Abstract
Insects are host to a diverse range of vertically transmitted micro-organisms, but while their bacterial symbionts are well-studied, little is known about their vertically transmitted viruses. We have found that two sigma viruses (Rhabdoviridae) recently discovered in Drosophila affinis and Drosophila obscura are both vertically transmitted. As is the case for the sigma virus of Drosophila melanogaster, we find that both males and females can transmit these viruses to their offspring. Males transmit lower viral titers through sperm than females transmit through eggs, and a lower proportion of their offspring become infected. In natural populations of D. obscura in the United Kingdom, we found that 39% of flies were infected and that the viral population shows clear evidence of a recent expansion, with extremely low genetic diversity and a large excess of rare polymorphisms. Using sequence data we estimate that the virus has swept across the United Kingdom within the past ∼11 years, during which time the viral population size doubled approximately every 9 months. Using simulations based on our lab estimates of transmission rates, we show that the biparental mode of transmission allows the virus to invade and rapidly spread through populations at rates consistent with those measured in the field. Therefore, as predicted by our simulations, the virus has undergone an extremely rapid and recent increase in population size. In light of this and earlier studies of a related virus in D. melanogaster, we conclude that vertically transmitted rhabdoviruses may be common in insects and that these host–parasite interactions can be highly dynamic.
INSECTS have a diverse range of vertically transmitted symbionts (Buchner 1965). Of these the best studied are bacteria, which are usually transmitted exclusively by females and have evolved a range of strategies to spread through host populations [such as distorting the sex ratio toward females or providing a metabolic benefit to their hosts (Douglas 1989; Hurst et al. 1993)]. Far less is known about vertically transmitted viruses in insects. Some viruses are both horizontally and vertically transmitted (Mims 1981; Bezier et al. 2009). Other species contain endogenous retroviruses or polydnaviruses that have integrated into the germline and are inherited with the host genome (Fleming and Summers 1991; Heredia et al. 2007; Bezier et al. 2009). However, very few free living and purely vertically transmitted viruses have been described in insects.
One such virus is the Drosophila melanogaster sigma virus (DMelSV), which infects ∼4% of wild flies (Brun and Plus 1980; Carpenter et al. 2007). DMelSV is a negative-sense RNA virus in the family Rhabdoviridae that is found in the cytoplasm of infected cells. Unlike bacterial symbionts, this virus is transmitted vertically through both sperm and eggs (Fleuriet 1988), so it is able to spread through populations despite being costly to infected flies (Seecof 1964; L’Heritier 1970; Fleuriet 1981). The pattern of DMelSV transmission differs between the sexes, with male flies transmitting at a lower rate than females (Brun and Plus 1980). Additionally, the transmission rate is reduced when the fly is infected by its father rather than its mother—if a female is infected by her father, her average transmission rate drops from ∼100% to a much lower rate (Brun and Plus 1980), and if a male is infected by his father, he does not transmit the virus at all. Therefore, the virus cannot be transmitted through males for two successive generations.
We have recently discovered two new sigma viruses in Drosophila obscura and Drosophila affinis—DObsSV and DAffSV (Longdon et al. 2010). Along with DMelSV, these viruses form a deep-branching clade in the Rhabdoviridae, which we have suggested be recognized as a new genus. However, important questions about their biology remain unanswered, including whether these new viruses are vertically transmitted. There is some evidence that DAffSV is: Williamson (1961) found that CO2 sensitivity was vertically transmitted in some lines of D. affinis in a way similar to that seen in DMelSV-infected flies [CO2 paralysis is a symptom of sigma viruses on their hosts (Longdon et al. 2010)]. We also do not know anything about the prevalence, population dynamics, or population genetics of these viruses. This article aims to address these questions by examining the transmission of these viruses in the lab and the dynamics of DObsSV in natural populations.
MATERIALS AND METHODS
Vertical transmission of viruses:
To test the mode of transmission of these newly discovered viruses, we carried out crosses between infected and uninfected virgin flies. The crosses used infected isofemale lines of D. affinis and D. obscura that were collected from Raleigh, North Carolina, and from Essex, United Kingdom, respectively, as described in Longdon et al. (2010). The crosses began with infected flies that had both an infected mother and an infected father (from a stock that was close to 100% infected). When both parents are infected, it has been shown for DMelSV that the viral type in the offspring is that of the mother (Brun and Plus 1980). The uninfected D. affinis isofemale lines were collected from the same location at the same time as the infected lines, and the uninfected D. obscura were isofemale lines collected during this study (see below). D. affinis flies were reared on a banana-malt-based Drosophila medium (see supporting information, Table S3), while D. obscura were reared on a cornmeal medium (Lewis 1960) with a piece of peeled mushroom (Agaricus bisporus) on the surface.
To test whether flies were infected with sigma virus, we exposed them to CO2 at 12° for 15 min and recorded flies dead or paralyzed 30 min later as infected. To confirm that CO2 sensitivity was linked to viral infection, we crossed infected males to uninfected females, carried out the CO2 assay on their offspring, and tested 15 paralyzed and 15 nonparalyzed/recovered offspring for sigma virus infection by quantitative real-time PCR (qRT-PCR) (40 cycles: 95° 15 sec, 60° 1 min) on an Applied Biosystems StepOnePlus system using a Power SYBR Green PCR Master-Mix (Applied Biosystems). Three technical replicates were carried out for each sample and primer pair, and samples were run in a blocked design across plates. The amount of virus was standardized to the housekeeping gene RpL32 (Rp49) to account for RNA extraction and reverse transcription efficiencies using the ▵▵ CT (critical threshold) method. Viral primers were designed to cross gene boundaries so only viral genomes were quantified (rather than mRNA); primer sequences are shown in Table S2. RpL32 endogenous control primers also crossed an intron–exon boundary to avoid amplifying genomic DNA contamination.
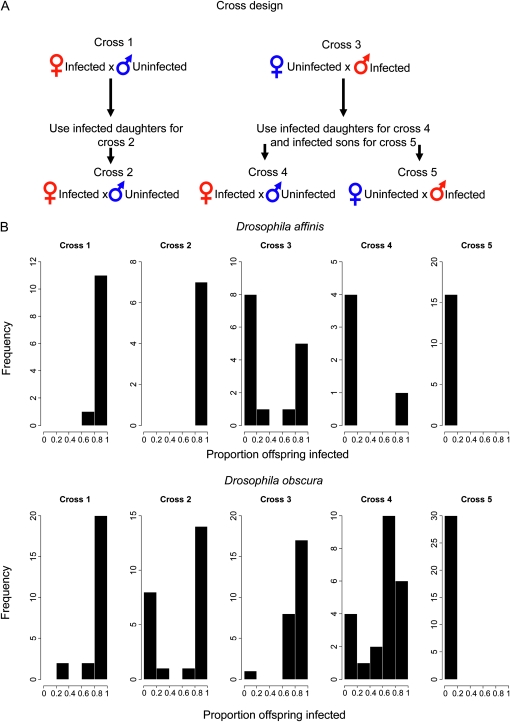
The following crosses were used to measure vertical transmission (Figure 1A) and to determine whether horizontal transmission occurred. In cross 1, infected females were crossed to uninfected males. Cross 2 took the daughters from cross 1 and crossed them to uninfected males. In cross 3, infected males were crossed to uninfected females. Cross 4 mated the daughters from cross 3 with uninfected males. Cross 5 mated the sons from cross 3 to uninfected females. Uninfected partners were assayed for infection to determine whether horizontal transmission had occurred.
Figure 1.—
(A) The cross diagram represents the fly crosses that were carried out to measure the transmission of each virus. (B) Histograms showing the proportion of infected offspring from each of the five crosses for D. affinis and D. obscura.
For D. affinis, multiple flies were placed in each vial, as the flies appear more likely to lay eggs when maintained at a higher stocking density. In cross 1, one to three females were placed in a vial with two to three males and allowed to lay eggs. For cross 3, two or three infected males were placed in a vial with one to three uninfected females. For crosses 4 and 5, the cross 3 offspring were placed individually in a vial with one or two uninfected flies of the opposite sex. Once eggs or larvae were visible, the adults were exposed to CO2 to confirm their infection status. In all crosses, uninfected partners were assayed for infection to test whether horizontal transmission had occurred.
For D. obscura, all crosses were carried out with a single pair of flies in each vial. Once eggs or larvae were visible, the parents were tested for DObsSV using a PCR assay on reverse-transcribed RNA (Longdon et al. 2010) (RT-PCR). PCR primers that amplify the RpL32 gene were used to check whether extractions were successful, and products were run on an ethidium bromide-stained 1% agarose gel. In crosses 1 and 3, the uninfected partners were also assayed for DObsSV to test whether horizontal transmission occurs. The emerging offspring were collected as virgins, aged, and mated as above to the appropriate uninfected lines. A mean of 25 replicates were set up for each cross, and a mean of three offspring were assayed by RT-PCR from each replicate. To examine whether males and females differ in their chances of being infected, we used a binomial test to examine whether the proportion of replicates where the majority of infected flies were female was significantly different from 50%.
To investigate the viral titers transmitted through eggs and sperm, the viral titer in early stage embryos was examined by qRT-PCR. Virgin females and males (with either the female or the male being infected) were placed together and allowed to lay eggs in bottles with a small amount of yeast paste on the surface of apple or grape juice agar. Embryos were collected twice daily and homogenized in Trizol (Invitrogen), using a microscope to ensure that the embryo was successfully crushed. Thirty embryos were collected for each cross. RNA was extracted and reverse-transcribed (see above), and qRT-PCR was used to measure the viral titer relative to an endogenous control (RpL32) using the delta delta CT method. If one or two of the technical replicates failed to amplify virus, these were given CT values of 40 for the statistical analysis. Any samples in which all three technical replicates failed to amplify using the viral PCR primers were classed as uninfected and excluded from the statistical analysis (i.e., the viral titers of infected embryos were compared). These samples are still present in Figure 2 and Figure 3. Two data points were removed from the DAffSV cross where the cDNA was of poor quality (RpL32 CT values >30). This did not affect the outcome of the analysis.
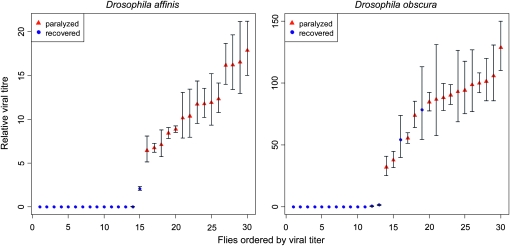
Figure 2.—
Viral titers in flies that were paralyzed or recovered after exposure to CO2. Titers were measured by quantitative RT-PCR on genomic viral RNA and are expressed relative to the copy number of the housekeeping gene RpL32. Error bars show the standard deviation of technical replicates.
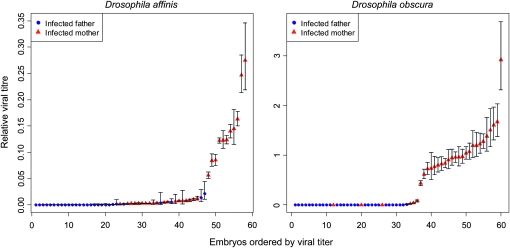
Figure 3.—
Viral titers in embryos that were infected either maternally or paternally. Titers were measured as in Figure 2. Error bars show the standard deviation of technical replicates.
Population samples of DObsSV:
D. obscura were collected from six woodland locations around the United Kingdom, with one to three sites at each location (longitude, latitude: Falmouth A 50.149411, −5.106007; Falmouth B 50.170063, −5.122495; Bristol A 51.455615, −2.639748; Bristol B 51.446629, −2.641035; Bristol C 51.340004, −2.782853; Essex 51.881352, 0.502710; Sussex 51.028827, −0.028390; Kent 51.099703, 0.164456 and 51.096517, 0.173151; and Derbyshire A 52.978411, −1.439769; Derbyshire B 52.883423, −1.398956). Flies were collected in the morning and evening from fruit baits. Males and females were separated, and females were placed in vials to establish isofemale lines. Flies from Kent were collected over two sites, using both bait traps and ground baits, and then combined. Flies at all other locations were collected using hanging bait traps. To examine whether the prevalence of the virus varied between sites, we used Fisher’s exact test and obtained P values by Monte Carlo simulation conditional on the row and column totals (10,000 replicates).
The wild-collected males and any females that did not lay eggs were tested for DObsSV infection by exposing them to CO2 as described above. The females that produced fertile eggs were not directly tested, but their infection status was inferred from whether their offspring were infected.
Fly identification:
D. obscura can be difficult to distinguish morphologically from Drosophila subobscura, and both species are common in the United Kingdom (Basden 1954; Shorrocks 1975). Therefore, all obscura group flies were collected and a diagnostic PCR assay was used to distinguish the species. RNA was extracted from flies paralyzed by the CO2 assay using Trizol reagent (Invitrogen, San Diego) in a chloroform–isopropanol extraction. RNA was then reverse-transcribed with M-MLV reverse transcriptase (Invitrogen) using random hexamer primers. DNA was extracted from flies that did not display CO2 sensitivity using chelex DNA extractions (Hurst et al. 2001). To confirm that nucleic acid extractions were successful, we amplified RpL32 from all samples. To identify the species, two sets of diagnostic PCR primers that amplify the mitochondrial cytochrome b (Cyt-b) gene and the nuclear alcohol dehydrogenase (Adh) gene (Table S2), were used. In both cases, a conserved forward primer was used. Two species-specific reverse primers were designed for Cyt-b and Adh by placing the 3′ end of the primer on a species-specific single nucleotide difference and on the penultimate 3′ base mismatching all of the available species sequences. Under suitably stringent PCR conditions (Table S1), these primers anneal to D. obscura or D. subobscura in different positions, resulting in different-sized products for the two species (Cyt-b and Adh give bands of 230 and 359 bp, respectively, for D. obscura and 575 and 194 bp for D. subobscura). The primers were designed such that they should not anneal to other common obscura group Drosophila found in the United Kingdom, and an agreement between assays was required for firm identification. To confirm reliability, we sequenced Cyt-b and/or COI from 28 wild flies (a mixture of D. obscura and other UK obscura group species), and in all cases the PCR test correctly identified the species.
Viral sequencing and sequence analysis:
To investigate the genetic diversity of DObsSV, we sequenced two regions of the virus, located in the N and L gene-coding sequence, of 634 and 648 bp, respectively. These genes were selected as they reside at opposite ends of the genome, so in the unlikely case of a recombination event (Chare et al. 2003), we would have more power to detect it. For the L gene, we used a variable region outside of the conserved motifs. These regions were amplified by PCR, and the PCR products were treated with exonuclease 1 and shrimp alkaline phosphatase to remove unused PCR primers and dNTPs and then sequenced directly using BigDye reagents (ABI, Carlsbad, CA) on an ABI 3730 capillary sequencer (provided by the Gene Pool Sequencing Facility, University of Edinburgh) in both directions. Sequences were edited in Sequencher (version 4.8; Gene Codes), and any polymorphisms were manually checked by eye. Direct sequencing of PCR products is expected to reduce the error rate to a negligible level (as compared to cloned sequences), and we confirmed this by repeating reverse transcription, PCR, and resequencing for 22 of the 67 SNPs (no errors were found). Any heterozygous sites (suggesting more than one viral infection in the host) were randomly assigned one of the possible base pairs (6/103 sequences contained a single ambiguity). Only one sequence contained more than one heterozygous site, and this was removed from the analysis as the phase of the haplotypes was unknown. If the heterozygous sites are removed from the Bayesian coalescent genealogy sampler (BEAST) or Tajima’s D analyses, this makes no difference to the conclusions (data not shown). The N and L gene sequences were concatenated, and median joining networks were created using the program Network (Bandelt et al. 1999). To assess whether Tajima’s D statistic (Tajima 1989) was significantly different from that expected under the standard neutral model, we produced a null distribution by recalculating the statistic from 1000 coalescent simulations conditional on the number of segregating sites observed in our data. We tested for recombination with a four-gamete test (Hudson and Kaplan 1985). To assess whether there was genetic differentiation between populations, we used the statistic KST [an analog of FST (Hudson et al. 1992)]. The statistical significance of KST was calculated by permuting the sequences across the populations and recalculating the statistic 10,000 times to produce a null distribution. These analyses were performed in DNA SP v5.0 (Librado and Rozas 2009).
To reconstruct past changes in the size of the viral population, we used BEAST (Drummond and Rambaut 2007). The substitution rate between viral sequences was assumed to be the same as in DMelSV (9.9 × 10−5 substitutions/site/year), which has been recently estimated from laboratory strains (L. Wilfert, unpublished data) and is similar to previous rate estimates for DMelSV (Carpenter et al. 2007), and other related viruses (Furio et al. 2005; Sanjuan et al. 2010). Carpenter et al. (2007) have previously shown that the lab-derived substitution rate in DMelSV does not differ significantly from that observed in the field. To account for uncertainty in this substitution rate estimate, we approximated its distribution with a truncated normal distribution (mean = 9.9 × 10−5, standard deviation = 3.6 × 10−5, lower limit = 1 × 10−10, upper limit = 1 substitutions/site/year), and this distribution was used as a fully informative prior. The model assumed a strict molecular clock model and an HKY85 substitution model (Hasegawa et al. 1985), which was selected after comparing Bayes factors with the more complex General Time Reversible model. Bayes factors were calculated from the marginal likelihoods by importance sampling, as implemented in Tracer (v1.4.1) (Rambaut and Drummond 2007), using the method of Suchard et al. (2001). Sites were partitioned into two categories by codon position (1+2, 3), and separate substitution rates were estimated for each category. Such codon partition models have been shown to be equivalent to more complex non codon-partitioned models (Shapiro et al. 2006). We first fitted a model of an exponentially expanding population (parameterized in terms of growth rate, rather than doubling time). This allowed us to exclude a constant population size, as the growth-rate parameter was significantly greater than zero (on the basis of the 95% highest posterior density interval). The population doubling time was calculated from the growth rate as ln(2)/growth rate. We also fitted a model that allows population size to vary freely over time (Bayesian skyline plot). Two runs of 500 million MCMC generations with sampling every 50,000 generations were run for each model, and a 10% burn-in was used for all parameter estimates. The two runs were combined and examined for convergence using Tracer (v1.4.1) (Rambaut and Drummond 2007). Posterior distributions were also examined using Tracer (v1.4.1) (Rambaut and Drummond 2007) to ensure an adequate number of independent samples. The 95% credible interval (C.I.) was taken as the region with the 95% highest posterior density. The two population size models were then compared by calculating Bayes factors as described above. Our analyses are based on both the combined sequences of two genes, but when each gene was analyzed independently, the results were very similar (data not shown).
RESULTS
CO2 paralysis and infection:
To examine whether DObsSV and DAffSV cause paralysis and death when infected flies are exposed to CO2, we crossed infected male flies to uninfected female flies and measured both viral titers and the effects of CO2 in the offspring. We found that in both species permanent paralysis is seen only in infected flies (Figure 2). Almost all flies contained some detectable virus, but flies that recovered after CO2 exposure had extremely low viral titers. In D. affinis, paralyzed flies had on average an 80.7 times greater viral titer (exact Wilcoxon rank sum test: W = 225, P < 0.0001), although 13 of 15 of flies that recovered after CO2 exposure also contained detectable amounts of virus. In D. obscura, the viral titer was on average 9.4 times greater in paralyzed flies (exact Wilcoxon rank sum test: W = 219, P < 0.0001), although all the flies contained detectable amounts of virus. Two of the flies that recovered after CO2 exposure had similar viral titers to the paralyzed flies.
Vertical transmission:
We found that DAffSV is transmitted in a similar way to DMelSV. In D. affinis, male flies transmit DAffSV to their offspring at a lower rate than females (Figure 1, cross 1 and cross 3; exact Wilcoxon rank sum test: W = 147.5, P < 0.001). Infected females transmitted the virus to 98% of their offspring over two successive generations (Figure 1, crosses 1 and 2), while males transmitted the virus to only 45% of their offspring (Figure 1, cross 3). The rate at which a fly transmits the virus to its offspring is also affected by whether the fly itself was infected by its mother or its father. If a female was infected by her father rather than her mother, then the average rate of transmission drops from 98% to 20% (cross 2 vs. cross 4, exact Wilcoxon rank sum test: W = 31, P = 0.01). If a male was infected by his father alone rather than his mother (and father), then the average rate of transmission drops from 45% to 0% (cross 3 vs. cross 5, exact Wilcoxon rank sum test: W = 208, P < 0.001). Therefore, the virus cannot be transmitted through males for two successive generations. None of the uninfected parental flies in the crosses were paralyzed by CO2, suggesting that horizontal transmission is either rare or absent.
We found that DObsSV in D. obscura is also vertically transmitted, but there are some important differences from DAffSV. As D. obscura can occasionally have a high viral titer yet recover from CO2 exposure (see above), we used RT-PCR rather than the CO2 assay to test flies for infection. Sex was not found to affect the likelihood of infection (binomial test: P = 0.35), so both sexes were analyzed together. Unlike in D. affinis, male flies transmit the virus to their offspring at a similar rate to females: infected females transmitted the virus to 92% of their offspring, while males transmitted the virus to 88% of their offspring (Figure 1, cross 1 and cross 3; exact Wilcoxon rank sum test: W = 357, P = 0.259). Furthermore, the rate at which a female transmits the virus to her offspring was not affected by whether she received the infection from her mother or her father (Figure 1, cross 2 and cross 4; exact Wilcoxon rank sum test: W = 3125.5, P = 0.435), with females infected by their mothers or fathers having transmission of 63% and 61%, respectively. Note that female transmission also declined from cross 1 to cross 2 (Figure 1, cross 1 and cross 2; exact Wilcoxon rank sum test: W = 376, P = 0.022) from 92% to 61%. If a male was infected by his father rather than his mother (and father), then the average rate of transmission dropped from 88% to 0% (Figure 1, cross 3 vs. cross 5; exact Wilcoxon rank sum test: W = 765, P < 0.001). Therefore, this virus cannot be transmitted through males for two successive generations. To check for any horizontal transmission, we also tested the uninfected parents in crosses 2 and 3. We found that horizontal transmission was rare or absent, as only one female had a very faint viral band, and this could be due to the presence of infected sperm.
Viral titers transmitted in eggs and sperm:
The different rates that males and females transmit to their offspring may be because eggs and sperm contain different numbers of virions. To investigate this hypothesis, we measured the viral titers of early stage embryos that had either an infected mother or an infected father. Considering only the embryos where there were detectable amounts of virus, in both species the embryonic viral titer was less when the virus was paternally transmitted than when it was maternally transmitted (Figure 3; DAffSV exact Wilcoxon rank sum test: W = 61, P < 0.0001; DObsSV exact Wilcoxon rank sum test: W = 17, P < 0.0001). In addition, more embryos contained no detectable virus after paternal transmission (47% of DObsSV and 4% of DAffSV paternally infected eggs and 3% of DObsSV and 0% of DAffSV maternally infected eggs had no detectable virus).
Population dynamics of DObsSV:
To examine whether the biparental pattern of vertical transmission that we have observed can explain the invasion and maintenance of the virus in populations, we simulated the spread of DObsSV on the basis of the transmission rates seen in our experiments. Pi is the prevalence in the adult population in generation i. We can calculate the prevalence among adults in generation i + 1 (Pi+1) from the proportion of infected and uninfected sperm (I♂ and U♂) and the proportion of infected and uninfected eggs (I♀ and U♀) produced in the previous generation. The frequency of infected and uninfected gametes can be calculated from the rate of vertical transmission and any change in the fertility of infected flies relative to uninfected flies (Table 1). Because male D. obscura that inherit the virus from their father do not transmit the virus to the next generation, we split the infected population into a fraction si that inherited the virus from their mother and a fraction (1 – si) that inherited the virus solely from their father.
TABLE 1.
Proportions of infected and uninfected eggs and sperm
| Gametes | Equation |
| Infected eggs | |
| Uninfected eggs | |
| Infected sperm | |
| Uninfected sperm |
Pi is the proportion of adults infected in generation i, which we assume to be equal for males and females. The virus is transmitted from mother to offspring at a rate t♀ and from father to offspring at rate t♂. We assume that both female and male fertility are changed by a factor C in infected flies relative to uninfected flies. We split the infected population in generation i into a fraction si that inherited the virus from their mother and a fraction (1 – si) that inherited the virus from their father. Male D. obscura that inherit the virus from their father do not transmit it to the next generation (i.e., they produce uninfected sperm). To obtain proportions, we divide by w♀, the sum of the numerators of I♀ and U♀, and w♂, the sum of the numerators of I♂ and U♂.
| (1) |
| (2) |
Using the transmission rates estimated in generation 1 (t♂ = 0.88, t♀ = 0.92), we found that the virus can rapidly invade a population and reach a high prevalence (Figure 4). Yampolsky et al. (1999) estimated that DMelSV reduces the fitness of D. melanogaster in the wild by ∼20–30%. If DObsSV causes a similar reduction in the fertility of infected flies, our simulations suggest that the virus can still rapidly invade a population (Figure 4). We repeated this analysis using the lower female transmission rate measured in the second generation (cross 2). This causes the virus to spread much more slowly, and it can invade only if the virus reduces the fertility of infected flies by <10% (data not shown).
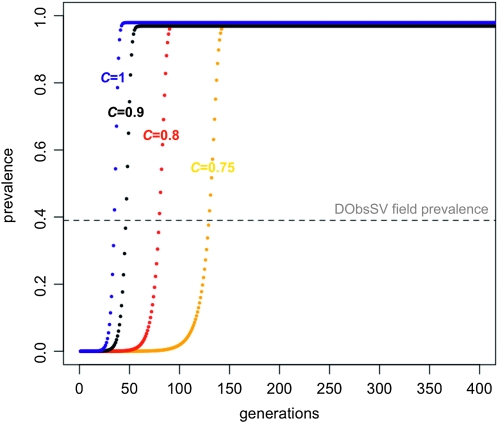
Figure 4.—
Simulations of DObsSV spreading through a population based on lab estimates of transmission rates and a range of possible fertility reductions. Colors represent the different fertilities of infected flies relative to uninfected flies (C), with blue, black, red, and yellow representing C = 1, 0.9, 0.8, and 0.75, respectively. The virus failed to invade if fertility is reduced by >25% (C < 0.75). The dashed horizontal line represents the mean prevalence of the virus in our samples. The transmission rates were t♂ = 0.88 and t♀= 0.92, and the starting frequency of infected flies was 10−6. In the United Kingdom, there are approximately three to four generations of D. obscura each year (Begon 1976).
Prevalence of DObsSV:
We tested 267 D. obscura collected from sites across the United Kingdom for infection with DObsSV using the CO2 assay and found that 103 (39%) were infected. The prevalence of DObsSV varied widely between sites (Table 2 and Figure S1; Fisher’s exact test, P = 0.0001). This is primarily caused by a low prevalence in Kent, but even if this location is excluded from the analysis, there is still significant variation between sites (Fisher’s exact test: p = 0.04). Because the results from the qRT-PCR linking CO2 paralysis and infection (see above) found that not all infected flies are CO2 sensitive, these values may be an underestimate of prevalence.
TABLE 2.
Percentage of flies infected and number of flies collected at each field site
| Site | Prevalence (%) | N |
| Bristol A | 33 | 33 |
| Bristol B | 62 | 21 |
| Bristol C | 50 | 2 |
| Derbyshire A | 48 | 66 |
| Derbyshire B | 73 | 11 |
| Kent | 22 | 83 |
| Sussex | 48 | 42 |
| Essex | 0 | 3 |
| Falmouth A | 0 | 5 |
| Falmouth B | 0 | 1 |
We confirmed that the CO2 assay accurately identifies infected flies by qRT-PCR, and of 105 lines that were paralyzed by CO2, 103 were infected. In all these samples, we sequenced two regions of the viral genome covering 634 bp of the N gene at the 3′ end of the viral genome and 648 bp of the L gene toward the 5′ end of the genome, and all the sequences were clearly DObsSV.
Viral sequence analysis:
The genetic diversity of DObsSV is very low. There were only 67 segregating sites over 1282 bp of sequence from all 103 viral isolates (30 in 634 bases of the N gene and 37 in 648 bases of the L gene with no obvious clustering of the SNPs within the sequences), and the average number of pairwise differences per site (π) was 0.002 across all sites and 0.006 at synonymous sites. We verified a third of our SNPs by repeating reverse transcription, PCR, and sequencing reactions and found no errors.
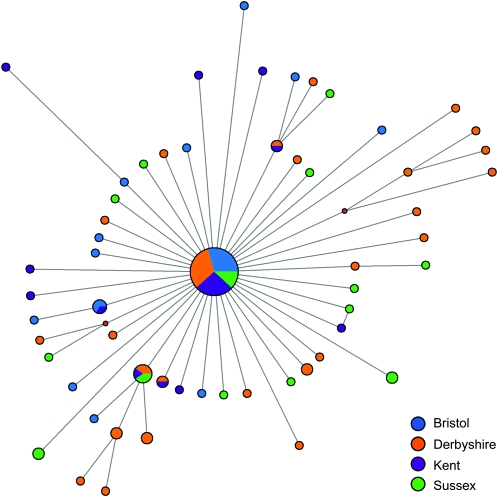
The phylogenetic network of the sequences is a star shape (Figure 5), suggesting a recent selective sweep or population expansion. This is caused by a large excess of rare variants in the data set—of the 67 segregating sites, 51 are singletons. For this reason, estimates of θW, [which are derived from the number of segregating sites and are insensitive to their frequency (Watterson 1975)] are greater than π (θW = 0.011 for all sites and θW = 0.036 for synonymous sites, compared to 0.002 and 0.006). This excess of rare polymorphisms is significantly greater than expected under the neutral model (Tajima’s D = −2.75; P < 0.001).
Figure 5.—
Phylogenetic network of DObsSV sequences. Nodes are color coded on the basis of location, and their size is proportional to the frequency of viral sequences. Branches are approximately sized to the number of mutations.
An excess of rare polymorphisms could result either from a recent sweep of the virus through the host population or from purifying selection on the SNPs in our data set. If the latter hypothesis were true, then we would expect the frequency of different functional classes of polymorphisms to be different as they are likely to have different effects on fitness. However, when analyzed independently, the N and L genes each had a significant excess of rare polymorphisms relative to the neutral expectation (Tajima’s D = −2.54, −2.68, respectively; P < 0.001 for each). Additionally, Tajima’s D differed very little between synonymous sites (−2.47, −2.55 for N and L, respectively; P < 0.001 for each) and nonsynonymous sites (−1.77 and −2.35, P < 0.001 for each) and was not more negative at nonsynonymous sites. It therefore seems unlikely that the departure from neutrality is driven by purifying selection.
There is very little genetic differentiation between viral sequences from different geographic locations (Figure 5). This is reflected in a low KST value of 0.015 (as mentioned above, KST is an analog of FST that measures the proportion of the genetic variation contained in subpopulations relative to the population as a whole). Despite the very low value of KST, it was significantly greater than zero (permutation test: P = 0.0013).
To check whether there might have been any recombination between our sequences, we used the four-gamete test. There was only a single pair of sites where all four gametes exist, suggesting that recombination is either very rare or absent. Given the apparent lack of recombination in negative-sense RNA viruses (Chare et al. 2003), this is most likely to result from homoplasy rather than recombination.
We reconstructed past changes in the size of the population using the coalescent sampler BEAST. A comparison of the Bayes factors indicates that the model of an exponentially expanding population was preferred over the skyline model in which the population size is free to vary through time (log10 Bayes factors averaged over two runs for each model was 54 in favor of exponential growth over the skyline coalescent model). Using the exponential model, we were able to reject a constant population size as the posterior distribution of the growth rate parameter does not include zero (P < 0.0001). We estimated that the effective population size of the virus has doubled approximately every 9 months (mean doubling time = 0.76 years; 95% C.I.: 0.24–1.51 years), and all the genotypes in our sample shared a common ancestor 11 years ago (95% C.I.: 4–19 years). When analyzed independently, the N and L genes gave similar estimates to each other and the combined analysis (data not shown).
These estimates are compatible with the very rapid invasion of the virus that is predicted by our simulations (Figure 4). Assuming that the host undergoes three to four generations per year in the United Kingdom (Begon 1976), the virus could reach its current prevalence of 39% within 12–16 years, even if infected hosts suffer a fertility reduction of 10% compared to uninfected individuals. The simulations also suggest that DObsSV may still be spreading in the United Kingdom, as the equilibrium prevalence in the simulations is higher than we have observed in the wild (Figure 4).
DISCUSSION
Vertical transmission:
We have found that two recently discovered rhabdoviruses from D. affinis and D. obscura are both vertically transmitted, and horizontal transmission is rare or absent over experimental timescales. To our knowledge, aside from viruses that are integrated into insect genomes, the only other obligately vertically transmitted virus that has been reported in animals is DMelSV from D. melanogaster. Our results suggest that sigma viruses may be common vertically transmitted insect symbionts.
If a vertically transmitted symbiont is transmitted solely by females, then anything less than perfect transmission is expected to lead to a decline in prevalence and ultimately extinction (L’Heritier 1970). Vertically transmitted bacteria use various different strategies to avoid this, including distorting the sex ratio toward females, spitefully reducing the fitness of uninfected individuals by causing cytoplasmic incompatibility (Hurst et al. 1993), or providing a fitness benefit to the host such as nutrients (Douglas 1989) or protection from pathogens (Hedges et al. 2008; Teixeira et al. 2008; Brownlie and Johnson 2009; Jaenike et al. 2010). An alternative strategy to spread through host populations is to be transmitted through both sperm and eggs. This is rarely seen in bacterial symbionts, probably because sperm contain little cytoplasm and hence few bacteria (Hurst 1990). However, although sigma viruses infect the cytoplasm of host cells, they have evolved biparental vertical transmission (L’heritier 1970); the sigma viruses may not be unique in this mode of transmission. Rhabdoviruses have been found in hemipteran sperm cells (Afzelius et al. 1989), and in Culex mosquitoes, CO2 sensitivity (a common phenotype of rhabdovirus infection that causes infected insects to become paralyzed after CO2 exposure) is inherited extra-chromosomally in a biparental manner (Shroyer and Rosen 1983). Furthermore, other virus-like particles have been found in the sperm of a range of different insects (Tandler 1972; Schrankel and Schwalm 1975; Degrugillier et al. 1991; Bao et al. 1996; Ferber et al. 1997; Wolf 1997; Longdon and Jiggins 2010). Together these results suggest that viruses may be transmitted vertically by both males and females much more often than is the case for bacterial symbionts.
The mode of transmission of the viruses that we studied is similar to that of DMelSV. In D. affinis, males transmit the virus at a lower rate than females, and the rate of transmission is reduced in flies that have inherited the virus from their father rather than from their mother (females have a reduced transmission rate and males do not transmit the virus at all). In D. obscura, males have comparable transmission rates to females, but males infected by their fathers do not transmit the virus. Curiously, we also found a reduction in transmission after two successive female generations, which may be due to the first generation of females being infected by both parents, or the uninfected flies being partially resistant. Although this high level of paternal transmission could potentially act to aid the spread of the virus, the reduced transmission by maternally infected daughters means that, over all, the virus is transmitted at a similar rate to DAffSV.
We found that DObsSV and DAffSV embryos that were infected by their fathers have a lower viral titer than those infected by their mother. It seems likely that the small size of the sperm and the fact that the cells that form one egg divide to form 64 sperm cells (Williamson and Lehman 1996) limits the amount of virus transmitted to offspring. This pattern has previously been observed in DMelSV (Brun and Plus 1980) and probably explains why paternal transmission is less efficient than maternal transmission in DMelSV and DAffSV. DObsSV has comparable paternal and maternal transmission rates (Figure 1, crosses 1 and 3) even though paternally infected embryos have much lower viral titers. We hypothesize that, although the low viral titer may not be limiting for one generation, over two generations this twofold dilution effect means that sons infected by their fathers do not transmit the virus. In DMelSV, the viral titers in paternally infected flies have recovered to normal levels, and yet flies infected from their father still transmit the virus at lower rates (Brun and Plus 1980). It has therefore been suggested that it is critical for the virus to infect the germline cells early in development if it is to be transmitted efficiently (Fleuriet 1988). This may also explain why DAffSV and DObsSV are transmitted less efficiently by flies that were infected by their father rather than by their mother.
Population dynamics:
Parasitic bacterial symbionts often have highly dynamic associations with their hosts, with new strains frequently spreading through host populations. Comparisons of insect and bacterial phylogenies have shown that symbionts rarely co-speciate with their hosts, but instead frequently switch between different host species (Werren et al. 1995; Weinert et al. 2009). These bacteria can spread very rapidly through populations. For example, Wolbachia spread at a rate of >100 km/year through uninfected populations in Drosophila simulans on the West Coast of the United States (Turelli and Hoffmann 1991). After a symbiont has invaded a population, co-evolution with the host can cause the turnover of strains within the population. For example, after Wolbachia had invaded U. S. populations of D. simulans, it evolved from a parasitic relationship toward a mutualistic one (Weeks et al. 2007). There can be similarly rapid evolution of the host population, where genes that make the host resistant to the pathogenic effects of symbionts can rapidly spread (Hornett et al. 2006).
Similar processes have occurred in D. melanogaster and its sigma virus DMelSV. A naturally occurring polymorphism in the Drosophila gene ref(2)P blocks the transmission of the virus through females, and natural selection has caused the resistant allele of this gene to spread through natural populations (Wayne et al. 1996; Bangham et al. 2008). In response, from the early 1980s to the early 1990s, DMelSV genotypes that are able to overcome this resistance were observed to sweep through two different European populations (Fleuriet et al. 1990; Fleuriet and Sperlich 1992). More recent molecular data confirm that the DMelSV type currently found in Europe has recently spread through the host population, with all the viral isolates in Europe sharing a common ancestor ∼200 years ago (Carpenter et al. 2007), which was either due to a selective sweep or due to D. melanogaster acquiring the virus from another species. This raises the question as to how common such sweeps of vertically transmitted parasites are in nature.
We found that DObsSV has very recently swept though British populations of D. obscura. This sweep has occurred in the past ∼11 years, with the frequency of this strain doubling every 9 months. Our model shows that the biparental transmission of the virus can explain these rapid changes in prevalence by creating a considerable drive through the host population. Furthermore, the virus can still rapidly spread even when it reduces fertility by up to 25% (although a reduction of ∼10% most closely matches the timescale of the sweep estimated using the sequence data).
As the virus does not recombine, we cannot tell whether the spread of the virus was caused by a selective sweep of an advantageous mutation through an existing viral population or by the spread of a new virus from a different species or population through an uninfected population. If this was a selective sweep, the new strain must have had a large selective advantage over existing viruses to spread so rapidly and must have almost totally replaced those viruses as there were no more divergent genotypes in our sample. To separate these hypotheses, we would need to discover either closely related viruses in other species or populations or remnants of a more diverse viral population that existed before a selective sweep.
In conclusion, our results suggest that vertically transmitted viruses may prove to be common in insect populations. Our simulations based on estimates of the transmission rates predict that this mode of transmission can drive very rapid changes in prevalence. In natural populations, we have found this to be the case, with DObsSV sweeping rapidly through populations over the past decade.
Acknowledgments
B.L. thanks everyone who provided fieldwork accommodation and lab space: Jonny Turner, Tom Price, Clare Stamper, Matthew Boyce, Sue and Keith Obbard, and Neil and Jacky Longdon. Thanks to Andrew Rambaut who provided numerous useful comments and suggestions. Many thanks to Jim Bull for his comments and suggestions, which greatly improved the manuscript. We also thank four anonymous reviewers for useful comments. B.L. is supported by a Biotechnology and Biological Sciences Research Council studentship. L.W. is supported by a Leverhulme trust grant. D.J.O. is funded by Wellcome Trust Research Career Development Fellowship 085064/Z/08/Z. F.M.J. is funded by a Royal Society University Research Fellowship and a Wellcome Trust Project Grant.
LITERATURE CITED
- Afzelius B. A., Alberti G., Dallai R., Godula J., Witalinski W., 1989. Virus-infected and Rickettsia-infected sperm cells in arthropods. J. Invertebr. Pathol. 53: 365–377 [Google Scholar]
- Bandelt H. J., Forster P., Rohl A., 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16: 37–48 [DOI] [PubMed] [Google Scholar]
- Bangham J., Kim K. W., Webster C. L., Jiggins F. M., 2008. Genetic variation affecting host-parasite interactions: different genes affect different aspects of sigma virus replication and transmission in Drosophila melanogaster. Genetics 178: 2191–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S. N., Kitajima E. W., Callaini G., Dallai R., 1996. Virus-like particles and Rickettsia-like organisms in male germ and cyst cells of Bemisia tabaci (Homoptera, Aleyrodidae). J. Invertebr. Pathol. 67: 309–311 [DOI] [PubMed] [Google Scholar]
- Basden E. B., 1954. The distribution and biology of Drosophilidae (Diptera) in Scotland, including a new species of Drosophila. Trans. R. Soc. Edinb. 62: 602–654 [Google Scholar]
- Begon M., 1976. Temporal variations in reproductive condition of Drosophila obscura fallen and Drosophila subobscura Collin. Oecologia 23: 31–47 [DOI] [PubMed] [Google Scholar]
- Bezier A., Annaheim M., Herbiniere J., Wetterwald C., Gyapay G., et al. , 2009. Polydnaviruses of Braconid wasps derive from an ancestral nudivirus. Science 323: 926–930 [DOI] [PubMed] [Google Scholar]
- Brownlie J. C., Johnson K. N., 2009. Symbiont-mediated protection in insect hosts. Trends Microbiol. 17: 348–354 [DOI] [PubMed] [Google Scholar]
- Brun G., Plus N., 1980. The viruses of Drosophila, pp. 625–702 in The Genetics and Biology of Drosophila, edited by Ashburner M., Wright T. R. F. Academic Press, New York [Google Scholar]
- Buchner P., 1965. Endosymbiosis of Animals With Plant Microorganisms. Interscience, New York [Google Scholar]
- Carpenter J. A., Obbard D. J., Maside X., Jiggins F. M., 2007. The recent spread of a vertically transmitted virus through populations of Drosophila melanogaster. Mol. Ecol. 16: 3947–3954 [DOI] [PubMed] [Google Scholar]
- Chare E. R., Gould E. A., Holmes E. C., 2003. Phylogenetic analysis reveals a low rate of homologus recombination in negative-sense RNA viruses. J. Gen. Virol. 84: 2691–2703 [DOI] [PubMed] [Google Scholar]
- Degrugillier M. E., Degrugillier S. S., Jackson J. J., 1991. Nonoccluded, cytoplasmic virus particles and Rickettsia-like organisms in testes and Spermathecae of Diabrotica virgifera. J. Invertebr. Pathol. 57: 50–58 [Google Scholar]
- Douglas A. E., 1989. Mycetocyte symbiosis in insects. Biol. Rev. Camb. Philos. Soc. 64: 409–434 [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A., 2007. BEAST: Bayesian evolutionary analysis by sampling trees. Bmc Evol. Biol. 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferber M. L., Rios A. F., Kuhl G., Comendador M. A., Louis C., 1997. Infection of the gonads of the SimES strain of Drosophila simulans by the hereditary reovirus DSV. J. Invertebr. Pathol. 70: 143–149 [DOI] [PubMed] [Google Scholar]
- Fleming J., Summers M. D., 1991. Polydnavirsus DNA is integrated in the DNA of its parasitoid wasp host. Proc. Natl. Acad. Sci. USA 88: 9770–9774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleuriet A., 1981. Comparison of various physiological traits in flies (Drosophila melanogaster) of wild origin, infected or uninfected by the hereditary Rhabdovirus sigma. Arch. Virol. 69: 261–272 [DOI] [PubMed] [Google Scholar]
- Fleuriet A., 1988. Maintenance of a hereditary virus—the Sigma—virus in populations of its host, Drosophila melanogaster. Evol. Biol. 23: 1–30 [Google Scholar]
- Fleuriet A., Sperlich D., 1992. Evolution of the Drosophila melanogaster sigma virus system in a natural population from Tubingen. Theor. Appl. Genet. 85: 186–189 [DOI] [PubMed] [Google Scholar]
- Fleuriet A., Periquet G., Anxolabehere D., 1990. Evolution of natural populations in the Drosophila melanogaster sigma virus system. 1. Languedoc (Southern France). Genetica 81: 21–31 [DOI] [PubMed] [Google Scholar]
- Furio V., Moya A., Sanjuan R., 2005. The cost of replication fidelity in an RNA virus. Proc. Natl. Acad. Sci. USA 102: 10233–10237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Kishino H., Yano T. A., 1985. Dating of the human ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22: 160–174 [DOI] [PubMed] [Google Scholar]
- Hedges L. M., Brownlie J. C., O'Neill S. L., Johnson K. N., 2008. Wolbachia and virus protection in insects. Science 322: 702. [DOI] [PubMed] [Google Scholar]
- Heredia F., Loreto E. L. S., Valente V. L. S., 2007. Distribution and conservation of the transposable element gypsy in Drosophilid species. Genet. Mol. Biol. 30: 133–138 [Google Scholar]
- Hornett E. A., Charlat S., Duplouy A. M. R., Davies N., Roderick G. K., et al. , 2006. Evolution of male-killer suppression in a natural population. PloS Biol. 4: 1643–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Kaplan N. L., 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Boos D. D., Kaplan N. L., 1992. A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 9: 138–151 [DOI] [PubMed] [Google Scholar]
- Hurst G. D. D., Hurst L. D., Majerus M. E. N., 1993. Altering sex ratios: the games microbes play. Bioessays 15: 695–697 [Google Scholar]
- Hurst G. D. D., Jiggins F. M., Robinson S. J. W., 2001. What causes inefficient transmission of male-killing Wolbachia in Drosophila? Heredity 87: 220–226 [DOI] [PubMed] [Google Scholar]
- Hurst L. D., 1990. Parasite diversity and the evolution of diploidy, multicellularity and anisogamy. J. Theor. Biol. 144: 429–443 [DOI] [PubMed] [Google Scholar]
- Jaenike J., Unckless R., Cockburn S. N., Boelio L. M., Perlman S. J., 2010. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329: 212–215 [DOI] [PubMed] [Google Scholar]
- Lewis E., 1960. A new standard food medium. Drosoph. Inf. Serv. 34: 117–118 [Google Scholar]
- L'Heritier P. H., 1970. Drosophila viruses and their role as evolutionary factors. Evol. Biol. 4: 185–209 [Google Scholar]
- Librado P., Rozas J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452 [DOI] [PubMed] [Google Scholar]
- Longdon B., Jiggins F. M., 2010. Paternally transmitted parasites. Curr. Biol. 20: R695–R696 [DOI] [PubMed] [Google Scholar]
- Longdon B., Obbard D. J., Jiggins F. M., 2010. Sigma viruses from three species of Drosophila form a major new clade in the rhabdovirus phylogeny. Proc. Biol. Sci. 277: 35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mims C. A., 1981. Vertical transmission of viruses. Microbiol. Rev. 45: 267–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Drummond A. J., 2007. Tracer v1.4. Available at http://beast.bio.ed.ac.uk/Tracer
- Sanjuan R., Nebot M. R., Chirico N., Mansky L. M., Belshaw R., 2010. Viral mutation rates. J. Virol. 84: 9733–9748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrankel K. R., Schwalm F. E., 1975. Viruslike particles in spermatids of Coelopa frigida (Diptera). J. Invertebr. Pathol. 26: 265–268 [Google Scholar]
- Seecof R. L., 1964. Deleterious effects on Drosophila development associated with sigma virus infection. Virology 22: 142 [Google Scholar]
- Shapiro B., Rambaut A., Drummond A. J., 2006. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 23: 7–9 [DOI] [PubMed] [Google Scholar]
- Shorrocks B., 1975. Distribution and abundance of woodland species of British Drosophila (Diptera-Drosophilidae). J. Anim. Ecol. 44: 851–864 [Google Scholar]
- Shroyer D. A., Rosen L., 1983. Extrachromosomal inheritance of carbon dioxide sensitivity in the mosquito Culex quinquefasciatus. Genetics 104: 649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard M. A., Weiss R. E., Sinsheimer J. S., 2001. Bayesian selection of continous-time Markov chain evolutionary models. Mol. Biol. Evol. 18: 1001–1013 [DOI] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandler B., 1972. Viruslike particles in testis of Drosophila virilis. J. Invertebr. Pathol. 20: 214. [DOI] [PubMed] [Google Scholar]
- Teixeira L., Ferreira A., Ashburner M., 2008. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PloS Biol. 6: 2753–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M., Hoffmann A. A., 1991. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature 353: 440–442 [DOI] [PubMed] [Google Scholar]
- Watterson G. A., 1975. Number of segregating sites in genetic models without recombination. Theor. Popul. Biol. 7: 256–276 [DOI] [PubMed] [Google Scholar]
- Wayne M. L., Contamine D., Kreitman M., 1996. Molecular population genetics of ref(2)P, a locus which confers viral resistance in Drosophila. Mol. Biol. Evol. 13: 191–199 [DOI] [PubMed] [Google Scholar]
- Weeks A. R., Turelli M., Harcombe W. R., Reynolds K. T., Hoffmann A. A., 2007. From parasite to mutualist: rapid evolution of Wolbachia in natural populations of Drosophila. PloS Biol. 5: 997–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert L. A., Werren J. H., Aebi A., Stone G. N., Jiggins F. M., 2009. Evolution and diversity of Rickettsia bacteria. BMC Biol. 7: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J. H., Zhang W., Guo L. R., 1995. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc R. Soc. Lond. Ser. B Biol. Sci. 261: 55–63 [DOI] [PubMed] [Google Scholar]
- Williamson A., Lehman R., 1996. Germ cell development in Drosophila. Annu. Rev. Cell Dev. Biol. 12: 365–391 [DOI] [PubMed] [Google Scholar]
- Williamson D., 1961. Carbon dioxide sensitivity in Drosophila affinis and Drosophila athabasca. Genetics 46: 1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K. W., 1997. Virus-like particles, bacteria and microsporidia affect spindle-associated membranes in spermatocytes of Lepidoptera species. Zygote 5: 21–30 [DOI] [PubMed] [Google Scholar]
- Yampolsky L. Y., Webb C. T., Shabalina S. A., Kondrashov A. S., 1999. Rapid accumulation of a vertically transmitted parasite triggered by relaxation of natural selection among hosts. Evol. Ecol. Res. 1: 581–589 [Google Scholar]