DamID of Gal4-DBD does not show any enrichment in RED chromatin.
RED chromatin resembles DBF binding hotspots that were previously discovered in a smaller-scale study in Drosophila cells (Moorman et al., 2006). Discrete genomic regions targeted by many DBFs have recently also been found in mouse ES cells (Chen et al., 2008), hence it is tempting to speculate that an equivalent of RED chromatin may also exist in mammalian cells. Housekeeping and dynamically regulated genes in budding yeast also exhibit a dichotomy in chromatin organization (Tirosh and Barkai, 2008) which may be related to our distinction between YELLOW and RED chromatin. The observations that RED chromatin is generally the earliest to replicate and strongly enriched in ORC binding, suggest that this chromatin type may be not only involved in transcriptional regulation but also in the control of DNA replication.
Chromatin types as guides for DBF targeting
Our analysis of DBF binding indicates that the five chromatin types together act as a guidance system to target DBFs to specific genomic regions. This system directs DBFs to certain genomic domains even though the DBF recognition motifs are more widely distributed. We propose that targeting specificity is at least in part achieved through interactions of DBFs with particular partner proteins that are present in some of the five chromatin types but not in others (Figure 7B). The observation that yeast Gal4-DBD binds its motifs with nearly equal efficiency in all five chromatin types suggests that differences in compaction among the chromatin types represent overall a minor factor in the targeting of DBFs. Although additional studies will be needed to further investigate the molecular mechanisms of DBF guidance, the identification of five principal types of chromatin provides a firm basis for future dissection of the roles of chromatin organization in global gene regulation.
MATERIAL & METHODS
Constructs
DamID constructs used for this study are listed in Supplementary Table 1. New constructs were cloned by TOPO cloning and GATEWAY recombination as described (Braunschweig et al., 2009) or by Cre-mediated recombination. For the latter we generated an acceptor vector containing the Hsp70 promoter upstream of myc-epitope tagged Dam, using the Creator Acceptor Vector Construction Kit (Clontech, 631618). Chromatin protein open reading frames from pDNR-Dual donor vectors (Drosophila Genomics Resource Center, Bloomington) were cloned into the acceptor vector using the Creator™ DNA Cloning Kit (Clontech PT3460-1). Nuclear localization was checked for all Dam-fusion proteins by immuno-fluorescence microscopy with the 9E10 anti-Myc antibody (Santa Cruz Biotechnology) after heat-shock induced expression as described (Greil et al., 2007). Only MNT, GRO and IAL gave weak nuclear signals but were not discarded because MNT and GRO were successfully mapped by DamID in previous studies (Bianchi-Frias et al., 2004; Orian et al., 2003) and IAL binds metaphase chromosomes (Giet and Glover, 2001).
DamID, ChIP and microarrays
DamID assays were carried out under standardized conditions as described previously (Moorman et al., 2006) with a minor modification: proteins were grouped in sets sharing the same Dam-only controls for hybridization purposes. For each group, 3-5 DamID assays on Dam alone were carried out in parallel, the product of which was pooled before labeling. ChIP and subsequent linear amplification reactions were done as described (Kind et al., 2008) using anti-H3K27me3 (07-449) and anti-H3K4me2 (07-030) from Upstate Biotechnology; anti-H3K9me2 (1220), and anti-H3 (1791) from Abcam; affinity-purified anti-H1 serum (Braunschweig et al., 2009); and anti-H3K79me3 (Schubeler et al., 2004) kindly provided by Fred van Leeuwen. Fluorescent labeling of DamID and ChIP samples and two-color hybridizations on custom-designed 385k NimbleGen arrays (Braunschweig et al., 2009) were performed according to NimbleGen’s array users guide, version 4.0. Arrays were scanned at 5 μm resolution, and raw data extracted using NimbleScan software. The identity of the hybridized material was tracked by the presence of unique oligonucleotide spikes in each sample. Furthermore, because the Dam-fusion expression vectors are produced in Dam-positive bacteria, small amounts of the transfected plasmids are co-amplified in the methylation-specific amplification protocol. This leads to a strong signal in the open reading frame of the mapped protein, which allows us to verify the identity of the used vector from the microarray data alone. This open reading frame was masked before further data analysis.
Digital gene expression
Total RNA was isolated from growing Kc cells using TriZOL (Invitrogen), and remaining DNA was degraded by shearing and DNaseI digestion. Poly(A) RNA tag sequencing was carried out on an Illumina Solexa GAII using the tag profiling kit with DpnII. Two RNA samples yielded 7.4 and 9.0 million reads. Tags were mapped by BLAST, requiring at most 2 mismatches and 11 consecutively matching bases. Only the tags mapping to the last GATC of a transcript (FlyBase release 5.8) were counted and represented 70.3% and 69.4% of the total number of reads, respectively. Counts were normalized to the total number of reads and replicates were averaged.
Data availability and analysis
DamID, ChIP and expression data, binarized DamID data and a list of the coordinates of all identified chromatin domains are available from NCBI’s Gene Expression Omnibus, accession number GSE22069. Computational methods are described in the Supplementary Methods.
Supplementary Material
Acknowledgments
We thank Francesco Russo for help with vector cloning; Marja Nieuwland and Arno Velds for help with RNA tag sequencing; Dirk Schübeler’s laboratory for sharing H3K36 methylation data prior to publication; Reuven Agami, Fred van Leeuwen, Wouter Meuleman, Ludo Pagie and Aleksey Pindyurin for helpful suggestions. Supported by an EMBO Long-term Fellowship to J.K.; National Institutes of Health grants T32GM008798, R01HG003008, and U54CA121852 to L.D.W. and H.J.B.; and grants from the Netherlands Genomics Initiative, NWO-ALW VICI and an EURYI Award to B.v.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Babenko VN, Makunin IV, Brusentsova IV, Belyaeva ES, Maksimov DA, Belyakin SN, Maroy P, Vasil’eva LA, Zhimulev IF. Paucity and preferential suppression of transgenes in late replication domains of the D. melanogaster genome. BMC Genomics. 2010;11:318. doi: 10.1186/1471-2164-11-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beato M, Eisfeld K. Transcription factor access to chromatin. Nucleic Acids Res. 1997;25:3559–3563. doi: 10.1093/nar/25.18.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell O, Schwaiger M, Oakeley EJ, Lienert F, Beisel C, Stadler MB, Schubeler D. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat Struct Mol Biol. 2010;17:894–900. doi: 10.1038/nsmb.1825. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bianchi-Frias D, Orian A, Delrow JJ, Vazquez J, Rosales-Nieves AE, Parkhurst SM. Hairy transcriptional repression targets and cofactor recruitment in Drosophila. PLoS Biol. 2004;2:E178. doi: 10.1371/journal.pbio.0020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Hogan GJ, Pagie L, van Steensel B. Histone H1 binding is inhibited by histone variant H3.3. Embo J. 2009;28:3635–3645. doi: 10.1038/emboj.2009.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci G, Rawson RB, Belloni G, Castrillon DH, Tudor M, Petrucci R, Goldberg ML, Wasserman SA, Gatti M. UbcD1, a Drosophila ubiquitin-conjugating enzyme required for proper telomere behavior. Genes Dev. 1997;11:863–875. doi: 10.1101/gad.11.7.863. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Collas P. The state-of-the-art of chromatin immunoprecipitation. Methods Mol Biol. 2009;567:1–25. doi: 10.1007/978-1-60327-414-2_1. [DOI] [PubMed] [Google Scholar]
- Crevel G, Huikeshoven H, Cotterill S. Df31 is a novel nuclear protein involved in chromatin structure in Drosophila melanogaster. J Cell Sci. 2001;114:37–47. doi: 10.1242/jcs.114.1.37. [DOI] [PubMed] [Google Scholar]
- Czermin B, Schotta G, Hulsmann BB, Brehm A, Becker PB, Reuter G, Imhof A. Physical and functional association of SU(VAR)3-9 and HDAC1 in Drosophila. EMBO Rep. 2001;2:915–919. doi: 10.1093/embo-reports/kve210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Greil F, van Steensel B. High-resolution mapping reveals links of HP1 with active and inactive chromatin components. PLoS Genet. 2007;3:e38. doi: 10.1371/journal.pgen.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14:377–392. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- Engstrom PG, Ho Sui SJ, Drivenes O, Becker TS, Lenhard B. Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome Res. 2007;17:1898–1908. doi: 10.1101/gr.6669607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauvarque MO, Laurenti P, Boivin A, Bloyer S, Griffin-Shea R, Bourbon HM, Dura JM. Dominant modifiers of the polyhomeotic extra-sex-combs phenotype induced by marked P element insertional mutagenesis in Drosophila. Genet Res. 2001;78:137–148. doi: 10.1017/s0016672301005274. [DOI] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foat BC, Morozov AV, Bussemaker HJ. Statistical mechanical modeling of genome-wide transcription factor occupancy data by MatrixREDUCE. Bioinformatics. 2006;22:e141–149. doi: 10.1093/bioinformatics/btl223. [DOI] [PubMed] [Google Scholar]
- Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 2005;19:942–954. doi: 10.1101/gad.1298905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R, Glover DM. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol. 2001;152:669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM. Replication timing and transcriptional control: beyond cause and effect. Curr Opin Cell Biol. 2002;14:377–383. doi: 10.1016/s0955-0674(02)00326-5. [DOI] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greil F, de Wit E, Bussemaker HJ, van Steensel B. HP1 controls genomic targeting of four novel heterochromatin proteins in Drosophila. Embo J. 2007;26:741–751. doi: 10.1038/sj.emboj.7601527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler AM, Harrell RA., 2nd Germline transformation of Drosophila melanogaster with the piggyBac transposon vector. Insect Mol Biol. 1999;8:449–457. doi: 10.1046/j.1365-2583.1999.00139.x. [DOI] [PubMed] [Google Scholar]
- Hediger F, Gasser SM. Heterochromatin protein 1: don’t judge the book by its cover! Curr Opin Genet Dev. 2006;16:143–150. doi: 10.1016/j.gde.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Kelley RL, Kuroda MI. The Drosophila roX1 RNA gene can overcome silent chromatin by recruiting the male-specific lethal dosage compensation complex. Genetics. 2003;164:565–574. doi: 10.1093/genetics/164.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Vaquerizas JM, Gebhardt P, Gentzel M, Luscombe NM, Bertone P, Akhtar A. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell. 2008;133:813–828. doi: 10.1016/j.cell.2008.04.036. [DOI] [PubMed] [Google Scholar]
- Laybourn PJ, Kadonaga JT. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991;254:238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- Lee JS, Shilatifard A. A site to remember: H3K36 methylation a mark for histone deacetylation. Mutat Res. 2007;618:130–134. doi: 10.1016/j.mrfmmm.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Lenz-Bohme B, Wismar J, Fuchs S, Reifegerste R, Buchner E, Betz H, Schmitt B. Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J Cell Biol. 1997;137:1001–1016. doi: 10.1083/jcb.137.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 2007;21:1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Oughtred R, Wing SS. Characterization of E3Histone, a novel testis ubiquitin protein ligase which ubiquitinates histones. Mol Cell Biol. 2005;25:2819–2831. doi: 10.1128/MCB.25.7.2819-2831.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Wontakal SN, Emelyanov AV, Morcillo P, Konev AY, Fyodorov DV, Skoultchi AI. Linker histone H1 is essential for Drosophila development, the establishment of pericentric heterochromatin, and a normal polytene chromosome structure. Genes Dev. 2009;23:452–465. doi: 10.1101/gad.1749309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, MacAlpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2010;20:201–211. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Tsukiyama T, Gdula D, Wu C. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc Natl Acad Sci U S A. 1998;95:132–137. doi: 10.1073/pnas.95.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman C, Sun LV, Wang J, de Wit E, Talhout W, Ward LD, Greil F, Lu XJ, White KP, Bussemaker HJ, et al. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc Natl Acad Sci U S A. 2006;103:12027–12032. doi: 10.1073/pnas.0605003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci U S A. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Hennetin J, Sun LV, Lavrov S, Bellis M, White KP, Cavalli G. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 2006;4:e170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian A, van Steensel B, Delrow J, Bussemaker HJ, Li L, Sawado T, Williams E, Loo LW, Cowley SM, Yost C, et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- Pindyurin AV, Moorman C, de Wit E, Belyakin SN, Belyaeva ES, Christophides GK, Kafatos FC, van Steensel B, Zhimulev IF. SUUR joins separate subsets of PcG, HP1 and B-type lamin targets in Drosophila. J Cell Sci. 2007;120:2344–2351. doi: 10.1242/jcs.006007. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annu Rev Biochem. 2009;78:245–271. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Schmiedeberg L, Skene P, Deaton A, Bird A. A temporal threshold for formaldehyde crosslinking and fixation. PLoS One. 2009;4:e4636. doi: 10.1371/journal.pone.0004636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M, Stadler MB, Bell O, Kohler H, Oakeley EJ, Schubeler D. Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes Dev. 2009;23:589–601. doi: 10.1101/gad.511809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevelyov YY, Lavrov SA, Mikhaylova LM, Nurminsky ID, Kulathinal RJ, Egorova KS, Rozovsky YM, Nurminsky DI. The B-type lamin is required for somatic repression of testis-specific gene clusters. Proc Natl Acad Sci U S A. 2009;106:3282–3287. doi: 10.1073/pnas.0811933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Kabbaj MH, Paik J, Gunjan A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol. 2009;11:925–933. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MB, Weiler KS. Drosophila D1 overexpression induces ectopic pairing of polytene chromosomes and is deleterious to development. Chromosoma. 2010;119:287–309. doi: 10.1007/s00412-010-0257-x. [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Tie F, Furuyama T, Prasad-Sinha J, Jane E, Harte PJ. The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development. 2001;128:275–286. doi: 10.1242/dev.128.2.275. [DOI] [PubMed] [Google Scholar]
- Tie F, Prasad-Sinha J, Birve A, Rasmuson-Lestander A, Harte PJ. A 1-megadalton ESC/E(Z) complex from Drosophila that contains polycomblike and RPD3. Mol Cell Biol. 2003;23:3352–3362. doi: 10.1128/MCB.23.9.3352-3362.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- Tomancak P, Berman BP, Beaton A, Weiszmann R, Kwan E, Hartenstein V, Celniker SE, Rubin GM. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2007;8:R145. doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, Meister P, Gasser SM. The nuclear envelope--a scaffold for silencing? Curr Opin Genet Dev. 2009;19:180–186. doi: 10.1016/j.gde.2009.01.006. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Delrow J, Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat Genet. 2001;27:304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP, Hayes JJ. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- Zhang P, Du J, Sun B, Dong X, Xu G, Zhou J, Huang Q, Liu Q, Hao Q, Ding J. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 2006;34:6621–6628. doi: 10.1093/nar/gkl989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhimulev IF, Belyaeva ES, Makunin IV, Pirrotta V, Volkova EI, Alekseyenko AA, Andreyeva EN, Makarevich GF, Boldyreva LV, Nanayev RA, et al. Influence of the SuUR gene on intercalary heterochromatin in Drosophila melanogaster polytene chromosomes. Chromosoma. 2003;111:377–398. doi: 10.1007/s00412-002-0218-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DamID, ChIP and expression data, binarized DamID data and a list of the coordinates of all identified chromatin domains are available from NCBI’s Gene Expression Omnibus, accession number GSE22069. Computational methods are described in the Supplementary Methods.