Abstract
We investigated whether nematodes contribute to the persistence, differentiation and amplification of Legionella species in soil, an emerging source for Legionnaires’ disease. Here we show that Legionella spp. colonize the intestinal tracts of Caenorhabditis nematodes leading to worm death. Susceptibility to Legionella is influenced by innate immune responses governed by the p38 mitogen-activated protein kinase and insulin/insulin growth factor-1 receptor signaling pathways. We also show that L. pneumophila colonizes the intestinal tract of nematodes cultivated in soil. To distinguish between transient infection and persistence, plate-fed and soil-extracted nematodes fed fluorescent strains of L. pneumophila were analyzed. Bacteria replicated within the nematode intestinal tract, did not invade surrounding tissue, and were excreted as differentiated forms that were transmitted to offspring. Interestingly, the ultrastructural features of the differentiated bacterial forms were similar to cyst-like forms observed within protozoa, amoeba and mammalian cell lines. While intestinal colonization of L. pneumophila dotA and icmT mutant strains did not alter the survival rate of nematodes in comparison to wild-type strains, nematodes colonized with the dot/icm mutant strains exhibited significantly increased levels of germline apoptosis. Taken together, these studies show that nematodes may serve as natural hosts for these organisms and thereby contribute to their dissemination in the environment and suggest that the remarkable ability of L. pneumophila to subvert host cell signaling and evade mammalian immune responses evolved through the natural selection associated with cycling between protozoan and metazoan hosts.
Keywords: Legionella pneumophila, Legionella longbeachae, Legionella/pathogenicity, Caenorhabditis elegans, Caenorhabditis briggsae, Caenorhabditis/*microbiology, soil, soil microbiology, virulence, disease reservoir, commensalism
Introduction
Legionella pneumophila and related species are facultative intracellular parasites of aquatic protozoa, a lifestyle that is both highly successful and not dependent on human infection. In fact, human infection represents a dead end for these organisms, as they are not transmitted from person to person [for reviews see (Cianciotto, 2001, Steinert et al., 2002, Molofsky et al., 2004)]. L. pneumophila, like close relative Coxiella burnetii, possesses a developmental cycle in which vegetative replicating intracellular forms (RFs) differentiate late in infection into cyst-like planktonic extracellular forms (CFs) that are metabolically dormant, resilient and hyperinfectious (Cirillo et al., 1994, Garduno et al., 2002, Molofsky et al., 2004, Bachman et al., 2004). CFs feature distinct morphological characteristics, including thickened cell wall, intracytoplasmic membranes, and inclusions of poly-β-hydroxybutyrate, that are easily differentiated from RFs (Garduno et al., 2002). The developmental cycle is partly controlled by transcriptional regulators (RpoS, LetA/LetS, PmrAB, IhfAB and OxyR) that activate genes whose products participate in the remodeling of the cell envelope and the delivery of effector molecules to host cells by the Dot/Icm type IV secretion system (Ninio et al., 2007, LeBlanc et al., 2008, Sahr et al., 2009, Rasis et al., 2009, Morash et al., 2009, Dalebroux et al., 2009). The ability of the secreted products of the Dot/Icm system to subvert host cell defences and abrogate phagolysosomal fusion is a hallmark of Legionnaires’ disease (Ninio et al., 2007, Isberg et al., 2009).
While L. pneumophila Philadelphia-1 serogroup 1 (sg1) strains are responsible for nearly 90% of cases of Legionnaires’ disease reported worldwide (Benin et al., 2002), Legionella longbeachae is the leading case of Legionnaires’ disease in Western Australia and Thailand and the third most common cause of Legionnaires’ disease (and second most common cause of community-acquired legionellosis) in the United States (Cameron et al., 1991, Doyle et al., 1998, Benin et al., 2002, Phares et al., 2007). Interestingly, both sporadic cases and outbreaks of Legionnaires’ disease due to L. longbeachae in Australia, Japan, the Netherlands, and the Pacific Northwest (USA) have been associated with exposure to potting soils or composted organic matter (Steele et al., 1990a, Koide et al., 1999, 2000, den Boer et al., 2007). A case of Legionnaires’ disease due to L. pneumophila sg1 has similarly been associated with exposure to potting soil (Wallis et al., 2005). Recently, L. longbeachae was shown to be dimorphic in amoebae, with ultrastructural characteristics similar to those described with L. pneumophila (Montanaro et al., 2005). In most clinical cases where garden or potting soils were suspected, the causative strain was successfully isolated from the soil samples (Steele et al., 1990a, Koide et al., 1999, den Boer et al., 2007, Wallis et al., 2005). However, none of these soil investigations were pursued further, so little is known of the soil ecology that enables these organisms to persist or become sufficiently abundant to be transmitted to humans.
The community of organisms within fertile soil is biologically diverse and includes a wide array of microbes (bacteria, fungi, and protozoa) and predatory invertebrates (Young et al., 2004). Free-living protozoa in moist soils have generally been equated with the species commonly studied in aquatic systems, including the amoeboid protozoa Hartmannella, Acanthamoeba and Naegleria, and ciliate protozoa of the genus Tetrahymena (Steele et al., 1996, Fields, 1996, Shadrach et al., 2005). More recent studies with the free-living social amoeba Dictyostelium discoideum, commonly found in soils, may indicate a greater diversity in potential hosts for species of Legionella than previously considered (Hilbi et al., 2007). While D. discoideum is a predator of Legionella, it can also be prey of C. elegans and presumably other microbiovorous nematodes (Kessin et al., 1996). Given the complex assemblages of organisms found in food webs of terrestrial ecosystems, we have investigated the possibility that nematodes might in some capacity be associated with persistence of Legionella species in soils.
Foraging invertebrates have potent immune defense mechanisms to guard against the constant threat of harm by bacterial pathogens present within the natural environment. C. elegans defense mechanisms include constitutive and inducible innate immune responses and pathogen avoidance behavior (Pujol et al., 2001, Zhang et al., 2005, Ewbank, 2006, Kim, 2008). Two evolutionarily conserved nematode signaling pathways – the NSY-1/SEK-1/PMK-1 p38 mitogen-activated protein kinase (MAPK) pathway and the DAF-2 insulin/IGF-1 receptor signaling pathway – play fundamental roles in innate immunity by acting in parallel to control expression of genes encoding a diverse array of antimicrobial products and, in the case of the p38 MAPK pathway, programmed cell death (PCD) of the germline during infection by Salmonella enterica serovar Typhimurium (Kim et al., 2002, Aballay et al., 2003, Sifri et al., 2003, Garsin et al., 2003, Troemel et al., 2006). Although C. elegans lacks known MyD88 and Rel/NFκB homologs, partial loss-of-function mutation of tol-1, the single Toll-like receptor (TLR) homolog, abrogates pathogen avoidance behavior towards Serratia marcescens and permits tissue invasion by S. Typhimurium (Pujol et al., 2001, Tenor et al., 2008).
While C. elegans can serve as a model system for a diverse array of human microbial pathogens, in few cases is the nematode considered a natural host or environmental reservoir (Anderson et al., 2006). Here we show that L. pneumophila can colonize and persist within the C. elegans digestive tract in laboratory assays and artificial soil environments without invading and replicating within host tissues. Nematodes colonized with L. pneumophila have a moderately shortened lifespan and do not exhibit pathogen avoidance behavior. Similar results were observed with L. longbeachae. Worm tolerance to infection was associated with innate immune responses, as PMK-1 p38 MAP kinase C. elegans mutants were more susceptible to Legionella killing, while stress resistant DAF-2 insulin signaling pathway mutants were less susceptible to infection. Our studies demonstrate that Legionella differentiate into highly infectious cyst forms within the nematode intestinal tract, which are then excreted by the worm. Furthermore, we show that nematodes colonized with icmT and dotA mutant strains have increased levels of germline apoptosis, suggesting the Dot/Icm type IV secretion system interacts with the C. elegans PCD pathway as it does with mammalian cell apoptosis pathways. We suggest that the unique association with metazoans may represent an intermediate in the evolution of host immune evasion traits attributed to the legionellae.
Results
While potting soils and composts have been shown to harbor species of Legionella (Steele et al., 1990a, Steele et al., 1990b, Hughes et al., 1994, Koide et al., 1999, 2000, den Boer et al., 2007, Wallis et al., 2005, Casati et al., 2009a, Casati et al., 2009b) and nutrient-rich organic matter appears to be a natural habitat for many Caenorhabditis species and other free-living nematodes (Barriere et al., 2005, Kiontke et al., 2006, Freckman, 1988), the role of nematodes as natural hosts for legionellae has not been investigated. With the objective of examining the predator-prey relationship between legionellae and a lower metazoan host, wild-type Bristol N2 (herein N2) C. elegans nematodes were fed lawns of L. pneumophila grown on BCYE agar plates. Over the course of 2-3 days, legionellae accumulated within (i.e. “colonized”) the nematode digestive tract but did not invade intestinal epithelial cells. To distinguish between a laboratory phenomenon and a biologically relevant relationship, we established several testable criteria that would define a bona fide host/parasite relationship: 1) bacterial infection shortens lifespan of the worm; 2) bacteria remain viable within the nematode; 3) bacteria alter their metabolism and/or cellular structure to adapt to the intraluminal environment; 4) bacteria are retained in the nematode when worms are fed other bacteria; 5) nematodes recycle bacteria in soils and transmit bacteria to offspring; and 6) the innate immune response of the nematode controls infection.
Legionella kill N2 C. elegans in vitro
To investigate whether Legionella had a detrimental effect on nematode health, an optimized laboratory survival assay was developed. N2 hermaphrodites of the fourth larval stage (L4) were placed on live or heat-killed Legionella-spotted BCYE assay plates and their survival was observed on a daily basis. Both L. pneumophila and L. longbeachae were examined, as the former is the preeminent pathogen of the genus and the latter is especially linked to organic potting composts, a favored environment of C. elegans (Barriere et al., 2005, Kiontke et al., 2006). The median survival time (time in which 50% of the population is alive) was six days for L. longbeachae sg1 strain 33152 and seven days for L. pneumophila Philadelphia-1 and its derivative Lp02 strain, which was considerably shorter than the lifespan of nematodes fed heat-killed Legionella (Fig. 1A). The median survival time of nematodes fed E. coli OP50 on BCYE and on NGM was approximately 8 and 10 days, respectively (data not shown), once again demonstrating that live E. coli is pathogenic to nematodes when grown on rich culture medium rather than minimal culture medium (Garsin et al., 2001).
Figure 1.
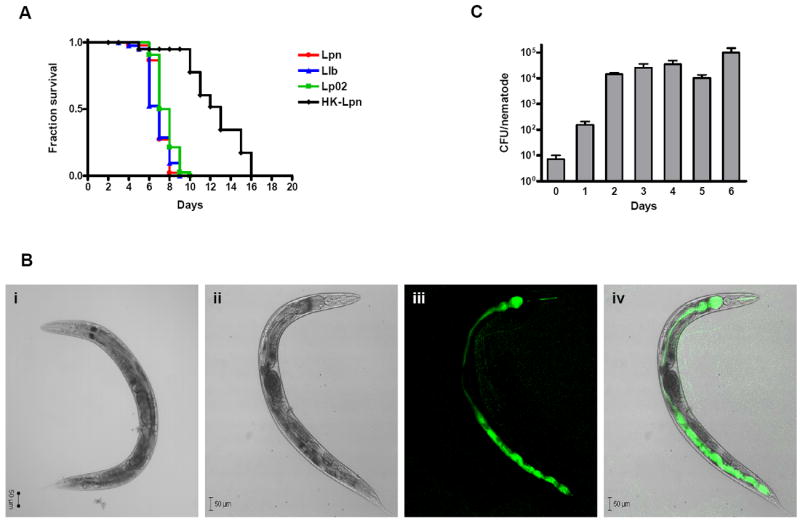
Survival of C. elegans fed on Legionella. (A) Kaplan-Meier survival plots of N2 nematodes fed L. pneumophila Philadelphia-1 (Lpn; circles, n=45), L. longbeachae ATCC 33462 (Llb; triangles, n=43), L. pneumophila Philadelphia-1 derivative Lp02 strain (squares; n=44), and heat-killed L. pneumophila Philadelphia-1 (HK-Lpn; diamonds, n=28). Nematode loss was not a significant consideration in the survival assay as the nematodes remained mostly within the bacterial lawn. P < 0.0001 by pairwise comparison by the log-rank test of the each of the strains (Lpn, Llb, Lp02) versus heat-killed. P ≤ 0.01 by pairwise comparison by the log-rank test for each of the strains Lpn and Llb vs. Lp02. Data is representative of one of three independent experiments. (B) Confocal microscopy of Legionella colonization and persistence in wild-type N2 nematodes. DIC images of wild-type N2 nematodes fed (i) heat-killed or (ii-iv) live GFP-expressed L. pneumophila KB130 for five days. (ii) Bright-field image using DIC optics; (iii) KB130 producing GFP colonizing the length of intestinal tract; and (iv) merged image of ii and iii. (C) Bacterial intestinal load of L. pneumophila KB130 within wild-type N2 nematodes (CFU/nematode) over a period of six days. Bacterial loads were determined in triplicate from approximately ten disrupted worms. Data represent mean ± SEM.
Multiple Caenorhabditis strains have shortened lifespans on Legionella
N2 is a wild-type, laboratory adapted C. elegans strain. To determine whether susceptibility to infection was restricted to the N2 nematode, several Caenorhabditis wild-type isolates from different regions around the world were examined in the survival assay. Four additional C. elegans strains and one C. briggsae strain also succumbed to infection by L. pneumophila and L. longbeachae (Fig. 2). C. elegans CB4555 and C. briggsae HK104 were moderately less susceptible to killing compared to the other strains surveyed. These results indicate that the detrimental effects of Legionella on worm survival are not limited to the N2 strain.
Figure 2.
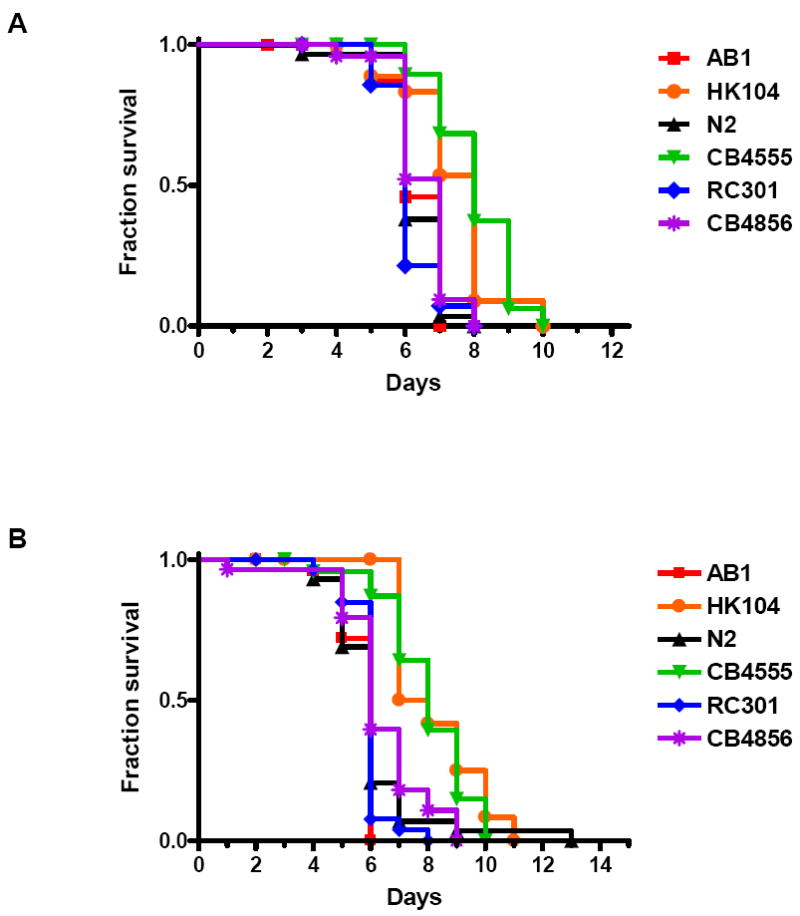
Survival of multiple Caenorhabditis nematodes fed Legionella. (A) Kaplan-Meier survival plot of C. elegans AB1 (squares, n=30), C. briggsae HK104 (circles, n=31), C. elegans N2 (triangles, n=30), C. elegans CB4555 (inverted triangles, n=24), C. elegans RC301 (diamonds, n=15), and C. elegans CB4856 (asterisks, n=26) fed L. pneumophila Philadelphia-1 type strain. (B) Kaplan-Meier survival plot of C. elegans AB1 (squares, n=26), C. briggsae HK104 (circles, n=27), C. elegans N2 (triangles, n=29), C. elegans CB4555 (inverted triangles, n=25), C. elegans RC301 (diamonds, n=30), and C. elegans CB4856 (asterisks, n=29) fed L. longbeachae ATCC 33462. P ≤ 0.0002 by pairwise comparison by the log-rank test of each of the nematode strains (HK104 and CB4555) versus wild-type N2 for both Legionella strains. All other nematode strain pairwise comparisons versus wild-type N2 were found to be statistically insignificant. Data is representative of one of three independent experiments.
Pathology
Previous studies have shown that nontoxogenic killing of C. elegans is typically associated with colonization of the nematode digestive tract with live bacterial or fungal pathogens (Sifri et al., 2005). Similarly, we observed marked distension of the intestinal tracts of all Caenorhabditis strains fed L. pneumophila or L. longbeachae due to the accumulation of large numbers of intact bacteria (Fig. 1B and data not shown). Distension was not observed in animals fed UV-killed L. pneumophila and was delayed for several days in those fed E. coli OP50 (Fig. 1B). We enumerated L. pneumophila bacteria in the digestive tract following mechanical disruption and serial dilution plating over a period of several days (Fig. 1C). On day one, intestinal tract colonization of L. pneumophila numbered at ~102 CFU/nematode and increased to ~104 CFU/nematode by day two and ~105 CFU/nematode by day six.
Several additional attributes of Legionella-infected worms are consistent with a pathogenic process. While some nematode deaths were unremarkable, a significant portion of nematodes developed excessive fluid retention leading to extrusion of their viscera through the vulva. Others died with the well-described “bagging” phenotype, in which eggs of a gravid hermaphroditic adult hatch internally and consume the parent (Sifri et al., 2005). This event appears to occur with more frequency in nematodes fed L. longbeachae than in nematodes fed L. pneumophila.
Do legionellae persist in worms?
Previous studies have demonstrated that digestive tract colonization may either be transient or persistent, depending on pathogen and the “chase” bacteria (Sifri et al., 2005). To investigate whether L. pneumophila transiently or persistently colonizes the nematode digestive tract, N2 nematodes were initially fed mCherry-producing L. pneumophila KB290 for three days (“pulsed”) and then transferred to lawns of GFP-producing L. pneumophila KB130 for an additional five days (“chased”), and persistence of KB290 was tracked by fluorescence microscopy (Table 2). As summarized in Table 2, the small population of mCherry-L. pneumophila KB290 observed in the nematode digestive tract 24 hr post transfer (p.t.) substantially increased by 72 hr p.t. and remained at that level up to 120 hr p.t. Images of a representative nematode 72 hr p.t. are shown in Figure 3. Expansion of KB290 population appears to be the result of intraluminal replication of KB290 and not to overgrowth of KB290 on the transfer lawns, since mCherry-producing bacteria were observed in vanishingly small amounts in the lawn of the chase plates, as is evident by the presence of only GFP-producing Legionella (i.e. no red fluorescent bacteria) in the pharynx of nematodes p.t. (Fig. 3, arrowhead). Likewise, GFP-producing KB130 expanded in the nematode digestive tract after worms were initially fed KB130 and then transferred to mCherry-producing KB290, ruling out the possibility that a difference in bacterial fitness due to GFP vs. mCherry production accounts for selective retention and/or expansion of a bacterial strain within the digestive tract. Surprisingly, bacterial replication appears to occur exclusively within the lumen of the nematode digestive tract, since fluorescent KB130 and KB290 were only rarely observed outside the nematode intestinal tract in antemortem nematodes (≤5 bacteria/worm in <10% of worms examined).
Table 2.
Persistence of mCherry-producing L. pneumophila KB290 in N2 nematodes “pulsed” with KB290 for three days before placement on “chase” bacterial strains for an additional five days.
| Number of hours post-exposure (h) | |||||
|---|---|---|---|---|---|
| Bacterial strain | 24a | 48 | 72 | 96 | 120 |
| E. coli OP50 | +++b | +++ | ++ | + | + |
| GFP-L. pneumophila KB130 | + | + | +++ | +++ | +++ |
Five worms were randomly selected from each bacterial strain for fluorescent microscopic analysis.
Qualitative observations of the fluorescence intensity level with + representing the least intensity and +++ representing the most intensity.
Figure 3.
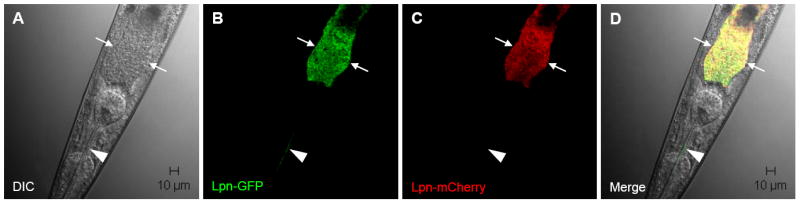
Confocal microscopy of Legionella colonization and persistence in wild-type N2 nematodes. Wild-type worms were “pulsed” with mCherry-producing KB290 for three days and “chased” with GFP-producing KB130. Representative images after three days of exposure to KB130 are shown. (A) Bright-field image using DIC optics; (B) KB130 producing GPF in the intestinal and pharyngeal lumens; (C) KB290 producing mCherry in the intestinal lumen; and (D) merged images of A, B and C. Arrows demarcate the intestinal tract lumen; arrowhead indicates intact KB130 producing GFP in the pharyngeal lumen.
By contrast, the number of KB290 or KB130 remained low but stable in nematodes initially exposed to the fluorescent L. pneumophila and then transferred to E. coli OP50. These results suggest that OP50, the laboratory food source for C. elegans, directly or indirectly alters the digestive tract environment to negatively influence L. pneumophila retention and/or replication. It should be noted that we found no evidence that OP50 was directly toxic to L. pneumophila in vitro. Other studies have also shown that feeding on OP50 can lead to the removal of most, but not all, of a persistent population of bacterial pathogens in the nematode digestive tract (Aballay et al., 2000).
Microscopic (fluorescence and TEM) analyses of Legionella within nematodes on BCYE assay plates indicate ultrastructural features similar to cyst-like forms
Fluorescent microscopic examination of nematodes fed KB130 or KB290 indicated that bacteria expelled from the anus via rhythmic peristalsis consisted of a mixture of motile short rods, characteristic of stationary phase forms, and motile coccoid forms, suggestive of terminally differentiated CFs. Of note, the fluorescent protein fusion gene fusion constructs of strains KB130 and KB290 employ the promoter of the magA gene, which is selectively induced during the later stages of intracellular growth of L. pneumophila (Hiltz et al., 2004). Since the L. pneumophila does not differentiate into CFs on BCYE agar (Garduno et al., 2002) we investigated the possibility that Legionella differentiation is induced within the worm intestinal tract. First, thin sections of BCYE agar grown L. pneumophila were prepared and analyzed by TEM. The bacteria exhibited a thin Gram negative cell wall structure and paucity of inclusion granules of poly-β-hydroxybutyrate (PHBA) characteristic of vegetative Legionella (Fig. 4A) (Garduno et al., 2002). By comparison, ingested L. longbeachae, viewed in a cross-section of the pharynx of a wild-type N2 nematode, is composed of a mixture of CFs, evident by abundant PHBA inclusions, as well as vegetative forms (Fig. 4B). TEM analysis of the intestinal tract of C. elegans fed L. pneumophila also revealed intraluminal bacteria with abundant PHBA granules (Fig. 4C-E). At higher magnification, the multiple laminar membranes observed in CFs that give the appearance of a thickened cell wall are clearly evident (Fig. 4F).
Figure 4.
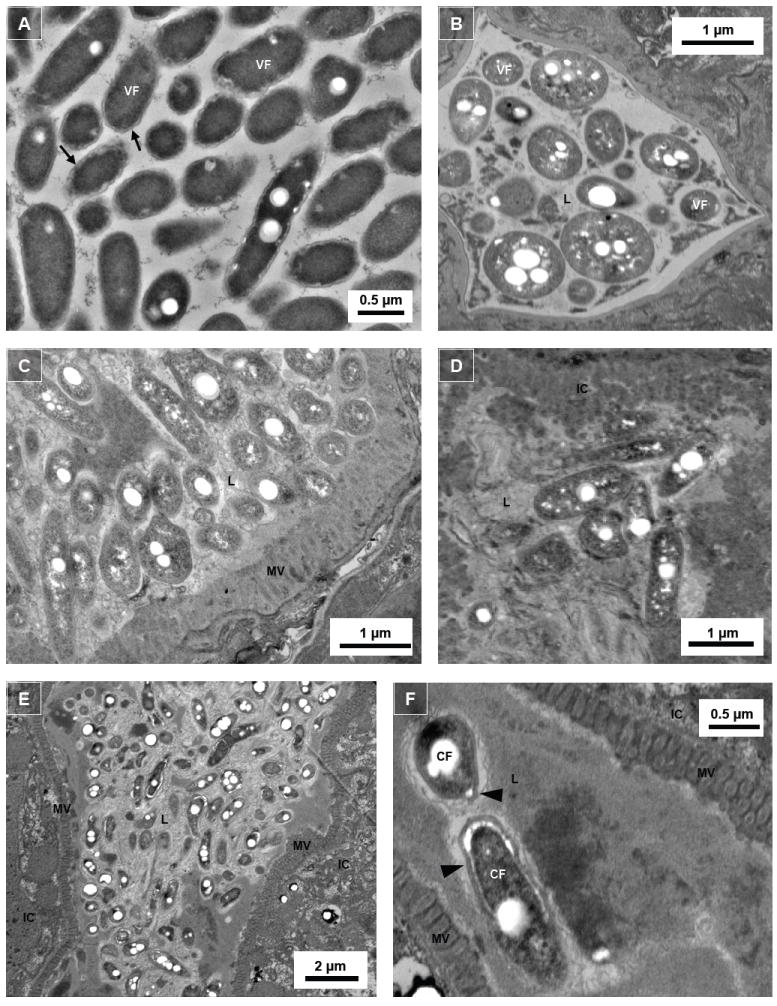
Intraluminal Legionella exhibit morphologically differentiated forms. Transmission electron microscopic images of (A) Legionella harvested from 6-day old assay plate culture without the presence of N2 nematodes; (B) cross-cut pharynx section of N2 nematodes colonized with L. longbeachae ATCC 33462 (note triangular shape of the grinder); (C) and (D) mid-body cross-cut digestive tract sections of N2 nematodes colonized with L. pneumophila Philadelphia-1 type strain; and (E) and (F) mid-body transverse-cut digestive tract sections of nematodes colonized with L. pneumophila SVir. Note that the thickened cell walls and the white circular spaces within the Legionella bacteria are poly-β-hydroxybutyrate (PHBA) granules. VF, vegetative form; CF, cyst-like form; L, intestinal lumen; IC, intestinal cell; MV, microvilli; arrow, thin cell wall of VF Legionella; arrowhead, thick cell wall of CF Legionella.
Legionella are viable for months in a soil environment populated by C. elegans
It is believed that legionellae are incapable of replicating extracellularly in the environment due to an obligate requirement for L-cysteine and other essential nutrients (Ewann et al., 2006). That these nutrients are not required for intracellular growth in protozoa or cell culture is prima facie evidence that Legionella acquires them from parasitized host cells. Morphologic differentiation and amplification of legionellae during plate-based Caenorhabditis assays, however, led us to hypothesize that the nematode digestive tract may be a previously unrecognized environmental niche for extracellular replication and development of Legionella. Indeed, the digestive tract of the nematode is both microaerobic and reducing (McGhee, 2007, Chavez et al., 2007), factors which will shift the equilibrium between cysteine and cystine and potentially make more cysteine available to the bacteria.
To replicate a more natural condition for nematode-Legionella interaction, we added mCherry-producing KB290 to hydrated autoclaved commercial potting soil without supplemental L-cysteine or other nutrients (‘simulated soil’). Using the transgenic worm strains PS3729, which expresses the marker AJM-1∷GFP brightly in the pharynx, and WS1904, which expresses YFP∷actin predominantly in intestinal cells, to facilitate recovery of fluorescent worms from the soil, ~150 L4 stage nematodes and KB290 were used to inoculate ~2.5 gm of simulated soil. Of note, plate-based assays showed that PS3729 and WS1904 nematodes were similarly susceptible to Legionella-mediated killing as wild-type N2 animals (data not shown). Approximately 30 days after initial inoculation of mCherry-producing KB290, brightly fluorescent bacteria were observed in the pharynx region of recovered L2-L4 and adult stage nematodes (Fig. 5). As L4 stage nematodes were initially plated in the soil assay, the presence of extracted young larval nematodes demonstrates that the nematode life cycle has repeated itself within the soil assay. In addition, the presence of fluorescent bacteria within the pharynx indicates recent ingestion of the fluorescent bacteria, as shown in Figure 5A. The fluorescent bacteria are considered viable, as killed bacteria lose fluorescence (data not shown). Figure 5B depicts a young adult nematode with a number of mCherry-producing bacteria in the lower digestive tract. At 60 days post inoculation, some nematodes displayed anal swelling, which appeared to coincide with the accumulation of mCherry-producing bacteria in the lower digestive tract, as shown in Figure 5C. Some nematodes in the soil assays were infected to high densities with mCherry-producing bacteria consistent with bacterial accumulation and/or multiplication in the worm intestinal tract. As seen in Figure 5D, it appears that individual bacteria are undergoing cell division in the upper digestive tract (arrowhead). Even after 90 days (the duration of the experiment), simulated-soil recovered nematodes contained fluorescent Legionella within their upper and lower intestinal tracts. In no instance was Legionella invasion of worm tissues observed, echoing observations made in the plate-based assays. In addition, no protozoa were recovered throughout the course of the experiment. Taken together, these studies suggest that the nematode intestinal tract can be durably colonized with Legionella in a moist soil environment, even when presented another food source, leading to persistence and amplification of L. pneumophila as well as their transmission to nematode offspring.
Figure 5.
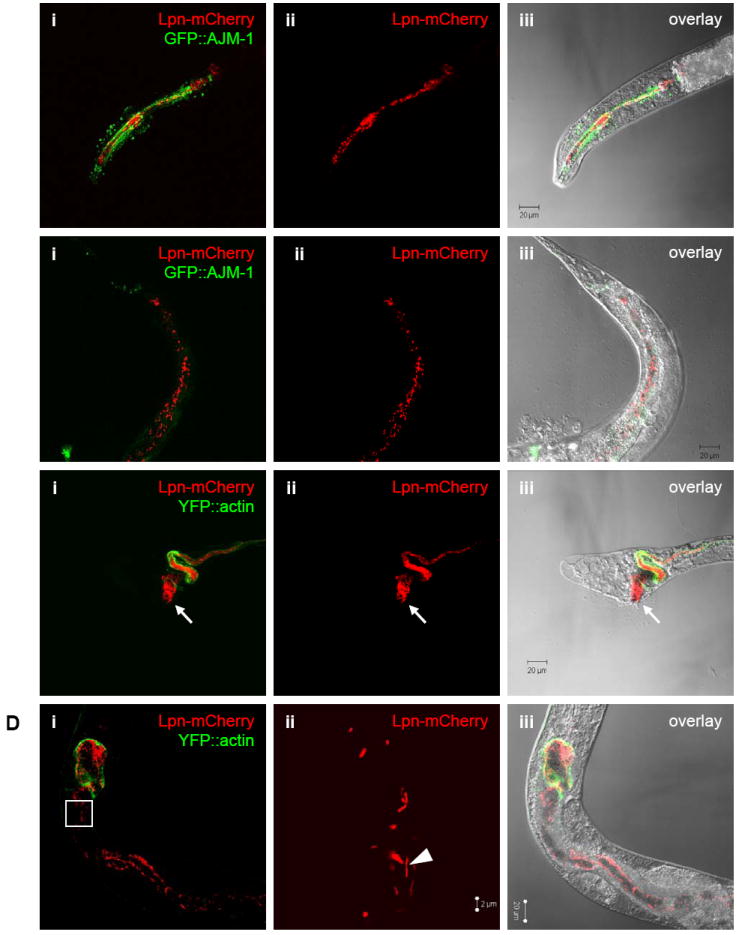
Presence of Legionella in nematodes cultivated in a soil environment. Confocal microscopic images of mCherry-producing L. pneumophila KB290 in C. elegans PS3729 (A) head of a L2 stage and (B) tail of a young adult 30 days after initial inoculation, and in WS1904 (C) tail of a L3 stage and (D) mid-body of an adult views 60 days after initial inoculation. Panels i-iii correspond (i) red and green channels showing fluorescent KB290 and nematode intestinal cells, (ii) red channel alone showing only KB290,and (iii) overlayed red channel, green channel, and DIC bright-field images. Green fluorescence indicates the apical tight junctions within the pharynx structure in PS3729 and outlines the intestinal tract in WS1904. Note that image (Dii) is at higher magnification and corresponds to the outlined inset box in (Di). Arrows denote a heavily colonized anus leading to anal swelling; arrowhead demonstrates bacilli that appear to have recently divided.
TEM analysis of Legionella within the intestinal tract of soil-extracted nematodes demonstrates ultrastructural features similar to CFs
Since confocal microscopy demonstrated that nematodes recovered from simulated soil are colonized with Legionella that may be replicating within the digestive tract, TEM analyses were conducted to determine whether the bacteria underwent morphologic differentiation as was observed in vitro. As shown in Figures 6, TEM of PS3729 nematodes extracted from simulated soil 90 days after initial exposure to KB290 reveals a mixed population of L. pneumophila and E. coli OP50 with in the digestive tract. As was observed in plate-fed nematodes, the L. pneumophila in the soil-extracted nematodes had ultrastructural features suggestive of CFs, including an electron-dense, multilaminar outer membrane and PHBA cytoplasmic inclusions.
Figure 6.
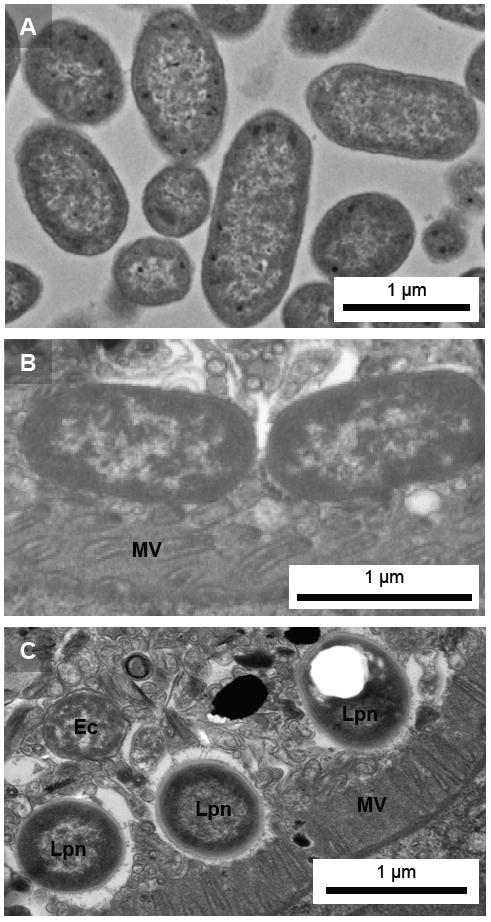
Ultrastructural analysis of bacteria within C. elegans digestive tract. Transmission electron microscopic images of (A) plate grown E. coli OP50, and transverse-cut sections of the digestive tracts of C. elegans PS3729 nematodes extracted from a soil environment 90 days after initial inoculation with intermittent supplementation of E. coli OP50 detail bacteria embedded within intestinal lumen lining the digestive tract; (B) E. coli OP50, and (C) mixed population of OP50 and Legionella. Note ultrastructural differences between OP50 and Legionella, in particular the thickened cell walls and the white spaces indicating the presence of PHBA in Legionella. Lpn, L. pneumophila; Ec, E. coli OP50; MV, microvilli.
C. elegans innate immune signaling controls Legionella infection
Recent studies using C. elegans as a surrogate model host for human microbial pathogens have identified several evolutionarily conserved signaling pathways of innate immunity that are shared across phyla. Prominent systems include the PMK-1 p38 MAPK and DAF-2 insulin/IGF-1 signaling pathways (Kim et al., 2002, Garsin et al., 2003, Troemel et al., 2006). These systems also play key roles in resistance to environmental stressors and natural nematode pathogens, including the nematophagous fungus Drechmeria coniospora and the nematocidal pore-forming toxin Cry5B of Bacillus thuringiensis (Birkenkamp et al., 2003, Huffman et al., 2004, Pujol et al., 2008). To explore whether Legionella elicits immune responses in C. elegans, nematodes with altered host defense function were exposed to L. pneumophila and L. longbeachae. As shown in Figure 7, loss-of-function sek-1(km4) and nsy-1(ag-3) mutants died more quickly than wild-type N2 nematodes when fed either L. pneumophila or L. longbeachae; by contrast, nsy-1(ag3) and sek-1(ag1) have normal lifespans on innocuous bacteria (Kim et al., 2002). NSY-1/MAPKKK and SEK-1/MAPKK are core components of the PMK-1 p38 MAPK pathway. Furthermore, stress and infection resistant loss-of-function daf-2(e1370) mutants were much less susceptible to L. pneumophila and L. longbeachae than wild-type nematodes (Fig. 8). While daf-2 mutants have enhanced longevity and stress resistance in general, the extension of lifespan observed during Legionella exposure (~2.5-3.5 fold) is, like other pathogens, disproportionate to that observed with exposure to innocuous bacteria (~1 fold) (Garsin et al., 2003, Begun et al., 2007, Evans et al., 2008). Signaling through the DAF-2 insulin/IGF-1 receptor inhibits the activity of the forkhead transcription factor DAF-16 by hindering its translocation to the nucleus, and prior studies have shown that the pathogen-resistance of daf-2(e1370) mutants is daf-16-dependent (Garsin et al., 2003, Begun et al., 2007). As shown in Figure 8, the extended lifespan observed in daf-2(e1370) mutants exposed to L. pneumophila or L. longbeachae are abolished in daf-2(e1370);daf-16(mgDf47) nematodes. Together, these data demonstrate that well characterized C. elegans innate immune responses restrict Legionella infection in vitro.
Figure 7.
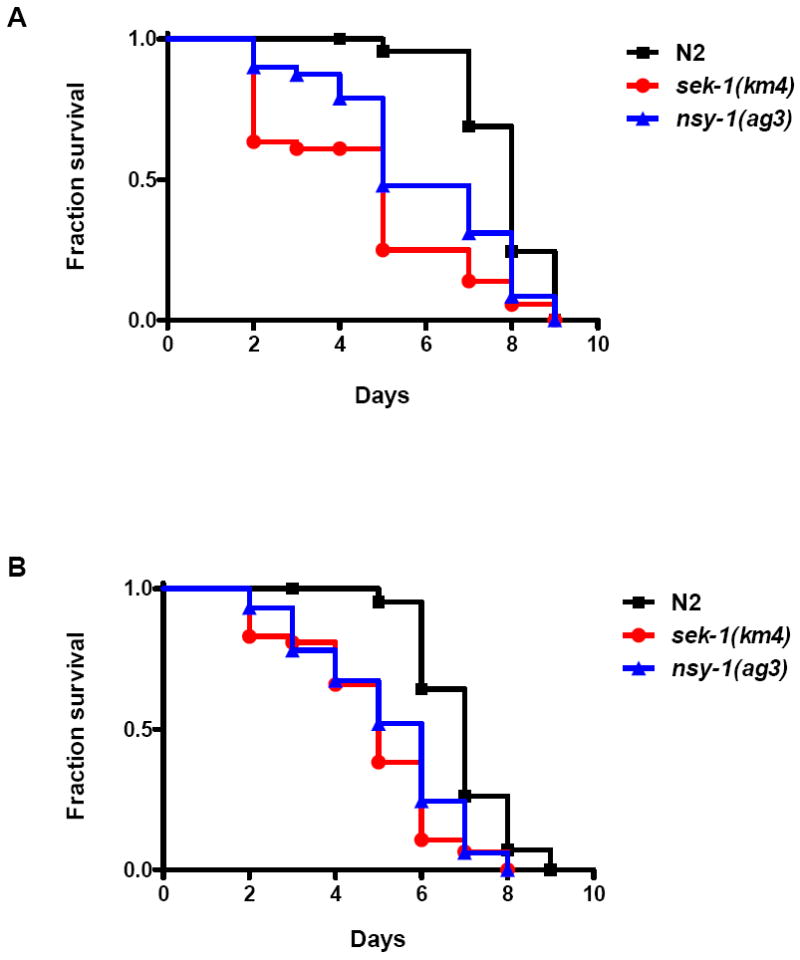
Survival of immunocompromised C. elegans fed Legionella. (A) Kaplan-Meier survival plot of C. elegans N2 (squares, n=45), sek-1(km4) (circles, n=44), and nsy-1(ag3) (triangles, n=45) fed L. pneumophila Philadelphia-1 type strain. (B) Kaplan-Meier survival plot of C. elegans N2 (squares, n=45), sek-1(km4) (circles, n=46), and nsy-1(ag3) (triangles, n=44) fed L. longbeachae type strain. P < 0.0001 by pairwise comparison by the log-rank test of each of the nematode strains [sek-1(km4) and nsy-1(ag3)] versus wild-type N2 for both Legionella strains. Data are representative of one of three independent experiments.
Figure 8.
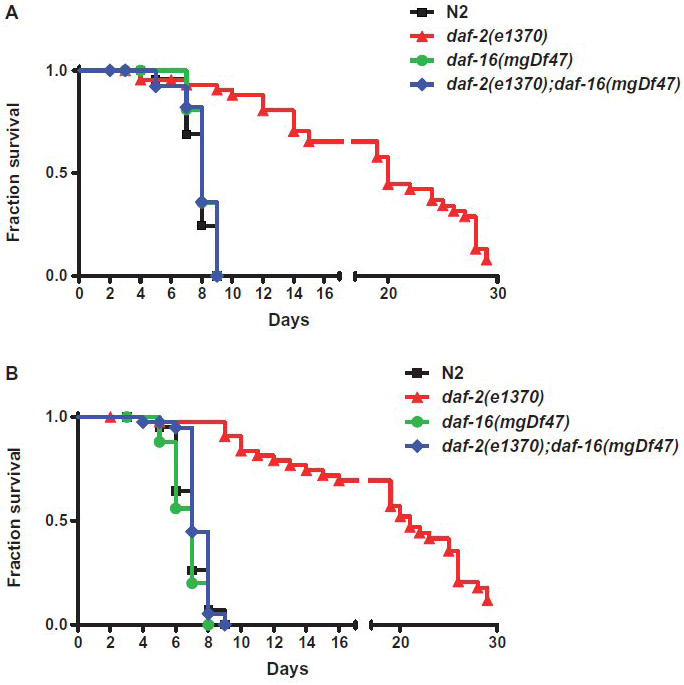
Survival of pathogen and stress-resistant C. elegans fed Legionella. (A) Kaplan-Meier survival plot of C. elegans N2 (squares, n=45), daf-2(e1370) (triangles, n=46), daf-16(mgDf47) (circles, n=46), and daf-2(e1370)/daf-16(mgDf47) (diamonds, n=45) fed L. pneumophila Philadelphia-1 type strain. (B) Kaplan-Meier survival plot of C. elegans N2 (squares, n=45), daf-2(e1370) (triangles, n=45), daf-16(mgDf47) (circles, n=27), and daf-2(e1370)/daf-16(mgDf47) (diamonds, n=41) fed on L. longbeachae type strain. P < 0.0001 by pairwise comparison by the log-rank test of daf-2 versus wild-type N2 for both Legionella strains. All other nematode strain pairwise comparisons versus wild-type N2 were found to be statistically insignificant. Data is representative of one of three independent experiments.
TOL-1, the single Toll-like receptor ortholog of C. elegans, also plays an important role in innate immunity against certain Gram negative pathogens. Partial loss-of-function tol-1(nr2033) mutants are defective in pathogen avoidance when exposed to S. marcescens and are markedly more susceptible to invasion of pharyngeal tissues when exposed to S. Typhimurium, compared to wild-type nematodes (Pujol et al., 2001, Tenor et al., 2008). To determine whether L. pneumophila invaded worm tissues antemortem, young adult tol-1(nr2033) nematodes were exposed to GFP-expressing L. pneumophila KB130 or GFP-expressing S. Typhimurium SL1344 on BCYE plates and monitored daily by DIC and fluorescence microscopy. While invasion of pharyngeal tissues is seen with S. enterica fluorescent SL1344, invasion by L. pneumophila fluorescent Lp02 was not observed (data not shown).
L. pneumophila Dot/Icm type IV secretion system inhibits programmed cell death in the C. elegans germline
Apoptosis occurs at two times during the life of the C. elegans nematode: during embryonic and larval development, when 12% of all somatic cells die, and during germ cell morphogenesis, when half of all pachytene cells (pre-oocytes) die (Lettre et al., 2006). In the gonad, apoptotic cell death can be induced by a number of factors, including genotoxic, oxidative, osmotic, and heat shock stresses and during infection by S. Typhimurium (Aballay et al., 2001, Aballay et al., 2003). Interestingly, induction of apoptosis appears to be pathogen-specific, as Pseudomonas aeruginosa does not elicit PCD within the C. elegans germline (Aballay et al., 2001). Since L. pneumophila induces caspase-3-mediated PCD in macrophages (Abu-Zant et al., 2007, Isberg et al., 2009), we investigated whether germline apoptosis was affected in nematodes colonized with L. pneumophila. We measured the number of gonadal apoptotic cells (i.e. corpse cells) in nematodes exposed to L. pneumophila Phildelphia-1, L. longbeachae ATCC 33462, S. Typhimurium SL1344 and E. coli OP50. As shown in Figure 9A, the number of apoptotic corpse cells observed in nematodes fed L. pneumophila and L. longbeachae were similar to those seen in nematodes fed S. Typhimurium and were significantly higher than those observed in OP50-fed nematodes.
Figure 9.
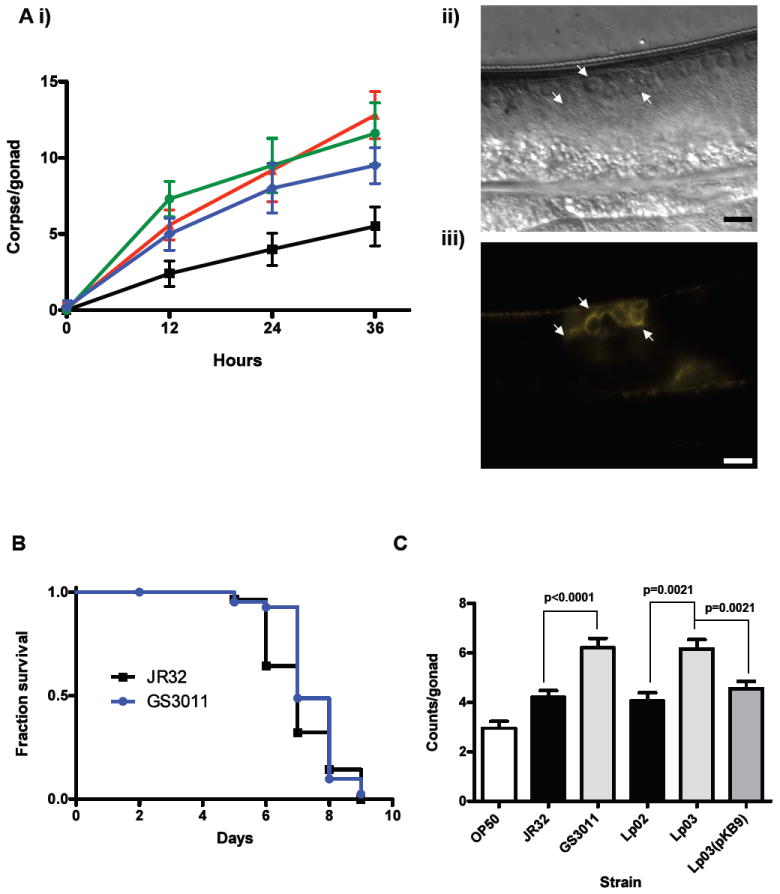
Elevated levels of germline apoptosis in C. elegans. (A) Germline apoptosis in C. elegans fed Legionella: (i) Number of corpse cells counted per gonad (n=10) plotted over time at 0, 12, 24, 36 hr in C. elegans N2 fed on E. coli OP50 (squares), L. pneumophila Philadelphia-1 type strain (triangles), L. longbeachae type strain (circles) and S. Typhimurium SL1344 type strain (diamonds); (ii) DIC image of corpse cells (indicated by arrows) in a gonad of a transgenic C. elegans WS2170 fed L. longbeachae for 36 hr with (iii) corresponding image of fluorescent actin filaments associated with corpse cells. Size marker is 10 μm. (B) Kaplan-Meier survival plot of C. elegans N2 fed L. pneumophila JR32 (squares, n=30) and the isogenic icmT mutant GS3011 (circles, n=44). P > 0.05 by pairwise comparison by the log-rank test. Data representative of three independent experiments. (C) Bar graph of mean corpse cell counts per gonad (n=20) in transgenic C. elegans WS2170 nematodes fed on the designated bacterial strain for 24 hr. P = 0.002 and P = 0.015 by pairwise comparison by the log-rank test of OP50 versus JR32 and Lp02, respectively.
While L. pneumophila induces apoptosis in macrophages, recent studies have also shown that PCD signaling is paradoxically inhibited early after phagocytosis of legionellae in a Dot/Icm-dependent manner (Santic et al., 2007). To investigate the contribution of the Dot/Icm system to disease in C. elegans, we assessed survival and germline apoptosis of nematodes colonized with L. pneumophila dot/icm mutant strains. We found that the icmT mutant strain GS3011 and the dotA mutant stain Lp03 were as virulent in the C. elegans plate-based survival assay as their isogenic parental strains JR32 and Lp02, respectively (Fig. 9B and data not shown). However, gonadal corpse cell counts for nematodes infected with GS3011 and Lp03 were significantly higher than those infected the respective parental strains (Fig. 9C). Moreover, germline apoptosis levels in nematodes colonized with the dotA-complemented strain Lp03(pKB9) were similar to those of the parental strain Lp02 (Fig. 9C). Taken together, these studies suggest that Dot/Icm type IV secretion system has an anti-apoptotic effect on PCD in C. elegans, mirroring observations of L. pneumophila in macrophage cells. How the Dot/Icm system inhibits apoptosis in C. elegans and whether this interaction influences the natural bacterium-host relationship remains to be determined.
Discussion
Legionella pneumophila and related species are considered obligate intracellular parasites of freshwater protozoa that can be grown in vitro on specialized complex medium supplemented with excess L-cysteine and ferric pyrophosphate (Ewann et al., 2006). This obligate nature is further underscored by the evolution of a complex type IV secretion system and family of secreted effector proteins that aid invasion and host cell remodeling to permit intracellular multiplication and differentiation of Legionella in protozoa, macrophages, D. discoideum, and a wide range of mammalian cell lines (Molmeret et al., 2004, Hilbi et al., 2007). Thus, our finding that Legionella spp. differentiate and multiply extracellularly within the intestinal tract of nematodes was unexpected. It follows that the nutrients necessary for Legionella survival and amplification must be available within the intestinal tract.
When fed Legionella ad libitum during plate-based survival assays, Caenorhabditis nematodes have a shortened lifespan that is dependent on both the species and strain of the bacterium and the nematode. Several observations suggest that this reduced longevity is the consequence of an infectious process. First, similar to many other microbial pathogens that kill C. elegans, large numbers of live Legionella colonize the digestive tract of nematodes during the course of infection, leading to marked intestinal distension and eventual worm death; however, colonization per se is not sufficient to kill worms (Garsin et al., 2001). Second, several distinctive abnormal phenotypes of nematode death – specifically bagging and the extrusion of viscera through the vulva – were observed during exposure to Legionella. While the former has been observed with other pathogens, to our knowledge the latter has not been reported in other Caenorhabditis-based infection models. Third, microscopic and ultrastructural analysis of intraluminal and defecated bacteria from plate-fed nematodes display phenotypes of differentiated Legionella, demonstrating that at least some Legionella responses to the intestinal environment of Caenorhabditis parallel those observed within terminally-infected mammalian and protozoan cells (Cirillo et al., 1994, Byrne et al., 1998, Berk et al., 1998, Garduno et al., 2002, Berk et al., 2008). A final reason to conclude that Legionella is virulent towards C. elegans is that nematodes with altered immune systems exhibit similar phenotypes when exposed to Legionella as they do with other pathogens. The immunocompromised p38 MAP kinase mutants nsy-1(ag3) and sek-1(ag1) are more susceptible to killing by either L. pneumophila or L. longbeachae. Conversely, the insulin/IGF-1 receptor mutant daf-2(e1370) is markedly resistant to Legionella infection, and daf-2(e1370) resistance to infection is daf-16 dependent. Based on these findings, we conclude that Legionella is pathogenic to C. elegans in plate-based survival assays. However, the virulence mechanisms that lead to demise of the nematode are not known at this time. Mutants of the Dot/Icm system fail to show differences in nematode survival in vitro, although they are significantly virulence-attenuated in host cell infection models (Molmeret et al., 2007, Isberg et al., 2009). These findings suggest that the factors mediating virulence within the intestinal tract of the nematode differ from those used to parasitize unicellular organisms and host cells. Further work will be needed to determine the role, if any, of other L. pneumophila virulence factors to disease in the nematode (as determined by plate-based survival assays), colonization of the nematode intestinal tract (as determined by soil-based co-culture experiments), and morphologic development (as determined by electron microscopy).
While a surfeit of Legionella is harmful to C. elegans in vitro, it is unlikely that free-living nematodes encounter Legionella in the amount or concentration present during plate-based assays in a natural aquatic or terrestrial environment. In natural freshwater habitats, Legionella concentrations have been reported to be 102 to 106 organisms per liter, depending on the specimen and detection method (Fliermans et al., 1981, Delgado-Viscogliosi et al., 2005, Declerck et al., 2007). Similarly, surveys of potting soils, organic composts and garden soils have recovered Legionella in concentrations of 103 to 108 CFU per gram soil (Steele et al., 1990b, Hughes et al., 1994, Casati et al., 2009a, Casati et al., 2009b). For this reason we developed simulated soil-based assays as a means to study Legionella-Caenorhabditis interactions in an environment that more closely approximates a natural condition.
We demonstrate that successive generations of nematodes extracted from sterile soil seeded with Legionella and C. elegans several months earlier harbor viable L. pneumophila in their digestive tract. Confirming findings of the plate-based assays, microscopic analysis of soil-extracted nematodes suggests that L. pneumophila undergoes bacterial division within the digestive tract, leading to a concentration of bacteria in this environmental niche. How Legionella is retained in the intestinal tract and resists antimicrobial defenses is not clear at this time. Morphological differentiation of organisms to hardier cyst-like forms within the nematode intestinal tract, observed during both the plate- and soil-based assays, represents one potential mechanism. A second possibility is that Legionella subverts host signaling pathways to improve bacterial survival or replication within the nematode intestinal tract. Intracellular survival of L. pneumophila within alveolar macrophages is dependent upon subverting host cell functions including abrogation of the phagosome-lysosome fusion pathway, evasion of activated innate immune defenses, successful establishment of the replicative niche and promotion of host cell survival (Horwitz, 1983, Swanson et al., 1995, Isberg et al., 2009). For example, activation of p38 and SAPK/JNK pathways were found to be necessary for L. pneumophila intracellular replication (Welsh et al., 2004, Shin et al., 2008). However, these regulatory pathways are absent or obscure in protozoan genomes, raising the question of how L. pneumophila evolved mechanisms to modulate these signaling pathways when humans and probably all other mammals appear to be dead end hosts (Katz et al., 1982). We show here that p38 MAP kinase and Daf pathways restrict Legionella infection in vitro, and induction of PCD within the nematode germline during Legionella infection is Dot/Icm-dependent. These results suggest a plausible evolutionary origin for the ability of Legionella to counteract and manipulate host apoptosis and immune signaling pathways in order to establish a replicative niche. Studies are currently underway to further define this paradigm.
In summary, we propose that the predator (host) – prey (pathogen) relationship between Legionella and Caenorhabditis modeled in these studies may reflect a natural ecological relationship between Legionella and free-living nematodes or other evolutionarily related bacteriovorus invertebrates in the environment. Consequently, predatory invertebrates may aid persistence, dissemination and amplification of Legionella in the natural environment, and thus may contribute to the acquisition of Legionnaires’ disease. Field studies will be needed to determine whether Legionella are parasitic or commensal organisms of lower metazoans in natural terrestrial or aquatic ecosystems. Such interactions may have profound implications into the evolution, natural ecology, life cycle, environmental distribution, and intrinsic virulence of Legionella.
Experimental procedures
Bacterial and nematode strains, oligonucleotides, media and general methods
Strains, plasmids and oligonucleotides used in this study are listed in Table 1. All Legionella strains were grown on buffered charcoal yeast extract agar (BCYE) or in buffered yeast extract (BYE) broth as previously described (Feeley et al., 1978), supplemented with thymidine (100 μg/ml), streptomycin (100 μg/ml), or kanamycin (25 μg/ml) when required. S. Typhimurium and E. coli DH5α, (used as a host strain for cloning strategies) was grown at 37°C on Luria-Bertani (LB) agar or in LB broth supplemented with kanamycin (25 ug/ml) or ampicillin (100 μg/ml). Background plasmid-encoded DotA expression levels in Lp03 (pKB9) were sufficient and did not require induction with 1 mM IPTG (Roy et al., 1998). Legionella, in BYE broth containing 10% dimethyl sulfoxide, and Salmonella and E. coli, in LB broth containing 15% glycerol, were stored at -75°C. All reagents, chemicals and antibiotics were purchased from Sigma-Aldrich (St. Louis, MO) or Thermo Fisher Scientific (Waltham, MA), and oligonucleotides synthesized by Invitrogen (Carlsbad, CA). Enzymes and Taq polymerases were purchased from New England Biolabs (Ipswich, MA) and Qiagen (Germantown, MD), respectively. DNA manipulations followed general protocols (Ausubel et al., 2007).
Table 1.
Strains and plasmids used in this study.
| Strain or plasmid | Description (genotype and/or phenotype) | Reference or sourcea |
|---|---|---|
| Strains | ||
| Legionella pneumophila | ||
| Phildelphia-1 | Type strain (ATCC 33152) | CDC, Atlanta |
| SVir | Spontaneous L. pneumophila Philaldephia-1 Smr mutant | (Hoffman et al., 1989) |
| Lp02 | Philadelphia-1 derivative, rpsL hsdR thyA (Smr) mutant | (Berger et al., 1993) |
| KB130 | Lp02 (pKB127), Thy+ | (Morash et al., 2009) |
| KB290 | Lp02 (pKB288), Thy+ | This work |
| Lp03 | dotA03, thyA, hsdR, rpsL | (Berger et al., 1994) |
| Lp02(pKB9) | dotA complement | This study |
| JR32 | NaCl-sensitive isolate of L. pneumophila AM511 | (Sadosky et al., 1993) |
| GS3011 | JR32 icmT3011∷Km | (Segal et al., 1998) |
| Legionella longbeachae | ||
| ATCC 33462 | Type strain | Yousef Abu Kwaik |
| Escherichia coli | ||
| DH5α | F- Ф80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1 | Clonetech |
| OP50 | Uracil auxotrophy | CGC, (Brenner, 1974) |
| Salmonella enterica serovar Typhimurium | ||
| SL1344 | Type strain | Lab collection |
| SM022 | SL1344(pGP704:rpsM∷gfp) | Alejandro Aballay, (Vazquez-Torres et al., 1999) |
| Caenorhabditis briggsae | ||
| HK104 | Wild isolate from Okayama, Japan | CGC |
| Caenorhabditis elegans | ||
| Bristol N2 | Wild-type isolate from Bristol, England | CGC, (Brenner, 1974) |
| AB1 | Wild isolate from Adelaide, Australia | CGC |
| CB4555 | Wild isolate from Pasadena, California, USA | CGC |
| RC301 | Wild isolate from Freiburg, Germany | CGC |
| CB4856 | Wild isolate from Hawaii, USA | CGC |
| KU4 | sek-1(km4) | (Tanaka-Hino et al., 2002) |
| AU3 | nsy-1(ag3) | (Kim et al., 2002) |
| CB1370 | daf-2(e1370) | (Garsin et al., 2003) |
| daf-16(mgDf47) | (Garsin et al., 2003) | |
| GR1309 | daf-2(e1370); daf-16(mgDf47) | (Garsin et al., 2003) |
| tol-1(nr2033) | CGC | |
| PS3729 | unc-119(ed4); syIs78[ajm-1∷gfp + unc-119(+)] | CGC |
| WS1904 | unc-119(ed3); opEx813[Pced-10∷yfp∷actin + unc-119(+)] | Kinchen and Hengartner, unpublished |
| WS2170 | unc-119(ed3);opIs110[Pced-1∷YFP∷act-5;unc-119(+)] | (Kinchen et al., 2005) |
| Plasmid | ||
| pKB9 | dotA expression plasmid | (Roy et al., 1998) |
| pBH6119 | RSF1010 ori, promoterless, gfpmut3 tdΔi ; Thy+, Ampr | (Hammer et al., 1999) |
| pKB127 | pBH6119∷magA 245-bp promoter region cloned into BamHI and XbaI sites | (Morash et al., 2009) |
| pmCherry | pUC19∷mCherry, Ampr | Roger Y. Tsien, (Shaner et al., 2004) |
| pKB288 | PCR-amplified mCherry inserted into XbaI and SphI sites of pKB127 replacing gfpmut3 | This work |
ATCC, American Type Culture Collection; CDC, Centers for Disease Control and Prevention; CGC, Caenorhabditis Genetics Center.
C. elegans strains were maintained at 15°C on nematode growth medium (NGM) plates spread with live or UV-light treated E. coli strain OP50 as a food source and were manipulated using established techniques (Brenner, 1974, Lewis et al., 1995).
Nematode survival and germline apoptosis assays
A loopful of Legionella bacteria was harvested from freshly streaked 48-72 hr BCYE plates, resuspended in 500 μl of BYE, and 20 μl of the resuspension was spread onto 3.5-cm-diameter assay plates containing BCYE supplemented with thymidine (100 μg/ml), streptomycin (100 μg/ml), or kanamycin (25 μg/ml) when appropriate, and incubated overnight at 37°C.
For survival assays, approximately 15 hermaphrodite nematodes in the fourth larval stage (L4) were placed on the assay plates (cooled to room temperature), and their survival was monitored over time at 25°C. Nematodes were transferred to fresh assay plates on the days two through five in order to separate subjects from progeny. Experiments were conducted in triplicate and repeated at least three times. For experiments with heat-treated Legionella, nematodes were transferred to fresh plates when necessary to ensure sufficient source of food. Nematodes were considered dead when they failed to respond to touch and pharyngeal pumping was no longer observed. Nematodes that died as a result of crawling off the plate were censored from the analysis. For each survival assay, nematode survival was calculated by the Kaplan-Meier method and survival differences were tested for significance using the log-rank test (GraphPad Prism, version 4.0).
For germline apoptosis assays, approximately 30 L4 hermaphrodite nematodes (N2 or WS2170) were placed on the assay plates and incubated at 25°C. For each time point, approximately 20 worms were removed and prepared for microscopic examination. Corpse cells in the gonad of N2 or WS2170 nematodes were enumerated using differential interference contrast or confocal fluorescence microscopy, respectively, as previously described (Kinchen et al. 2005).
For tissue invasion assays, approximately 15-20 young adult hermaphrodite tol-1(nr2033) nematodes were placed on the assay plates and incubated at 25°C. Nematodes were monitored on a daily basis for tissue invasion via fluorescent microscopic analysis.
Quantification of nematode colonization
For quantification of bacterial colonization of C. elegans, nematodes were allowed to feed on L. pneumophila under standard survival assay conditions. On a daily basis for a six day period, approximately 30 nematodes were transferred manually from the survival plates and divided into three Eppendorf tubes containing 100 μl of ice-cold M9 buffer (22 mM KH2PO4, 42 mM Na2HPO4, 86 mM NaCl, 1 mM MgSO4). The nematodes were centrifuged in a microcentrifuge at 3000 rpm for 30 sec and decanted to remove residual loose bacteria. The nematodes were washed four more times with 500 μl of ice-cold M9 buffer and resuspended in 250 μl of ice-cold M9 buffer. Nematodes were homogenized by adding approximately 200 μl of 1 mm-diameter silicone carbide beads (BioSpec Products, Bartlesville, OK) and vortexing for 1 min. Serial decimal dilutions of the supernatant before and after vortexing were made to determine the net number of colony forming units (CFU) per nematode.
Persistence assays
Persistence of bacterial colonization of C. elegans was evaluated using fluorescent strains of L. pneumophila Lp02. Construction of the GFP-expressing strain KB130 has been described previously (Morash et al., 2009). mCherry (red fluorescent protein) – producing Lp02 was constructed by PCR-amplifying mCherry from plasmid pmCherry with primers PFMCHERRYXBAI (5’-GCGATATCTAGAATGGTGAGCAAGGGCGA-3’) and PFMCHERRYSPHI (5’- GCGATAGCATGCTTACTTGTACAGCTCGTCCA-3’). These primers contain sequences for the restrictions enzymes XbaI and SphI (underlined). The XbaI and SphI-restricted amplicon was ligated into plasmid pKB127 that had been digested with the same restriction enzymes, replacing excised gfpmut3. The resulting plasmid pKB288 was then eletroporated into Lp02, yielding strain KB290.
To evaluate persistent bacterial colonization in agar plate-based assays, L4 nematodes were allowed to feed on the initial (pulse) strain of L. pneumophila for 72 hr and were then transferred to a second (chase) bacterial lawn of L. pneumophila or OP50. Each day prior to and after transfer, five nematodes were selected for fluorescent microscopic analysis and qualitatitve assessments of fluorescence intensity were made.
Soil assay
Commercial potting soil (Hyponex Corporation, Ohio) was sterilized by autoclaving, and an aliquot (~2.5 g) was placed in a 3.5-cm Petri plate. A loopful of mCherry-producing L. pneumophila KB290 was harvested from a freshly-streaked BCYE plate, washed three times in 1 mL sterile dH2O, resuspended in 1 mL sterile dH2O and added to the soil assay plate. Approximately 150 L4 stage nematodes (PS3729 or WS1904) were removed from a standard NGM propragation plate, washed three times and resuspended in M9 buffer and deposited onto the soil assay plate. These transgenic nematode strains produce green fluorescent protein (GFP), which aided in their re-isolation from the soil. Plates were kept in an aerobic humid chamber at 25°C. Overnight broth cultures of OP50 (3 ml) were concentrated, washed with sterile M9 buffer, and added to the soil every 1-2 weeks to ensure a sufficient source of food for maintenance of the nematode colony and progeny development. On a weekly basis, the nematodes were extracted for microscopic analysis and when necessary, additional M9 buffer was added to maintain moisture level. Nematodes were extracted from the soil using the NGM agar capture method, as previously described (Barriere et al., 2006, Sifri et al., 2008). Every several weeks soil samples (~0.1 gm) were resuspended in a 10x volume of PBS and aliquots were plated on BCYE agar plates supplemented with streptomycin and on LB agar plates. In each instance all recovered colonies of L. pneumophila remained fluorescent while recovered colonies of E. coli were nonfluorescent, demonstrating that pKB288 was retained by L. pneumophila and not transferred by conjugation or transformation to E. coli.
Differential interference contrast (DIC) and confocal microscopy
Nematodes were mounted on 2% agarose pads in M9, anesthetized with 3-5 mM levamisole, sandwiched under a coverslip, and examined by differential interference contrast microscopy with Nomarski optics using an Axiovert 200 microscope (Carl Zeiss AG, Oberkochen, Germany) equipped with a Qicam 12-bit Fast 1394 (QImaging Corp, Surrey, British Columbia, Canada) digital camera and fluorescence illuminator Xcite 120 (EXFO Life Sciences, Mississauga, Ontario, Canada). Images were captured with Image Pro Plus software (Media Cybernetics, Bethesda, MD). Confocal fluorescene microscopy was performed using a LSM 510 confocal microscope with accompanying LSM Image Browser software (Zeiss). Images were edited using Photoshop CS (Adobe).
Transmission electron microscopy
Worms exposed to Legionella on BCYE assay plates or extracted from soil were washed three times in sterile PBS and resuspended in 200 μl of 4% glutaraldehyde in PBS and stored at 4°C until processing. Intact Legionella-exposed worms were fixed overnight at 4°C in modified Karnovsky’s fixative (0.1 M phosphate buffer (PB) containing 4% (w/v) paraformaldehyde and 2.5% (w/v) glutaraldehyde, pH 7.4). All subsequent processing was carried out at room temperature unless otherwise noted. Following primary fixation, the worms were washed three times for a total of 45 min with PB and post-fixed for 1 hr in 1.0% osmium tetroxide. The worms were then washed in distilled water, dehydrated employing a graded ethanol series, embedded in Embed 812 epoxy resin (Electron Microscopy Sciences, Hatfield, PA), and polymerized for two days at 60°C. Ultrathin sections approximately 70 nm in thickness, obtained with a diamond knife (Diatome, Hatfield, PA) on a Ultracut UCT ultramicrotome (Leica, Richmond, IL), were collected on 200 mesh copper grids, contrast stained with uranyl acetate and lead citrate according to routine procedures, and examined in a JEM-1230 transmission electron microscope (JEOL, Tokyo, Japan). Images were acquired using an SIA L3-C digital camera (Scientific Instruments and Applications, Duluth, GA).
Acknowledgments
We thank Alejandro Aballay, Hubert Hilbi, Ralph Isberg, Yousef Abu Kwaik and Roger Y. Tsien for generously providing strains, plasmids and reagents for this work. Several Caenorhabditis strains were obtained from the Caenorhabditis Genetics Center (CGC), which is supported by the National Institutes of Health National Center of Research Resources.
This work was supported by a Howard Hughes Medical Institute Early Career Award to C.D.S., NIAID grant R01 AI066058 to P.S.H., and a Canadian Institutes for Health Research post-doctoral research fellowship to A.K.C.B.
References
- Aballay A, Ausubel FM. Programmed cell death mediated by ced-3 and ced-4 protects Caenorhabditis elegans from Salmonella typhimurium-mediated killing. Proc Natl Acad Sci U S A. 2001;98:2735–2739. doi: 10.1073/pnas.041613098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aballay A, Drenkard E, Hilbun LR, Ausubel FM. Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr Biol. 2003;13:47–52. doi: 10.1016/s0960-9822(02)01396-9. [DOI] [PubMed] [Google Scholar]
- Aballay A, Yorgey P, Ausubel FM. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr Biol. 2000;10:1539–1542. doi: 10.1016/s0960-9822(00)00830-7. [DOI] [PubMed] [Google Scholar]
- Abu-Zant A, Jones S, Asare R, Suttles J, Price C, Graham J, Kwaik YA. Anti-apoptotic signalling by the Dot/Icm secretion system of L. pneumophila. Cell Microbiol. 2007;9:246–264. doi: 10.1111/j.1462-5822.2006.00785.x. [DOI] [PubMed] [Google Scholar]
- Anderson GL, Kenney SJ, Millner PD, Beuchat LR, Williams PL. Shedding of foodborne pathogens by Caenorhabditis elegans in compost-amended and unamended soil. Food Microbiol. 2006;23:146–153. doi: 10.1016/j.fm.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, G SJ, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 2007. [Google Scholar]
- Bachman MA, Swanson MS. Genetic evidence that Legionella pneumophila RpoS modulates expression of the transmission phenotype in both the exponential phase and the stationary phase. Infect Immun. 2004;72:2468–2476. doi: 10.1128/IAI.72.5.2468-2476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriere A, Felix MA. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr Biol. 2005;15:1176–1184. doi: 10.1016/j.cub.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Barriere A, Felix MA. Isolation of C. elegans and related nematodes. WormBook. 2006:1–9. doi: 10.1895/wormbook.1.115.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun J, Gaiani JM, Rohde H, Mack D, Calderwood SB, Ausubel FM, Sifri CD. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 2007;3:e57. doi: 10.1371/journal.ppat.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benin AL, Benson RF, Besser RE. Trends in legionnaires disease, 1980-1998: declining mortality and new patterns of diagnosis. Clin Infect Dis. 2002;35:1039–1046. doi: 10.1086/342903. [DOI] [PubMed] [Google Scholar]
- Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- Berger KH, Merriam JJ, Isberg RR. Altered intracellular targeting properties associated with mutations in the Legionella pneumophila dotA gene. Mol Microbiol. 1994;14:809–822. doi: 10.1111/j.1365-2958.1994.tb01317.x. [DOI] [PubMed] [Google Scholar]
- Berk SG, Faulkner G, Garduno E, Joy MC, Ortiz-Jimenez MA, Garduno RA. Packaging of live Legionella pneumophila into pellets expelled by Tetrahymena spp. does not require bacterial replication and depends on a Dot/Icm-mediated survival mechanism. Appl Environ Microbiol. 2008;74:2187–2199. doi: 10.1128/AEM.01214-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk SG, Ting RS, Turner GW, Ashburn RJ. Production of respirable vesicles containing live Legionella pneumophila cells by two Acanthamoeba spp. Appl Environ Microbiol. 1998;64:279–286. doi: 10.1128/aem.64.1.279-286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenkamp KU, Coffer PJ. FOXO transcription factors as regulators of immune homeostasis: molecules to die for? J Immunol. 2003;171:1623–1629. doi: 10.4049/jimmunol.171.4.1623. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne B, Swanson MS. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun. 1998;66:3029–3034. doi: 10.1128/iai.66.7.3029-3034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron S, Roder D, Walker C, Feldheim J. Epidemiological characteristics of Legionella infection in South Australia: implications for disease control. Aust N Z J Med. 1991;21:65–70. doi: 10.1111/j.1445-5994.1991.tb03007.x. [DOI] [PubMed] [Google Scholar]
- Casati S, Conza L, Bruin J, Gaia V. Compost facilities as a reservoir of Legionella pneumophila and other Legionella species. Clin Microbiol Infect. 2009a doi: 10.1111/j.1469-0691.2009.03009.x. [DOI] [PubMed] [Google Scholar]
- Casati S, Gioria-Martinoni A, Gaia V. Commercial potting soils as an alternative infection source of Legionella pneumophila and other Legionella species in Switzerland. Clin Microbiol Infect. 2009b;15:571–575. doi: 10.1111/j.1469-0691.2009.02742.x. [DOI] [PubMed] [Google Scholar]
- Chavez V, Mohri-Shiomi A, Maadani A, Vega LA, Garsin DA. Oxidative stress enzymes are required for DAF-16-mediated immunity due to generation of reactive oxygen species by Caenorhabditis elegans. Genetics. 2007;176:1567–1577. doi: 10.1534/genetics.107.072587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciotto NP. Pathogenicity of Legionella pneumophila. Int J Med Microbiol. 2001;291:331–343. doi: 10.1078/1438-4221-00139. [DOI] [PubMed] [Google Scholar]
- Cirillo JD, Falkow S, Tompkins LS. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect Immun. 1994;62:3254–3261. doi: 10.1128/iai.62.8.3254-3261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux ZD, Edwards RL, Swanson MS. SpoT governs Legionella pneumophila differentiation in host macrophages. Mol Microbiol. 2009;71:640–658. doi: 10.1111/j.1365-2958.2008.06555.x. [DOI] [PubMed] [Google Scholar]
- Declerck P, Behets J, van Hoef V, Ollevier F. Detection of Legionella spp. and some of their amoeba hosts in floating biofilms from anthropogenic and natural aquatic environments. Water Res. 2007;41:3159–3167. doi: 10.1016/j.watres.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Delgado-Viscogliosi P, Simonart T, Parent V, Marchand G, Dobbelaere M, Pierlot E, et al. Rapid method for enumeration of viable Legionella pneumophila and other Legionella spp. in water. Appl Environ Microbiol. 2005;71:4086–4096. doi: 10.1128/AEM.71.7.4086-4096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boer JW, Yzerman EP, Jansen R, Bruin JP, Verhoef LP, Neve G, van der Zwaluw K. Legionnaires’ disease and gardening. Clin Microbiol Infect. 2007;13:88–91. doi: 10.1111/j.1469-0691.2006.01562.x. [DOI] [PubMed] [Google Scholar]
- Doyle RM, Steele TW, McLennan AM, Parkinson IH, Manning PA, Heuzenroeder MW. Sequence analysis of the mip gene of the soilborne pathogen Legionella longbeachae. Infect Immun. 1998;66:1492–1499. doi: 10.1128/iai.66.4.1492-1499.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans EA, Chen WC, Tan MW. The DAF-2 insulin-like signaling pathway independently regulates aging and immunity in C. elegans. Aging Cell. 2008;7:879–893. doi: 10.1111/j.1474-9726.2008.00435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewann F, Hoffman PS. Cysteine metabolism in Legionella pneumophila: characterization of an L-cystine-utilizing mutant. Appl Environ Microbiol. 2006;72:3993–4000. doi: 10.1128/AEM.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewbank JJ. Signaling in the immune response. WormBook. 2006:1–12. doi: 10.1895/wormbook.1.83.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeley JC, Gorman GW, Weaver RE, Mackel DC, Smith HW. Primary isolation media for Legionnaires disease bacterium. J Clin Microbiol. 1978;8:320–325. doi: 10.1128/jcm.8.3.320-325.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields BS. The molecular ecology of legionellae. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- Fliermans CB, Cherry WB, Orrison LH, Smith SJ, Tison DL, Pope DH. Ecological distribution of Legionella pneumophila. Appl Environ Microbiol. 1981;41:9–16. doi: 10.1128/aem.41.1.9-16.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freckman DW. Bacterivorous nematodes and organic-matter decomposition. Agric Ecosyst Environ. 1988;24:195–217. [Google Scholar]
- Garduno RA, Garduno E, Hiltz M, Hoffman PS. Intracellular growth of Legionella pneumophila gives rise to a differentiated form dissimilar to stationary-phase forms. Infect Immun. 2002;70:6273–6283. doi: 10.1128/IAI.70.11.6273-6283.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV, Murray BE, et al. A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci U S A. 2001;98:10892–10897. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, et al. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- Hammer BK, Swanson MS. Co-ordination of legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol Microbiol. 1999;33:721–731. doi: 10.1046/j.1365-2958.1999.01519.x. [DOI] [PubMed] [Google Scholar]
- Hilbi H, Weber SS, Ragaz C, Nyfeler Y, Urwyler S. Environmental predators as models for bacterial pathogenesis. Environ Microbiol. 2007;9:563–575. doi: 10.1111/j.1462-2920.2007.01238.x. [DOI] [PubMed] [Google Scholar]
- Hiltz MF, Sisson GR, Brassinga AK, Garduno E, Garduno RA, Hoffman PS. Expression of magA in Legionella pneumophila Philadelphia-1 is developmentally regulated and a marker of formation of mature intracellular forms. J Bacteriol. 2004;186:3038–3045. doi: 10.1128/JB.186.10.3038-3045.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman PS, Butler CA, Quinn FD. Cloning and temperature-dependent expression in Escherichia coli of a Legionella pneumophila gene coding for a genus-common 60-kilodalton antigen. Infect Immun. 1989;57:1731–1739. doi: 10.1128/iai.57.6.1731-1739.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz MA. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med. 1983;158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman DL, Abrami L, Sasik R, Corbeil J, Van Der Goot FG, Aroian RV. Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins. Proc Natl Acad Sci U S A. 2004;101:10995–11000. doi: 10.1073/pnas.0404073101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MS, Steele TW. Occurrence and distribution of Legionella species in composted plant materials. Appl Environ Microbiol. 1994;60:2003–2005. doi: 10.1128/aem.60.6.2003-2005.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg RR, O’Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz SM, Habib WA, Hammel JM, Nash P. Lack of airborne spread of infection by Legionella pneumophila among guinea pigs. Infect Immun. 1982;38:620–622. doi: 10.1128/iai.38.2.620-622.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessin RH, Gundersen GG, Zaydfudim V, Grimson M. How cellular slime molds evade nematodes. Proc Natl Acad Sci U S A. 1996;93:4857–4861. doi: 10.1073/pnas.93.10.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. Studying host-pathogen interactions and innate immunity in Caenorhabditis elegans. Dis Model Mech. 2008;1:205–208. doi: 10.1242/dmm.000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- Kinchen JM, Cabello J, Klingele D, Wong K, Feichtinger R, Schnabel H, et al. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature. 2005;434:93–99. doi: 10.1038/nature03263. [DOI] [PubMed] [Google Scholar]
- Kiontke K, Sudhaus W. Ecology of Caenorhabditis species. WormBook. 2006:1–14. doi: 10.1895/wormbook.1.37.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide M, Saito A, Okazaki M, Umeda B, Benson RF. Isolation of Legionella longbeachae serogroup 1 from potting soils in Japan. Clin Infect Dis. 1999;29:943–944. doi: 10.1086/520470. [DOI] [PubMed] [Google Scholar]
- LeBlanc JJ, Brassinga AK, Ewann F, Davidson RJ, Hoffman PS. An ortholog of OxyR in Legionella pneumophila is expressed postexponentially and negatively regulates the alkyl hydroperoxide reductase (ahpC2D) operon. J Bacteriol. 2008;190:3444–3455. doi: 10.1128/JB.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G, Hengartner MO. Developmental apoptosis in C. elegans: a complex CEDnario. Nat Rev Mol Cell Biol. 2006;7:97–108. doi: 10.1038/nrm1836. [DOI] [PubMed] [Google Scholar]
- Lewis JA, Fleming JT. Basic culture methods. Methods Cell Biol. 1995;48:3–29. [PubMed] [Google Scholar]
- McGhee JD. The C. elegans intestine. WormBook. 2007:1–36. doi: 10.1895/wormbook.1.133.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molmeret M, Santic M, Asare R, Carabeo RA, Abu Kwaik Y. Rapid escape of the dot/icm mutants of Legionella pneumophila into the cytosol of mammalian and protozoan cells. Infect Immun. 2007;75:3290–3304. doi: 10.1128/IAI.00292-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molmeret M, Zink SD, Han L, Abu-Zant A, Asari R, Bitar DM, Abu Kwaik Y. Activation of caspase-3 by the Dot/Icm virulence system is essential for arrested biogenesis of the Legionella-containing phagosome. Cell Microbiol. 2004;6:33–48. doi: 10.1046/j.1462-5822.2003.00335.x. [DOI] [PubMed] [Google Scholar]
- Molofsky AB, Swanson MS. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol Microbiol. 2004;53:29–40. doi: 10.1111/j.1365-2958.2004.04129.x. [DOI] [PubMed] [Google Scholar]
- Montanaro JC, Meyer W, Gilbert D. Ultrastructural investigations show a dimorphic infection cycle of virulent Legionella longbeachae in amoebae. 6th International Conference on Legionella; Chicago, IL. 2005. [Google Scholar]
- Morash MG, Brassinga AK, Warthan M, Gourabathini P, Garduno RA, Goodman SD, Hoffman PS. Reciprocal expression of integration host factor and HU in the developmental cycle and infectivity of Legionella pneumophila. Appl Environ Microbiol. 2009;75:1826–1837. doi: 10.1128/AEM.02756-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio S, Roy CR. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 2007;15:372–380. doi: 10.1016/j.tim.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Phares CR, Wangroongsarb P, Chantra S, Paveenkitiporn W, Tondella ML, Benson RF, et al. Epidemiology of severe pneumonia caused by Legionella longbeachae, Mycoplasma pneumoniae, and Chlamydia pneumoniae: 1-year, population-based surveillance for severe pneumonia in Thailand. Clin Infect Dis. 2007;45:e147–155. doi: 10.1086/523003. [DOI] [PubMed] [Google Scholar]
- Prevention, C.f.D.C.a. Legionnaires’ Disease associated with potting soil--California, Oregon, and Washington, May-June 2000. MMWR Morb Mortal Wkly Rep. 2000;49:777–778. [PubMed] [Google Scholar]
- Pujol N, Cypowyj S, Ziegler K, Millet A, Astrain A, Goncharov A, et al. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr Biol. 2008;18:481–489. doi: 10.1016/j.cub.2008.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan M, et al. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- Rasis M, Segal G. The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol Microbiol. 2009;72:995–1010. doi: 10.1111/j.1365-2958.2009.06705.x. [DOI] [PubMed] [Google Scholar]
- Roy CR, Berger KH, Isberg RR. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol Microbiol. 1998;28:663–674. doi: 10.1046/j.1365-2958.1998.00841.x. [DOI] [PubMed] [Google Scholar]
- Sadosky AB, Wiater LA, Shuman HA. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun. 1993;61:5361–5373. doi: 10.1128/iai.61.12.5361-5373.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahr T, Bruggemann H, Jules M, Lomma M, Albert-Weissenberger C, Cazalet C, Buchrieser C. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol Microbiol. 2009 doi: 10.1111/j.1365-2958.2009.06677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic M, Asare R, Doric M, Abu Kwaik Y. Host-dependent trigger of caspases and apoptosis by Legionella pneumophila. Infect Immun. 2007;75:2903–2913. doi: 10.1128/IAI.00147-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G, Shuman HA. Intracellular multiplication and human macrophage killing by Legionella pneumophila are inhibited by conjugal components of IncQ plasmid RSF1010. Mol Microbiol. 1998;30:197–208. doi: 10.1046/j.1365-2958.1998.01054.x. [DOI] [PubMed] [Google Scholar]
- Shadrach WS, Rydzewski K, Laube U, Holland G, Ozel M, Kiderlen AF, Flieger A. Balamuthia mandrillaris, free-living ameba and opportunistic agent of encephalitis, is a potential host for Legionella pneumophila bacteria. Appl Environ Microbiol. 2005;71:2244–2249. doi: 10.1128/AEM.71.5.2244-2249.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shin S, Case CL, Archer KA, Nogueira CV, Kobayashi KS, Flavell RA, et al. Type IV secretion-dependent activation of host MAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila. PLoS Pathog. 2008;4:e1000220. doi: 10.1371/journal.ppat.1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifri CD, Begun J, Ausubel FM. The worm has turned - microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 2005;13:119–127. doi: 10.1016/j.tim.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Sifri CD, Begun J, Ausubel FM, Calderwood SB. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect Immun. 2003;71:2208–2217. doi: 10.1128/IAI.71.4.2208-2217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifri CD, Brassinga AK, Flohr T, Kinchen JM, Hazen KC, Sawyer RG, et al. Moraxella osloensis bacteremia in a kidney transplant recipient. Transpl Int. 2008;21:1011–1013. doi: 10.1111/j.1432-2277.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- Steele TW, Lanser J, Sangster N. Isolation of Legionella longbeachae serogroup 1 from potting mixes. Appl Environ Microbiol. 1990a;56:49–53. doi: 10.1128/aem.56.1.49-53.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele TW, McLennan AM. Infection of Tetrahymena pyriformis by Legionella longbeachae and other Legionella species found in potting mixes. Appl Environ Microbiol. 1996;62:1081–1083. doi: 10.1128/aem.62.3.1081-1083.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele TW, Moore CV, Sangster N. Distribution of Legionella longbeachae serogroup 1 and other legionellae in potting soils in Australia. Appl Environ Microbiol. 1990b;56:2984–2988. doi: 10.1128/aem.56.10.2984-2988.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert M, Hentschel U, Hacker J. Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol Rev. 2002;26:149–162. doi: 10.1111/j.1574-6976.2002.tb00607.x. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Isberg RR. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun. 1995;63:3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Hino M, Sagasti A, Hisamoto N, Kawasaki M, Nakano S, Ninomiya-Tsuji J, et al. SEK-1 MAPKK mediates Ca2+ signaling to determine neuronal asymmetric development in Caenorhabditis elegans. EMBO Rep. 2002;3:56–62. doi: 10.1093/embo-reports/kvf001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenor JL, Aballay A. A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep. 2008;9:103–109. doi: 10.1038/sj.embor.7401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Torres A, Jones-Carson J, Baumler AJ, Falkow S, Valdivia R, Brown W, et al. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature. 1999;401:804–808. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- Wallis L, Robinson P. Soil as a source of Legionella pneumophila serogroup 1 (Lp1) Aust N Z J Public Health. 2005;29:518–520. doi: 10.1111/j.1467-842x.2005.tb00242.x. [DOI] [PubMed] [Google Scholar]
- Welsh CT, Summersgill JT, Miller RD. Increases in c-Jun N-terminal kinase/stress-activated protein kinase and p38 activity in monocyte-derived macrophages following the uptake of Legionella pneumophila. Infect Immun. 2004;72:1512–1518. doi: 10.1128/IAI.72.3.1512-1518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young IM, Crawford JW. Interactions and self-organization in the soil-microbe complex. Science. 2004;304:1634–1637. doi: 10.1126/science.1097394. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438:179–184. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]


