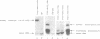Abstract
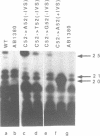
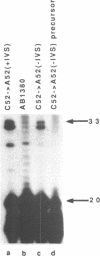
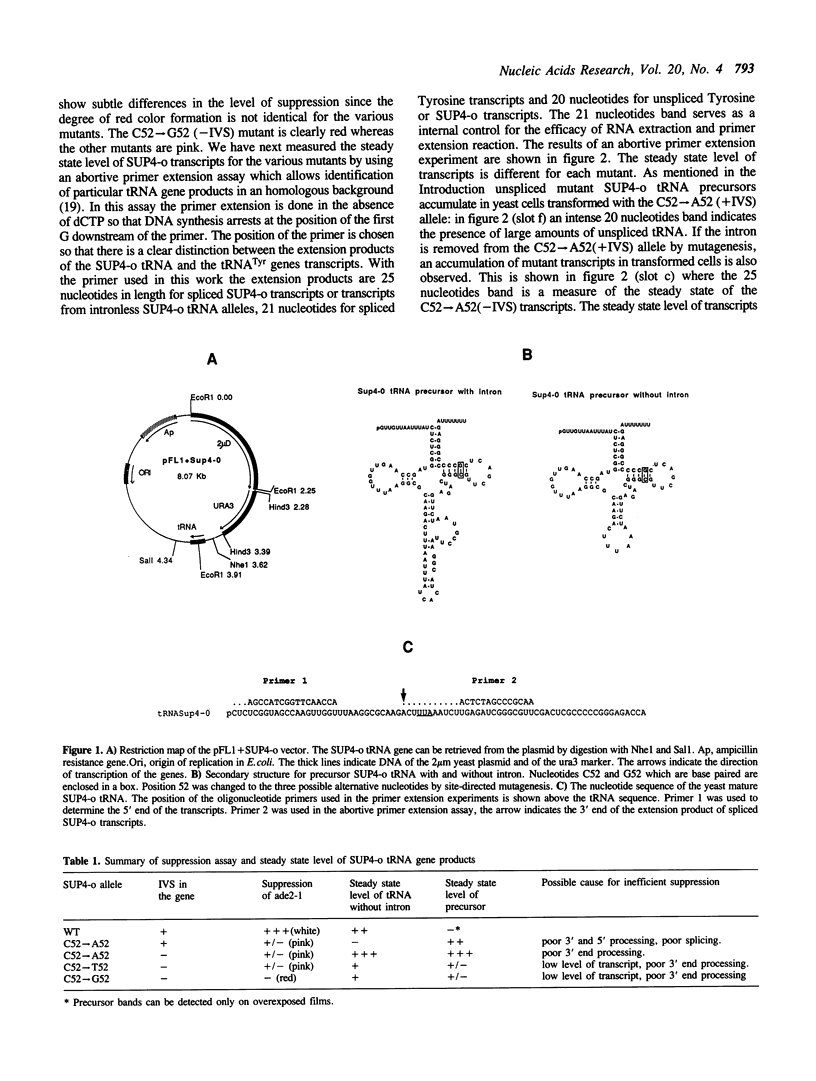
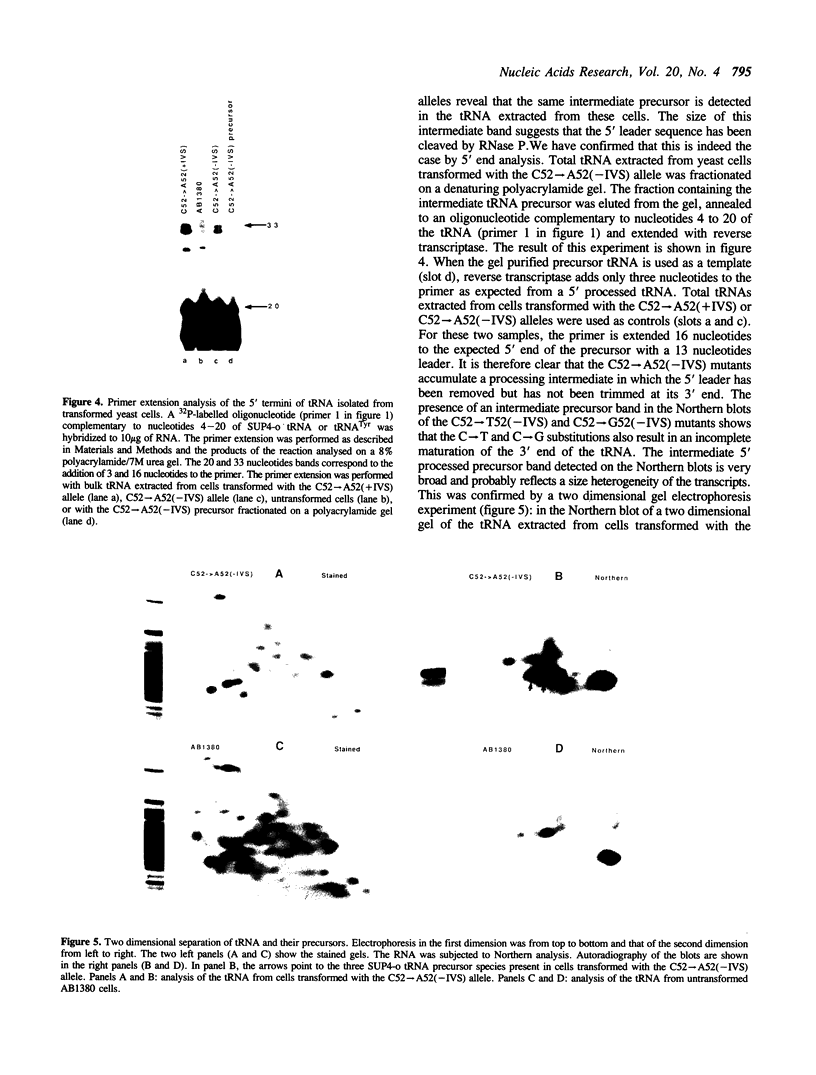
The expression of mutant tyrosine-inserting ochre suppressor SUP4-o tRNA genes in vivo in S. cerevisiae was examined as a basis for further studies of tRNA transcription and processing. In vivo yeast precursor tRNAs have been identified by filter hybridization and primer extension analysis. We have previously shown that a mutant SUP4-o tRNA gene with a C52----A52 transversion at positive 52 (C52----A52(+IVS) allele) was transcribed but that the primary transcript was not processed correctly. We show here that 5' and 3' end processing as well as splicing are defective for this mutant but that the 5' end processing is restored when the intron is removed from the gene by oligonucleotide directed mutagenesis (C52----A52(-IVS) allele). Our results imply that the C52----A52 transversion by itself cannot account for the lack of susceptibility to RNase P cleavage but that the overall tertiary structure of the mutant tRNA precursor is destabilized by the intron/anticodon stem. A second consequence of the C52----A52 transversion is to prevent complete maturation of the tRNA precursor at its 3' end since intermediates containing incompletely processed 3' trailers accumulate in the yeast cells transformed with the C52----A52(-IVS) allele. A correct structure of the T stem might therefore define a structural feature required for the recognition of the 3' processing activity.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chevallier M. R., Bloch J. C., Lacroute F. Transcriptional and translational expression of a chimeric bacterial-yeast plasmid in yeasts. Gene. 1980 Oct;11(1-2):11–19. doi: 10.1016/0378-1119(80)90082-7. [DOI] [PubMed] [Google Scholar]
- De Robertis E. M., Olson M. V. Transcription and processing of cloned yeast tyrosine tRNA genes microinjected into frog oocytes. Nature. 1979 Mar 8;278(5700):137–143. doi: 10.1038/278137a0. [DOI] [PubMed] [Google Scholar]
- Dingermann T., Nerke K. Primer extension analysis of tRNA gene transcripts synthesized in vitro and in vivo. Anal Biochem. 1987 May 1;162(2):466–475. doi: 10.1016/0003-2697(87)90422-2. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Gegenheimer P., Abelson J. Nucleolytic processing of a tRNAArg-tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae. J Biol Chem. 1985 Jan 25;260(2):1271–1279. [PubMed] [Google Scholar]
- Fradin A., Gruhl H., Feldmann H. Mapping of yeast tRNAs by two-dimensional electrophoresis on polyacrylamide gels. FEBS Lett. 1975 Feb 1;50(2):185–189. doi: 10.1016/0014-5793(75)80485-6. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Dingermann T., Cooley L., Söll D. Processing of precursor tRNAs in Drosophila. Processing of the 3' end involves an endonucleolytic cleavage and occurs after 5' end maturation. J Biol Chem. 1985 Jan 10;260(1):449–454. [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A. K. Genetic methods for study of trans-acting genes involved in processing of precursors to yeast cytoplasmic transfer RNAs. Methods Enzymol. 1990;181:400–421. doi: 10.1016/0076-6879(90)81139-l. [DOI] [PubMed] [Google Scholar]
- Hopper A. K., Kurjan J. tRNA synthesis: identification of in vivo precursor tRNAs from parental and mutant yeast strains. Nucleic Acids Res. 1981 Feb 25;9(4):1019–1029. doi: 10.1093/nar/9.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. D., Ogden R., Johnson P., Abelson J., Dembeck P., Itakura K. Transcription and processing of a yeast tRNA gene containing a modified intervening sequence. Proc Natl Acad Sci U S A. 1980 May;77(5):2564–2568. doi: 10.1073/pnas.77.5.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle D., Wehmeyer U., Krupp G. Substrate recognition by RNase P and by the catalytic M1 RNA: identification of possible contact points in pre-tRNAs. EMBO J. 1990 Jun;9(6):1929–1937. doi: 10.1002/j.1460-2075.1990.tb08320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klekamp M. S., Weil P. A. Specific transcription of homologous class III genes in yeast-soluble cell-free extracts. J Biol Chem. 1982 Jul 25;257(14):8432–8441. [PubMed] [Google Scholar]
- Kurjan J., Hall B. D., Gillam S., Smith M. Mutations at the yeast SUP4 tRNATyr locus: DNA sequence changes in mutants lacking suppressor activity. Cell. 1980 Jul;20(3):701–709. doi: 10.1016/0092-8674(80)90316-5. [DOI] [PubMed] [Google Scholar]
- Kurjan J., Hall B. D. Mutations at the Saccharomyces cerevisiae SUP4 tRNA(Tyr) locus: isolation, genetic fine-structure mapping, and correlation with physical structure. Mol Cell Biol. 1982 Dec;2(12):1501–1513. doi: 10.1128/mcb.2.12.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis N., DaLio A., Strobel M., Engelke D. Effects of tRNA-intron structure on cleavage of precursor tRNAs by RNase P from Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Mar 25;16(6):2537–2552. doi: 10.1093/nar/16.6.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathison L., Winey M., Soref C., Culbertson M. R., Knapp G. Mutations in the anticodon stem affect removal of introns from pre-tRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Oct;9(10):4220–4228. doi: 10.1128/mcb.9.10.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Cortese R. Transcription of cloned tRNA genes and the nuclear partitioning of a tRNA precursor. Cell. 1979 Dec;18(4):1165–1172. doi: 10.1016/0092-8674(79)90229-0. [DOI] [PubMed] [Google Scholar]
- Melton D. A., De Robertis E. M., Cortese R. Order and intracellular location of the events involved in the maturation of a spliced tRNA. Nature. 1980 Mar 13;284(5752):143–148. doi: 10.1038/284143a0. [DOI] [PubMed] [Google Scholar]
- Nishikura K., De Robertis E. M. RNA processing in microinjected Xenopus oocytes. Sequential addition of base modifications in the spliced transfer RNA. J Mol Biol. 1981 Jan 15;145(2):405–420. doi: 10.1016/0022-2836(81)90212-6. [DOI] [PubMed] [Google Scholar]
- Nishikura K., Kurjan J., Hall B. D., De Robertis E. M. Genetic analysis of the processing of a spliced tRNA. EMBO J. 1982;1(2):263–268. doi: 10.1002/j.1460-2075.1982.tb01157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor J. P., Peebles C. L. In vivo pre-tRNA processing in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jan;11(1):425–439. doi: 10.1128/mcb.11.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden R. C., Beckman J. S., Abelson J., Kang H. S., Söll D., Schmidt O. In vitro transcription and processing of a yeast tRNA gene containing an intervening sequence. Cell. 1979 Jun;17(2):399–406. doi: 10.1016/0092-8674(79)90166-1. [DOI] [PubMed] [Google Scholar]
- Thomann H. U., Schmutzler C., Hüdepohl U., Blow M., Gross H. J. Genes, variant genes and pseudogenes of the human tRNA(Val) gene family. Expression and pre-tRNA maturation in vitro. J Mol Biol. 1989 Oct 20;209(4):505–523. doi: 10.1016/0022-2836(89)90590-1. [DOI] [PubMed] [Google Scholar]
- Thurlow D. L., Shilowski D., Marsh T. L. Nucleotides in precursor tRNAs that are required intact for catalysis by RNase P RNAs. Nucleic Acids Res. 1991 Feb 25;19(4):885–891. doi: 10.1093/nar/19.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M. L., Wilhelm F. X., Ebel J. P. Analysis of mutant tRNA gene transcripts in vivo in Saccharomyces cerevisiae by abortive primer extension. Anal Biochem. 1991 Jul;196(1):156–160. doi: 10.1016/0003-2697(91)90132-d. [DOI] [PubMed] [Google Scholar]
- Zasloff M., Santos T., Hamer D. H. TRNA precursor transcribed from a mutant human gene inserted into a SV40 vector is processed incorrectly. Nature. 1982 Feb 11;295(5849):533–535. doi: 10.1038/295533a0. [DOI] [PubMed] [Google Scholar]
- Zasloff M., Santos T., Romeo P., Rosenberg M. Transcription and precursor processing of normal and mutant human tRNAiMet genes in a homologous cell-free system. J Biol Chem. 1982 Jul 10;257(13):7857–7863. [PubMed] [Google Scholar]
- Zennaro E., Francisci S., Ragnini A., Frontali L., Bolotin-Fukuhara M. A point mutation in a mitochondrial tRNA gene abolishes its 3' end processing. Nucleic Acids Res. 1989 Jul 25;17(14):5751–5764. doi: 10.1093/nar/17.14.5751. [DOI] [PMC free article] [PubMed] [Google Scholar]