Abstract
The chlamydia-specific hypothetical protein CT311 was detected both inside and outside of the chlamydial inclusions in C. trachomatis-infected cells. The extra-inclusion CT311 molecules were distributed in the host cell cytoplasm with a pattern similar to that of CPAF, a known Chlamydia-secreted protease. The detection of CT311 was specific since the anti-CT311 antibody labeling was only removed by absorption with CT311 but not CPAF fusion proteins. In addition, both anti-CT311 and anti-CPAF antibodies only detected their corresponding endogenous proteins without cross-reacting with each other or any other antigens in the whole cell lysates of C. trachomatis-infected cells. Although both CT311 and CPAF proteins were first detected 12h after infection, localization of CT311 into host cell cytosol was delayed until 24h while CPAF secretion into host cell cytosol was already obvious by 18h after infection. The host cell cytosolic localization of CT311 was further confirmed in human primary cells. CT311 was predicted to contain a N-terminal secretion signal sequence and the CT311 signal sequence directed secretion of PhoA into bacterial periplasmic region in a heterologous assay system, suggesting that a sec-dependent pathway may play a role in the secretion of CT311 into host cell cytosol. This hypothesis is further supported by the observation that secretion of CT311 in Chlamydia-infected cells was blocked by a C16 compound known to inhibit signal peptidase I. These findings have provided important molecular information for further understanding the C. trachomatis pathogenic mechanisms.
Keywords: Chlamydia trachomatis, hypothetical CT311, secretion
1. Introduction
Urogenital tract infection with C. trachomatis is a leading cause of sexually transmitted bacterial diseases (Centers for Disease Control and Prevention, November 2009; Peterman et al., 2006; Sherman et al., 1990). Although the pathogenic mechanisms of C. trachomatis-induced diseases remain unknown, it is thought that intracellular survival and replication of the C. trachomatis organisms may significantly contribute to the inflammatory pathologies during C. trachomatis infection (Chen et al., 2010b; Cheng et al., 2008; Stephens, 2003; Zhong, 2009). The C. trachomatis organisms undergo an intracellular growth cycle with distinct biphasic stages (Hackstadt et al., 1997). An infectious elementary body (EB) initiates the intracellular infection by invading an epithelial cell. The internalized EB rapidly develops into a noninfectious but metabolically active reticulate body (RB) that is able to undergo biosynthesis and multiply. The accumulation of progeny RBs in the inclusions triggers the differentiation of RBs back into EBs for spreading to new cells. All chlamydial biosynthesis and metabolism activities are restricted within a cytoplasmic vacuole known as inclusion (Hackstadt et al., 1997).
To establish and maintain a successful intracellular infection, the C. trachomatis organisms have evolved the ability to secrete proteins into host cells. The secreted proteins may modify host cellular processes and facilitate C. trachomatis invasion, intracellular survival/replication and spreading to new cells. For example, the C. trachomatis EB organisms can inject preexisting proteins into epithelial cells to induce endocytosis (Clifton et al., 2004; Engel, 2004) so that the EBs can rapidly enter the host cells that are normally inefficient in taking up particles. Some of the injected preexisting proteins may further modulate host cell cytoskeletal structures and endocytic pathways (Hower et al., 2009) so that the chlamydial organism-laden vacuoles cannot be fused with host lysosomes (Scidmore et al., 2003). Some of the newly synthesized proteins by RBs are secreted into the inclusion membrane (Li et al., 2008a; Rockey et al., 2002) and host cell cytoplasm (Fields et al., 2003; Valdivia, 2008; Zhong, 2009). These secretion proteins may facilitate the intracellular chlamydial organisms to both take up nutrients and energy from host cells (Cocchiaro et al., 2008; Hackstadt et al., 1995; McClarty, 1994; Su et al., 2004) and prevent the infected host cells from undergoing apoptosis and host immune detection and attack (Zhong, 2009). For example, CPAF, a chlamydial protease/proteasome-like activity factor, is secreted into host cell cytosol (Zhong et al., 2001). CPAF is a serine protease (Chen et al., 2009; Huang et al., 2008) that can degrade a wide array of host proteins including cytokeratins for assisting chlamydial inclusion expansion (Dong et al., 2004; Kumar & Valdivia, 2008; Scidmore, 2008), transcriptional factors required for MHC antigen expression for evading immune responses (Zhong et al., 1999; Zhong et al., 2000) and BH3-only domain proteins for inhibiting apoptosis (Fan et al., 1998; Pirbhai et al., 2006).
Identification of Chlamydia-secreted proteins has greatly facilitated our understanding of the molecular mechanisms of chlamydial pathogenesis (Chellas-Gery et al., 2007; Clifton et al., 2004; Dong et al., 2006; Gong S, 2011; Hobolt-Pedersen et al., 2009; Hower et al., 2009; Li et al., 2008b; Misaghi et al., 2006; Qi et al., 2011a; Qi et al., 2011b; Subtil et al., 2005; Valdivia, 2008; Vandahl et al., 2005; Zhong et al., 2001). Using an anti-fusion protein antibody approach, we have systematically evaluated the localization of endogenous proteins for identifying C. trachomatis-secreted proteins (CtSPs; ref:(Li et al., 2008a; Sharma et al., 2006; Wang et al., 2010). We found that the anti-CT311 antibodies detected signals both inside and outside of the inclusions. The extra-inclusion signals of CT311 displayed a distribution pattern similar to CPAF, suggesting that CT311 is secreted into host cell cytosol during chlamydial infection. The specificity of CT311 detection was confirmed using an absorption approach both in immunofluorescence and Western blot assays and the secretion of CT311 into host cell cytosol was also confirmed in human primary cells. Although both CT311 and CPAF proteins were first detected at 12h after infection and both molecules carry an N-terminal secretion signal sequence, the secretion of CT311 into host cell cytosol was delayed until 24h while CPAF secretion was already obvious at 18h after infection, suggesting that CT311 and CPAF may fulfill different functions during chlamydial intracellular infection.
2. Results
2.1. The hypothetical protein CT311 is detected both inside and outside of C. trachomatis inclusions
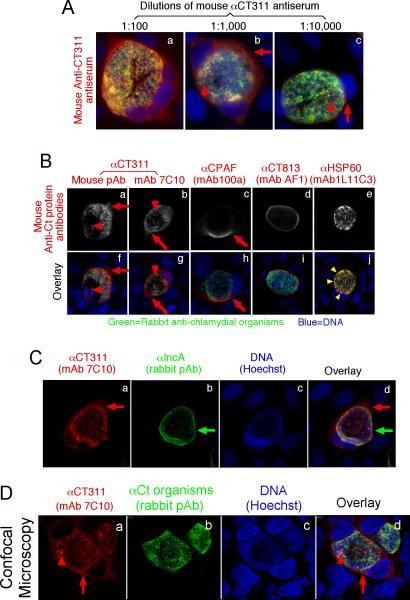
When a mouse antiserum raised with GST-CT311 fusion protein was used to localize the endogenous protein in C. trachomatis-infected HeLa cells in an immunofluorescence assay, we found that CT311 was detected both inside and outside of the chlamydial inclusions (Fig. 1A). The cytosolic labeling pattern of the extra-inclusional CT311 molecules was similar to that of CPAF, a known Chlamydia-secreted protease, but distinct from those of CT813, a chlamydial inclusion membrane protein (Chen et al., 2006), and HSP60, a chlamydial cytosolic protein (Fig. 1B). Co-labeling of CT311 and the inclusion membrane further confirmed that CT311 was indeed detected outside of the chlamydial inclusions and in the host cell cytosol (Fig. 1C). These observations have demonstrated that CT311 is at least partially secreted into host cell cytosol. However, unlike CPAF, CT311 was also significantly detected inside the chlamydial inclusions (Fig. 1A, panel b & 1B, panel g). Interestingly, the intra-inclusion granular signals labeled with the anti-CT311 antibodies did not overlap with the anti-C. trachomatis organism labeling under confocal microscopy (Fig. 1D), suggesting that the intra-inclusional CT311 molecules might also be secreted out of chlamydial organisms (possibly in organism-free secretion vesicles) but were still trapped in the inclusion lumen. These results together demonstrated that CT311 was secreted into both intra-inclusional space and the host cell cytosol.
FIG. 1.
Immunofluorescence detection of CT311 in the cytoplasm of C. trachomatis-infected cells. HeLa cells infected with C. trachomatis organisms were processed 40h post infection for co-staining with mouse antibodies visualized with a goat anti-mouse IgG conjugated with Cy3 (red), rabbit antibodies visualized with a Cy2-conjugated goat anti-rabbit IgG (green) and the DNA dye Hoechst (blue). The mouse antibodies included an anti-CT311 antiserum at various dilutions (A, panels a-c) and at 1:1000 dilution (B, panels a & f) as well as a mAb (clone 7C10; b & g) both raised with GST-CT311 fusion protein, a mAb (100a) against CPAF (c & h), a mAb (AF1) against GST-CT813 (a C. trachomatis inclusion membrane protein, d & i), and a mAb (1L11C3) against chlamydial HSP60 (e & j). The co-staining of the mouse anti-CT311 (red, panel a) with the rabbit anti-IncA antibodies (green, b) was also carried out (C). The co-staining of the anti-CT311 antibody (panel a) with the anti-chlamydial organism antibody (b) was visualized under a confocal microscope (D). Note that the anti-CT311 antibodies detected signals in both the chlamydial inclusions (but without overlapping with the chlamydial organisms, red arrowheads) and host cell cytosol (red arrows) while the anti-CPAF antibody mainly detected signals in the host cell cytosol. The anti-HSP60 antibody labeling overlapped with the chlamydial organisms (yellow arrowheads). Images are representative of at least 3 independent experiments.
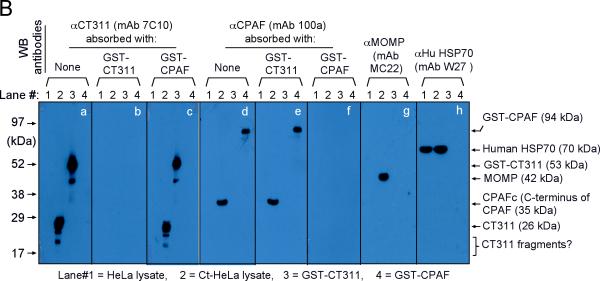
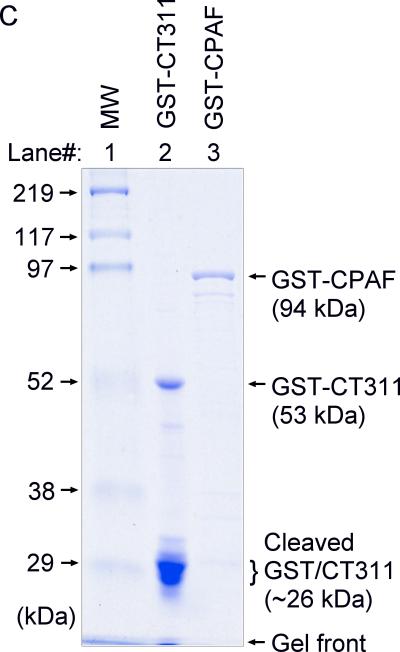
We next confirmed the antibody labeling specificity by using an absorption procedure (Fig. 2A). Both the granular staining inside inclusion and the diffused staining in the host cell cytosol labeled by the anti-CT311 antiserum or mAb 7C10 were removed by absorption with GST-CT311 but not GST-CPAF fusion proteins. The same was true for the anti-CPAF antibody stainings, demonstrating that both anti-CT311 and anti-CPAF antibodies specifically labeled the corresponding endogenous proteins without cross-reacting with each other. On a Western blot (Fig. 2B), the mouse anti-CT311 mAb only reacted with the endogenous CT311 and the GST-CT311 fusion protein without cross-reacting with any other proteins from the C. trachomatis-infected cells or unrelated fusion proteins. The anti-CPAF mAb 100a detected both the GST-CPAF fusion protein and the C-terminal fragment of CPAF (CPAFc) in chlamydia-infected cells as demonstrated previously (Zhong et al., 2001). In addition, the anti-CT311 mAb also detected a fast migrating protein band below the full length of endogenous CT311 or GST-CT311 fusion protein bands, suggesting that CT311 may be processed. The fact that absorption of mAb 7C10 with GST-CT311 but not GST-CPAF fusion proteins efficiently removed the labeling to both the full length CT311 and fast migrating bands confirmed that the fast migrating band was processed from CT311. As loading controls, the anti-MOMP antibody detected MOMP in the infected cell sample while the anti-human HSP70 antibody recognized HSP70 in both normal and infected HeLa samples. Both the GST-CT311 and GST-CPAF fusion protein preps used for the Western blot contained adequate amounts of full-length fusion proteins although cleavage bands were also detected in the GST-CT311 fusion protein lane (Fig. 2C). These results further confirmed that both anti-CT311 and anti-CPAF antibodies only specifically detected the corresponding endogenous proteins without cross-reacting with each other or any other chlamydial or host cell proteins. Thus, we can conclude that the cytosolic signals labeled by the anti-CT311 and anti-CPAF antibodies revealed under the immunofluorescence microscope represent the corresponding endogenous proteins.
Fig. 2.
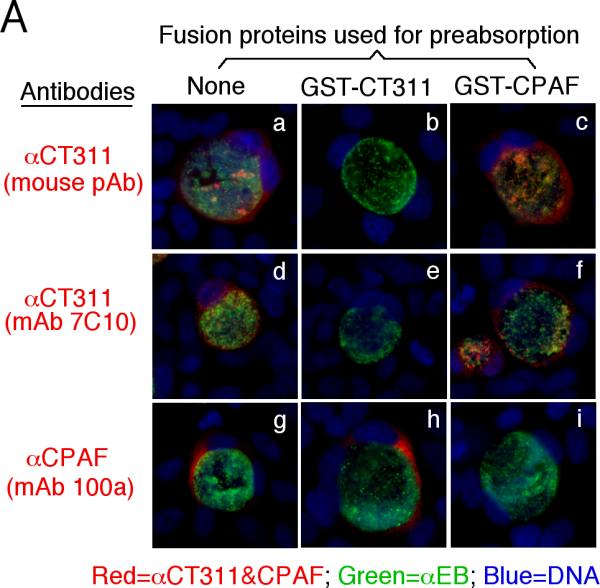
Specific detection of endogenous CT311 by the anti-CT311 antibodies. (A) The anti-CT311 mouse antiserum (panels a-c) and mAb 7C10 (d-f), the anti-CPAF mAb 100a (g-i) with (panels b, c, e, f, h & i) or without (a, d & g) pre-absorption with the corresponding or heterologous GST fusion proteins were used to detect the endogenous proteins in C. trachomatis-infected cells (red). Note that both the anti-CT311 and anti-CPAF antibody labelings were removed by pre-absorption with the corresponding (panels b, e & i) but not heterologous (c, f &h) fusion proteins. (B) Antigens including HeLa lysates (lanes 1), C. trachomatis-infected HeLa (Ct-HeLa) lysates (lanes 2) and fusion proteins GST-CT311 (lanes 3) and GST-CPAF (lanes 4) were resolved in SDS-polyacrylamide gels and blotted onto nitrocellulose membrane for detection with antibodies against CT311 (mAb 7C10, panels a-c), CPAF (mAb 100a, d-f), MOMP (mAb MC22, g) and human HSP70 (mAb W27, h) with (b, c, e & f) or without (a, d, g & h) absorption with corresponding (b & f) or heterologous (c & e) fusion proteins. Note that the mouse anti-CT311 mAb only reacted with the endogenous CT311 and the GST-CT311 without cross reacting with any other GST fusion proteins or any other proteins from the C. trachomatis-infected cells. CPAFc represents the C-terminal fragment of CPAF generated as a result of CPAF processing occurring in chlamydia-infected cells. The epitope recognized by mAb 100a is located in the C-terminal fragment. In addition, the anti-CT311 mAb detected fast migrating protein bands besides the full length of CT311 or GST-CT311, suggest that CT311 may be processed. (C) The quality of fusion proteins used in the Western blot assay was monitored in a Coomassie blue staining gel. Full length GST-CT311 and GST-CPAF were detected although degradation fragments from GST-CT311 were also observed. All Fig. 2 experiments were repeated 3 times.
2.2. Characterization of CT311 secretion
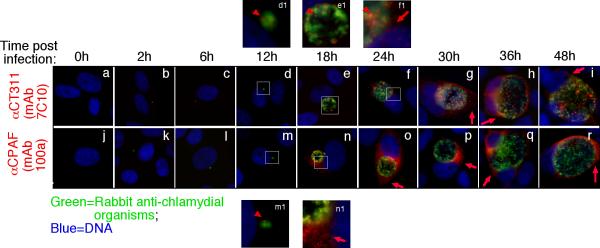
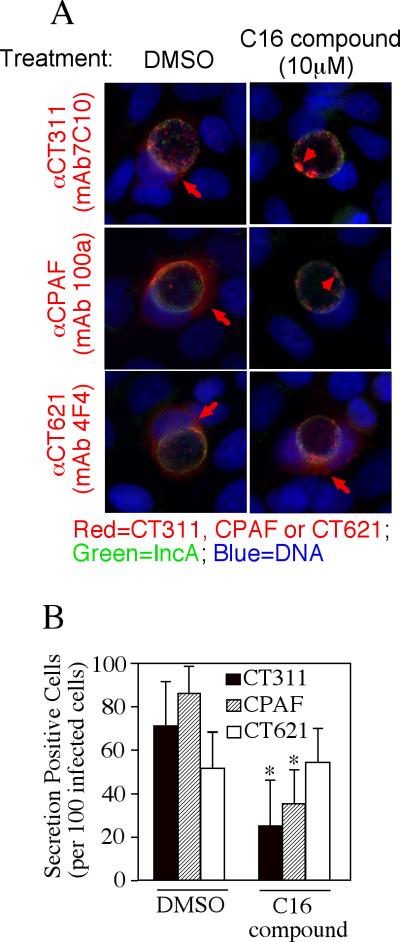
We further used the specific anti-CT311 mAb (7C10) to monitor the biosynthesis and secretion of CT311 at single cell level (Fig. 3). CT311 was first detected 12h post infection. Clear secretion into host cell cytosol was detected 24h after infection. Although CPAF protein was also first detected 12h after infection, its secretion was much more rapid and became very obvious 18h after infection. In addition to secretion into host cell cytosol, some CT311 molecules were obviously accumulated in organism-free granules in the lumen of chlamydial inclusions throughout the infection course while CPAF secretion into host cell cytosol was more complete without any significant accumulation in the lumen of inclusions. To test whether the secretion of CT311 is unique to HeLa cells, we monitored the intracellular distribution of CT311 in C. trachomatis-infected primary human cells (Fig. 4). Both CT311 and CPAF were significantly secreted into the cytosol of MS74 and THSEC, immortalized human endometrial epithelial and stroma cells respectively, HUVEC, primary human umbilical vein endothelial cell, and HOF, primary human ovarian fibroblast. These results suggest that secretion of CT311 into host cell cytosol may be a biologically relevant event. The next question is how CT311 is secreted into host cell cytosol. When the CT311 sequence was analyzed with a signal peptide prediction software, a putative gram-negative bacterial signal peptide covering the first 20 amino acids was identified (Fig. 5A). We then evaluated the functionality of the signal sequence in a PhoA-E. coli system (Chen et al., 2010a). The CT311 signal sequence (CT311ss) directed translocation of mature PhoA across the inner membrane to reach to the E. coli periplasmic space as evidenced by detection of PhoA activity (Fig. 5B). These results have demonstrated that the putative CT311 signal sequence is functional and may be able to direct CT311 across the chlamydial inner membrane to enter the periplasmic region. Secretion of CT311 into the periplasmic space may be necessary for chlamydial organisms to secrete CT311 into host cell cytosol. This hypothesis is further supported by the observation that a C16 compound known to inhibit signal peptidase I (Chen et al., 2010a) blocked the secretion of CT311 in Chlamydia-infected cells (Fig. 6). As controls, C16 also blocked secretion of CPAF (known to require a sec-dependent pathway for secretion; ref: (Chen et al., 2010a)) but not CT621 (known to depend on a Type III secretion pathway for secretion; ref: (Gong S, 2011; Hobolt-Pedersen et al., 2009)).
Fig.3.
Time course expression and secretion of CT311 protein during C. trachomatis infection. The C. tracomatis-infected HeLa cell samples were processed at various times after infection (as indicated on the top) for immunofluorescence staining as described in the legend to Fig. 1. The anti-CT311 (mAb 7C10; panels a to i), anti-CPAF (mAb 100a; j to r) were visualized with a goat anti-mouse IgG conjugated with Cy3 (red) while the chlamydial organisms were visualized with a rabbit anti-chlamydia antibody plus a goat anti-rabbit IgG-Cy2 conjugate (green). Images amplified in separate panels were marked with white squares and the corresponding amplified images were labeled the same letters followed by the number 1. Note that both CT311 (d & d1) and CPAF (m & m1) proteins were first detected associated with the chlamydial inclusions at 12h (red arrowheads) while secretion out of the inclusions was first detected at 24h (for CT311, panels f & f1) and 18h (for CPAF, n & n1) post infection respectively (red arrows). The secreted CT311 and CPAF remained in the infected cells throughout the infection cycle. The experiments were repeated 3 times.
Fig.4.
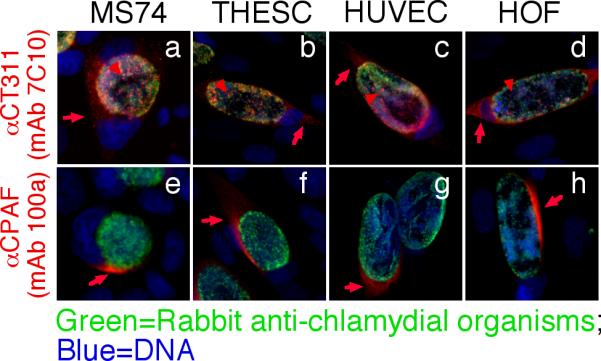
Secretion of CT311 into the cytosol of immortalized and primary cells. Immortalized human endometrial epithelial cell MS74, immortalized human endometrial stroma cell THESC, primary Human Umbilical Vein Endothelial cell (HUVEC), and primary Human ovarian fibroblast (HOF) as indicated on top of each image were infected with C. trachomatis and processed 40h after infection for immunofluorescence labeling as described in the Fig. 1 legend. Both the anti-CT311 (7C10, a-d) and anti-CPAF (100a, e-h) mAbs detected significant signals in the cytosol of all infected cells. Red arrows indicate proteins secreted into the cytosol while red arrowheads indicate signals detected inside chlamydial inclusions but not overlapping with chlamydial organisms. The experiments were repeated twice.
Fig.5.
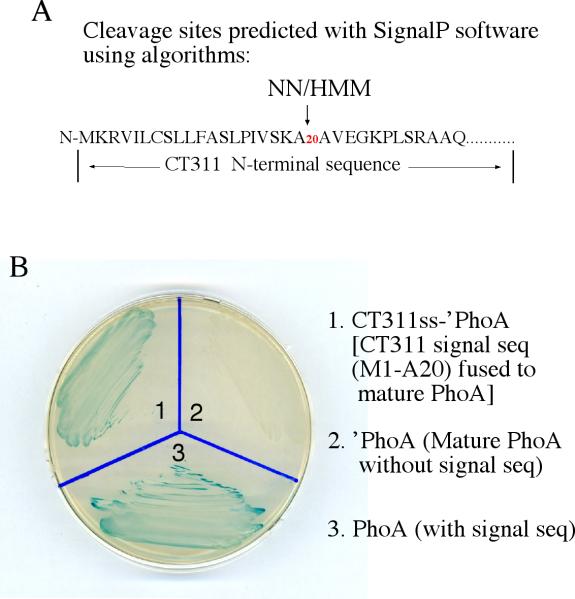
Prediction and evaluation of N-terminal secretion signal sequence in CT311. The SignalP software was used to identify type II secretion signal peptides (A). Both the NN and HMM algorithms predicted the N-terminal region covering the first 20 amino acids as a signal sequence. The DNA coding for the CT311 signal sequence (CT311ss) was then used to replace the native signal peptide-coding region of the PhoA gene. The chimeric CT311ss-‘PhoA construct was transformed into E. coli for detecting the translocation of PhoA into periplasmic space where PhoA enzymatic activity can be measured in X-gal blue plate. Blue indicates that PhoA has crossed the inner membrane and reached to the periplasm. Note that the bacteria transformed with the full length PhoA (plate slot 1) but not the ‘PhoA missing the secretion sequence (slot 2) turned blue. The CT311ss-‘PhoA chimeric construct-transformed bacteria also turned blue (slot 3), indicating that the CT311 N-terminal 20 amino acid sequence is functional at least in directing the translocation of PhoA to cross the inner membrane. The experiments were repeated twice.
Fig.6.
Inhibition of CT311 secretion in Chlamydia by a signal peptidase I inhibitor. (A) Hela cells infected with C.trachomatis were treated with DMSO or 10mM C16 compound at 6 hours post infection and processed for immunofluorescence microscopy analysis 30 hours after treatment. The mouse monoclonal antibody (mAb 7C10) against CT311, CT621 (mAb 4F4) or CPAF (mAb 100a) was respectively co-stained with a rabbit anti-IncA antibody (green) and the DNA dye Hoechst (blue). Red arrows indicate proteins secreted outside of the inclusions while arrowheads indicate proteins still inside the inclusions. (B) The secretion-positive cells were quantitated by counting 100 infected cells randomly from each coverslip and expressed as % of total infected cells. The data presented came from 3 independent experiments with duplicate coverslips in each experiment. Note that both CT311 and CPAF but not CT621 secretion-positive cells were significantly reduced after C16 treatment (*P<0.05, two-tailed Student t-test).
3. Discussion
Secreting proteins into host cell cytosol represents one of the most effective strategies for C. trachomatis organisms to constantly improve their intracellular living. Identifying chlamydia-secreted proteins will be essential for uncovering the mysteries of the chlamydial intracellular life and understanding the molecular mechanisms of C. trachomatis pathogenesis. In the current manuscript, we have presented convincing evidence that the hypothetical protein CT311 is secreted into host cell cytosol during C. trachomatis infection. First, both the anti-CT311 polyclonal antiserum and mAb detected CT311 both inside and outside of the chlamydial inclusions in C. trachomatis-infected cells. The intra-inclusion CT311 molecules were labeled in granules free of chlamydial organisms while the extra-inclusion CT311 molecules were in the host cell cytoplasm just like the chlamydia-secreted protease CPAF, demonstrating that at least a portion of CT311 was completely secreted into the host cell cytosol. Second, the detection of the endogenous CT311 was specific. The anti-CT311 antibody labeling observed under the fluorescence microscope was only removed by absorption with CT311 but not CPAF fusion proteins. Furthermore, both anti-CT311 and anti-CPAF antibodies only detected their corresponding endogenous proteins without cross-reacting with each other or any other antigens in the whole cell lysates of C. trachomatis-infected cells. Third, like CPAF, CT311 also contains a N-terminal signal sequence and the signal sequence is functional in directing a heterologous protein across the inner membrane, suggesting that a sec-dependent pathway may contribute to the secretion of both CT311 and CPAF into host cell cytosol. Finally, despite the many similarities between CT311 and CPAF, the secretion kinetics of CT311 seemed to be distinct from that of CPAF. Although both CT311 and CPAF proteins were first detected 12h after infection, secretion of CT311 into host cell cytosol was delayed until 24h while CPAF secretion was already very obvious by 18h after infection, indicating that CPAF secretion was rapid, which is consistent with what was reported previously (Gong S, 2011; Heuer et al., 2003; Hobolt-Pedersen et al., 2009). The secretion of CPAF also seemed to be more complete since once the protein was detectable, most CPAF was detected outside of the inclusions while there were a lot of CT311 signals retained in the inclusion throughout the infection course. The distinct secretion kinetics suggests that the chlamydial organisms may use CT311 to accomplish a function that is different from what CPAF does.
The next question is how CT311 is exported into host cell cytosol during chlamydial infection. This is a common question faced by the entire chlamydia research fields. The observations that the CT311 N-terminal signal sequence directed the translocation of mature PhoA into E. coli periplasm and secretion of CT311 in Chlamydia-infected cells was inhibited by a signal peptidase I inhibitor strongly suggest that a sec-dependent pathway may play an important role in the secretion of CT311 into host cell cytosol. However, the sec-dependent pathway can only translocate its cargoes into the periplasmic region. How is the periplasmic CT311 further exported out of the chlamydial organisms and into host cell cytoplasm? The C. trachomatis organisms do express homologs of the general secretion proteins (Gsps) in the outer membrane, including CT570-572 for GspF, E & D respectively. However, these outer membrane secretion proteins export periplasmic proteins out of the organisms in free form. It is hard to imagine how soluble proteins once dumped into the inclusion lumen can further cross the inclusion membrane to enter host cell cytosol in a controlled/regulated manner. Since CT311-laden vesicule-like structures free of chlamydial organisms were detected in the lumen of chlamydial inclusions, we like to propose that the chlamydial organisms may use an outer membrane vesicular (OMV) budding mechanism to export the periplasmic cargoes into host cell cytosol. This hypothesis is also consistent with various previous observations that the chlamydial RB outer membrane was induced to undergo vesiculation (Matsumoto & Manire, 1970a; Matsumoto & Manire, 1970b) and chlamydial organism-free vesicles were detected both inside (Jorgensen & Valdivia, 2008) and outside of inclusion membrane (Giles et al., 2006). The vesiculized CT311 may further enter host cell cytosol by vesicle fusing with or passing through the inclusion membrane. Although OMVs have been recognized as an essential means for gram-negative bacteria to secrete virulence factors (Ellis & Kuehn, 2010; Ellis et al., 2010; Parker et al., 2010; Unal et al., 2010), the precise mechanisms on how OMVs are regulated remain unknown (Haurat et al., 2010), which has led some scientists to question whether OMVs can really represent a specific mechanism for protein secretion. The C. trachomatis-infected cells may provide a unique model system for dissecting the molecular pathways of OMVs.
Regardless of how CT311 is secreted into host cell cytosol, a more important question is what role the secreted CT311 may play during C. trachomatis infection. CT311 is categorized as a Chlamydia-specific hypothetical protein with a total of 235 amino acids and a pI of 9.75 (http://stdgen.northwestern.edu). CT311 is conserved among all chlamydial genomes sequenced so far, suggesting that it is essential for maintaining the chlamydial intracellular parasitism. However, little is known about how CT311 works. No putative conserved domains have been detected using bioinformatics programs (http://toolkit.tuebingen.mpg.de/hhpred; http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi). Indeed, identifying the secretion pathway and determining the functionality of Chlamydia trachomatis-secreted proteins (CtSPs) have been the common challenges faced by modern chlamydial researchers. As more CtSPs are identified, more knowledge will be accumulated and more tools will be available for figuring out biological significance of CtSPs. While we continue our path to identify new CtSPs, we are also using approaches such as yeast-two hybrid and co-precipitation to gain in-depth knowledge on the already identified CtSPs.
4. Materials and methods
4.1. Chlamydial infection
The C. trachomatis L2/LGV-434/Bu organisms used in the current study were propagated, purified, aliquoted and stored as described previously (Greene & Zhong, 2003; Greene et al., 2004). The chlamydial organisms were routinely checked for mycoplasma contamination. The human cells used in the current study include HeLa cells (human cervical carcinoma epithelial cells, ATCC cat# CCL2), MS74 cells (immortalized human endometrial epithelial cells, ref: (Greene et al., 2004)), THESC cells (immortalized human endometrial stroma cells), HOF cells (human ovarian fibroblast, cat NO# 7330, Sciencell research laboratories, Carlsbad, CA) and HUVEC (human umbilical vein endothelial cells, Cambrex Bio Science Rockland, Inc., East Rutherford, N.J.). For C. trachomatis infection, human cells were grown in either 24 well plates with coverslips or tissue culture flasks containing DMEM (GIBCO BRL, Rockville, MD) for immortalized cell lines, fibroblast medium (Sciencell research laboratories) for the primary HOF cells or complete EGM medium (Cambrex) for the primary HUVEC cells all with 10% fetal calf serum (GIBCO BRL) at 37°C in an incubator supplied with 5% CO2. The human cells were inoculated with chlamydial organisms at an MOI of 0.5 or as indicated in individual experiments. The infected cultures were processed at various time points after infection for either immunofluorescence assays or Western blot analyses as described below. For some immunofluorescence experiments, the infected cultures were treated with arylomycin C16, a compound known to inhibit signal peptidase I (Chen et al., 2010a).
4.2. Chlamydial gene cloning, fusion protein expression and antibody production
The ORF CT311 from C. trachomatis serovar D genome (http://stdgen.northwestern.edu) was cloned into pGEX vectors (Amersham Pharmacia Biotech, Inc., Piscataway, NJ) using the following primers: forward primer, 5’-CGC-GGATCC (BamHI)-ATGAAAAGAGTTATCCTCTGCT-3’, back primer, 5’-TTTTCCTTTTGCGGCCGC(NotI)-CTATTTTCCATTTTGCAGATCTTT-3’; The gene ct311 was expressed as fusion proteins with glutathione-s-transferase (GST) fused to the N-terminus as previously described (Sharma et al., 2006). Expression of the fusion protein was induced with isopropyl-beta-D-thiogalactoside (IPTG; Invitrogen, Carlsbad, CA) and the fusion proteins were extracted by lysing the bacteria via sonication in a Triton-X100 lysis buffer (1%TritonX-100, 1mM PMSF, 75 units/ml of Aprotinin, 20 mM Leupeptin and 1.6 mM Pepstatin, all from Sigma). After a high-speed centrifugation to remove debris, the fusion protein-containing supernatants were further purified using glutathione-conjugated agarose beads (Pharmacia) and the purified proteins were used to immunize mice for producing antibodies as described previously (Sharma et al., 2006). Besides collecting antiserum from the immunized mice, the mouse splenocytes were also harvested for generating hybridomas and producing monoclonal antibodies (mAb) as described previously (Zhong et al., 1997). A total of three independent hybridoma clones were generated and their corresponding mAbs were designated as 5H8, 7C10 & 7E3. All three mAbs displayed the same IgG2a isotype and detected signals in the host cell cytosol of C. trachomatis-infected cells. The mAb 7C10 was used in the current study.
4.3. Immunofluorescence assay
The immunofluorescence assay was carried out as described previously (Fan et al., 1998; Zhong et al., 2006). Briefly, HeLa, MS74, THESC, HUVEC or HOF cells with or without C. trachomatis infection grown on coverslips were fixed with 2% paraformaldehyde (Sigma, St. Luis, MO) dissolved in PBS (phosphate-buffer saline solution, pH 7.6) for 30 min at room temperature, followed by permeabilization with 2% saponin (Sigma) for an additional 30 min. After washing and blocking, the cell samples were subjected to antibody and chemical staining. Hoechst (blue, Sigma) was used to visualize DNA. A rabbit anti-chlamydial organism antibody (R1L2, raised with C. trachomatis L2 organisms, unpublished data) or anti-IncA [kindly provided by Ted Hackstadt. Laboratory of Intracellular Parasites, Rocky Mountain Laboratories, NIAID, NIH, Hamilton, Montana; ref: (Rockey et al., 1995)] plus a goat anti-rabbit IgG secondary antibody conjugated with Cy2 (green; Jackson ImmunoResearch Laboratories, Inc) was used to visualize chlamydial organisms or inclusion membrane. The various mouse antibodies plus a goat anti-mouse IgG conjugated with Cy3 (red; Jackson ImmunoResearch) were used to visualize the corresponding antigens. The mouse antibodies included: polyclonal antibodies (pAbs) made against GST-CT311, mAb 7C10 against CT311 and mAb 100a against CPAF (Zhong et al., 2001). All primary antibodies were preabsorbed with a bacterial lysate containing GST alone before use. In addition, for some experiments, the primary antibodies were further absorbed with either the corresponding or heterologous fusion proteins immobilized onto glutathione-conjugated agarose beads (Pharmacia) prior to staining, which was used to prove the antibody binding specificities. The absorption was carried out by incubating the antibodies with bead-immobilized antigens for 1h at room temperature (RT) or overnight at 4°C followed by pelleting the beads. The remaining supernatants were used for immunostaining. The immunofluorescence images were acquired using an Olympus AX-70 fluorescence microscope equipped with multiple filter sets and Simple PCI imaging software (Olympus, Melville, NY) as described previously (Fan et al., 1998; Zhong et al., 2001). The images were processed using Adobe Photoshop (Adobe Systems, San Jose, CA).
4.4. Western blot assay
The Western blot assay was carried out as described elsewhere (Wang et al., 2010). Briefly, HeLa cells with or without C. trachomatis infection and GST fusion proteins were solublized in 2% SDS sample buffer and loaded onto SDS polyacrylamide gel wells. After electrophoresis, the resolved protein bands were transferred to nitrocellulose membranes for antibody detection. The primary antibodies were: mAb 7C10 against CT311 (current study) and mAb 100a against CPAF (Zhong et al., 2001), mAb MC22 against chlamydial major outer membrane protein (MOMP; ref: (Zhong et al., 2001) and mAb W27 against host cell HSP70 (cat#Sc-24, Santa Cruz Biotechnology, CA). The anti-MOMP antibody was used to ensure that all lanes with chlamydial organism-containing samples had equivalent amounts of the organisms loaded while the lanes without chlamydial organism samples should be negative for MOMP. The anti-HSP70 antibody was used to make sure that an equal amount of normal HeLa and chlamydia-infected HeLa samples were loaded. To prove the specificities of the anti-CT311 and anti-CPAF antibodies, these antibodies were absorbed with their corresponding or heterologous fusion proteins immobilized onto glutathione-conjugated agarose beads (Pharmacia) prior to staining parallel blots. The primary antibody binding was probed with an HRP (horse radish peroxidase)-conjugated goat anti-mouse IgG secondary antibody (Jackson ImmunoResearch) and visualized with an enhanced chemiluminescence (ECL) kit (Santa Cruz Biotech).
4.5. BCIP Assay
The constructs were constructed and BCIP assay was carried out as described previously (Chen et al., 2010a). The E. coli phoA gene with or without the signal peptide region was inserted into the XhoI/KpnI sites of pFLAG-CTC (cat#E8408, Sigma) to create the recombinant plasmids pFLAG-PhoA (expressing full length precursor PhoA) and pFLAG-’PhoA (expressing mature PhoA without the signal peptide). To construct the plasmid pFLAG-CT311ss-’PhoA, a 60bp DNA sequence coding for the CT311 signal peptide (M1-A20, designated as CT311ss, with restriction enzyme sites of XhoI/BamHI) was amplified from Chlamydia trachomatis serovar D genome and 1400bp DNA sequence coding for ’PhoA (BamHI/KpnI) was amplified from pFLAG-’PhoA plasmid, both the 60bp CT311 and 1400bp ’PhoA were inserted into the XhoI/KpnI sites of the plasmid pFLAG-CTC. The resultant recombinant plasmid pFLAG-CT311ss-‘PhoA together with pFLAG-PhoA and pFLAG-’PhoA were transformed into the E. coli DH5a strain (Invitrogen, Carlsbad, CA) for the BCIP assay. Briefly, the transformed bacterial cells were grown in LB supplemented with the corresponding selection antibiotics at 37°C overnight. The overnight cultures were streaked onto LB agar containing the same selection antibiotics and 50μg/ml 5-bromo-4-chloro-3-indolyl phosphate (BCIP, cat# B6149, Sigma) and the plates were incubated at 30°C for 2 days. The bacterial colonies that are capable of exporting mature PhoA into periplasm turn blue while the colonies incapable of doing so remain white.
4.6. Statistic analysis
A two-tailed student t-test was used to compare each two groups in the current study.
Acknowledgements
This work was supported in part by grants (to G. Zhong) from the US National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Centers for Disease Control and Prevention, C . In: Sexually Transmitted Disease Surveillance, 2008. U. S. D. o. H. a. H. Services, editor. Atlanta, GA: Nov, 2009. http://www.cdc.gov/std/stats08/toc.htm. [Google Scholar]
- Chellas-Gery B, Linton CN, Fields KA. Human GCIP interacts with CT847, a novel Chlamydia trachomatis type III secretion substrate, and is degraded in a tissue-culture infection model. Cell Microbiol. 2007;9:2417–2430. doi: 10.1111/j.1462-5822.2007.00970.x. [DOI] [PubMed] [Google Scholar]
- Chen C, Chen D, Sharma J. The hypothetical protein CT813 is localized in the Chlamydia trachomatis inclusion membrane and is immunogenic in women urogenitally infected with C. trachomatis. Infect Immun. 2006;74:4826–4840. doi: 10.1128/IAI.00081-06. other authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Chai J, Hart PJ, Zhong G. Identifying catalytic residues in CPAF, a Chlamydia-secreted protease. Arch Biochem Biophys. 2009;485:16–23. doi: 10.1016/j.abb.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Lei L, Lu C, Flores R, DeLisa D, Roberts TC, Romesberg FE, Zhong G. Secretion of the Chlamydial Virulence Factor CPAF Requires Sec-Dependent Pathway. Microbiology. 2010a June 3; doi: 10.1099/mic.0.040527-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Lei L, Chang X, Li Z, Lu C, Zhang X, Wu Y, Yeh IT, Zhong G. Mice deficient in MyD88 Develop a Th2-dominant response and severe pathology in the upper genital tract following Chlamydia muridarum infection. J Immunol. 2010b;184:2602–2610. doi: 10.4049/jimmunol.0901593. [DOI] [PubMed] [Google Scholar]
- Cheng W, Shivshankar P, Li Z, Chen L, Yeh IT, Zhong G. Caspase-1 contributes to Chlamydia trachomatis-induced upper urogenital tract inflammatory pathologies without affecting the course of infection. Infect Immun. 2008;76:515–522. doi: 10.1128/IAI.01064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. A chlamydial type III translocated protein is tyrosinephosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci U S A. 2004;101:10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchiaro JL, Kumar Y, Fischer ER, Hackstadt T, Valdivia RH. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc Natl Acad Sci U S A. 2008;105:9379–9384. doi: 10.1073/pnas.0712241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Su H, Huang Y, Zhong Y, Zhong G. Cleavage of host keratin 8 by a Chlamydia-secreted protease. Infect Immun. 2004;72:3863–3868. doi: 10.1128/IAI.72.7.3863-3868.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Flores R, Chen D, Luo J, Zhong Y, Wu Z, Zhong G. Localization of the hypothetical protein Cpn0797 in the cytoplasm of Chlamydia pneumoniae-infected host cells. Infect Immun. 2006;74:6479–6486. doi: 10.1128/IAI.00855-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev. 2010;74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis TN, Leiman SA, Kuehn MJ. Naturally produced outer membrane vesicles from Pseudomonas aeruginosa elicit a potent innate immune response via combined sensing of both lipopolysaccharide and protein components. Infect Immun. 2010;78:3822–3831. doi: 10.1128/IAI.00433-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J. Tarp and Arp: How Chlamydia induces its own entry. Proc Natl Acad Sci U S A. 2004;101:9947–9948. doi: 10.1073/pnas.0403633101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan T, Lu H, Hu H, Shi L, McClarty GA, Nance DM, Greenberg AH, Zhong G. Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J Exp Med. 1998;187:487–496. doi: 10.1084/jem.187.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields KA, Mead DJ, Dooley CA, Hackstadt T. Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol Microbiol. 2003;48:671–683. doi: 10.1046/j.1365-2958.2003.03462.x. [DOI] [PubMed] [Google Scholar]
- Giles DK, Whittimore JD, LaRue RW, Raulston JE, Wyrick PB. Ultrastructural analysis of chlamydial antigen-containing vesicles everting from the Chlamydia trachomatis inclusion. Microbes Infect. 2006;8:1579–1591. doi: 10.1016/j.micinf.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Gong S. Chlamydia trachomatis secretion of hypothetical protein CT622 into host cell cytoplasm via a secretion pathway that can be inhibited by a C1 compound. Microbiology. 2011 doi: 10.1099/mic.0.047746-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene W, Zhong G. Inhibition of host cell cytokinesis by Chlamydia trachomatis infection. J Infect. 2003;47:45–51. doi: 10.1016/s0163-4453(03)00039-2. [DOI] [PubMed] [Google Scholar]
- Greene W, Xiao Y, Huang Y, McClarty G, Zhong G. Chlamydia-infected cells continue to undergo mitosis and resist induction of apoptosis. Infect Immun. 2004;72:451–460. doi: 10.1128/IAI.72.1.451-460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T, Scidmore MA, Rockey DD. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc Natl Acad Sci U S A. 1995;92:4877–4881. doi: 10.1073/pnas.92.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T, Fischer ER, Scidmore MA, Rockey DD, Heinzen RA. Origins and functions of the chlamydial inclusion. Trends Microbiol. 1997;5:288–293. doi: 10.1016/S0966-842X(97)01061-5. [DOI] [PubMed] [Google Scholar]
- Haurat MF, Aduse-Opoku J, Rangarajan M, Dorobantu L, Gray MR, Curtis MA, Feldman MF. Selective sorting of cargo proteins into bacterial membrane vesicles. J Biol Chem. 2010 doi: 10.1074/jbc.M110.185744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer D, Brinkmann V, Meyer TF, Szczepek AJ. Expression and translocation of chlamydial protease during acute and persistent infection of the epithelial HEp-2 cells with Chlamydophila (Chlamydia) pneumoniae. Cell Microbiol. 2003;5:315–322. doi: 10.1046/j.1462-5822.2003.00278.x. [DOI] [PubMed] [Google Scholar]
- Hobolt-Pedersen AS, Christiansen G, Timmerman E, Gevaert K, Birkelund S. Identification of Chlamydia trachomatis CT621, a protein delivered through the type III secretion system to the host cell cytoplasm and nucleus. FEMS Immunol Med Microbiol. 2009;57:46–58. doi: 10.1111/j.1574-695X.2009.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hower S, Wolf K, Fields KA. Evidence that CT694 is a novel Chlamydia trachomatis T3S substrate capable of functioning during invasion or early cycle development. Mol Microbiol. 2009;72:1423–1437. doi: 10.1111/j.1365-2958.2009.06732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Feng Y, Chen D. Structural Basis for Activation and Inhibition of the Secreted Chlamydia Protease CPAF. Cell Host Microbe. 2008;4:529–542. doi: 10.1016/j.chom.2008.10.005. other authors. [DOI] [PubMed] [Google Scholar]
- Jorgensen I, Valdivia RH. Pmp-like proteins Pls1 and Pls2 are secreted into the lumen of the Chlamydia trachomatis inclusion. Infect Immun. 2008;76:3940–3950. doi: 10.1128/IAI.00632-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar Y, Valdivia RH. Actin and intermediate filaments stabilize the Chlamydia trachomatis vacuole by forming dynamic structural scaffolds. Cell Host Microbe. 2008;4:159–169. doi: 10.1016/j.chom.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chen C, Chen D, Wu Y, Zhong Y, Zhong G. Characterization of fifty putative inclusion membrane proteins encoded in the Chlamydia trachomatis genome. Infect Immun. 2008a;76:2746–2757. doi: 10.1128/IAI.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chen D, Zhong Y, Wang S, Zhong G. The chlamydial plasmid-encoded protein pgp3 is secreted into the cytosol of Chlamydia-infected cells. Infect Immun. 2008b;76:3415–3428. doi: 10.1128/IAI.01377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Manire GP. Electron microscopic observations on the effects of penicillin on the morphology of Chlamydia psittaci. J Bacteriol. 1970a;101:278–285. doi: 10.1128/jb.101.1.278-285.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Manire GP. Electron Microscopic Observations on the Fine Structure of Cell Walls of Chlamydia psittaci. J Bacteriol. 1970b;104:1332–1337. doi: 10.1128/jb.104.3.1332-1337.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClarty G. Chlamydiae and the biochemistry of intracellular parasitism. Trends Microbiol. 1994;2:157–164. doi: 10.1016/0966-842x(94)90665-3. [DOI] [PubMed] [Google Scholar]
- Misaghi S, Balsara ZR, Catic A, Spooner E, Ploegh HL, Starnbach MN. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol Microbiol. 2006;61:142–150. doi: 10.1111/j.1365-2958.2006.05199.x. [DOI] [PubMed] [Google Scholar]
- Parker H, Chitcholtan K, Hampton MB, Keenan JI. Uptake of Helicobacter pylori outer membrane vesicles by gastric epithelial cells. Infect Immun. 2010;78:5054–5061. doi: 10.1128/IAI.00299-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterman TA, Tian LH, Metcalf CA. High incidence of new sexually transmitted infections in the year following a sexually transmitted infection: a case for rescreening. Ann Intern Med. 2006;145:564–572. doi: 10.7326/0003-4819-145-8-200610170-00005. other authors. [DOI] [PubMed] [Google Scholar]
- Pirbhai M, Dong F, Zhong Y, Pan KZ, Zhong G. The secreted protease factor CPAF is responsible for degrading pro-apoptotic BH3-only proteins in Chlamydia trachomatis-infected cells. J Biol Chem. 2006;281:31495–31501. doi: 10.1074/jbc.M602796200. [DOI] [PubMed] [Google Scholar]
- Qi M, Gong S, Lei L, Liu Q, Zhong G. A Chlamydia trachomatis OmcB C-terminal fragment is released into host cell cytoplasm and is immunogenic in humans. Infect Immun. 2011a doi: 10.1128/IAI.00003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M, Lei L, Gong S, Liu Q, Delisa MP, Zhong G. Chlamydia trachomatis Secretion of an Immunodominant Hypothetical Protein (CT795) into Host Cell Cytoplasm. J Bacteriol. 2011b;193:2498–2509. doi: 10.1128/JB.01301-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockey DD, Heinzen RA, Hackstadt T. Cloning and characterization of a Chlamydia psittaci gene coding for a protein localized in the inclusion membrane of infected cells. Mol Microbiol. 1995;15:617–626. doi: 10.1111/j.1365-2958.1995.tb02371.x. [DOI] [PubMed] [Google Scholar]
- Rockey DD, Scidmore MA, Bannantine JP, Brown WJ. Proteins in the chlamydial inclusion membrane. Microbes Infect. 2002;4:333–340. doi: 10.1016/s1286-4579(02)01546-0. [DOI] [PubMed] [Google Scholar]
- Scidmore M. Chlamydia weave a protective cloak spun of actin and intermediate filaments. Cell Host Microbe. 2008;4:93–95. doi: 10.1016/j.chom.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Scidmore MA, Fischer ER, Hackstadt T. Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect Immun. 2003;71:973–984. doi: 10.1128/IAI.71.2.973-984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma J, Zhong Y, Dong F, Piper JM, Wang G, Zhong G. Profiling of human antibody responses to Chlamydia trachomatis urogenital tract infection using microplates arrayed with 156 chlamydial fusion proteins. Infect Immun. 2006;74:1490–1499. doi: 10.1128/IAI.74.3.1490-1499.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman KJ, Daling JR, Stergachis A, Weiss NS, Foy HM, Wang SP, Grayston JT. Sexually transmitted diseases and tubal pregnancy. Sex Transm Dis. 1990;17:115–121. doi: 10.1097/00007435-199007000-00001. [DOI] [PubMed] [Google Scholar]
- Stephens RS. The cellular paradigm of chlamydial pathogenesis. Trends Microbiol. 2003;11:44–51. doi: 10.1016/s0966-842x(02)00011-2. [DOI] [PubMed] [Google Scholar]
- Su H, McClarty G, Dong F, Hatch GM, Pan ZK, Zhong G. Activation of Raf/MEK/ERK/cPLA2 signaling pathway is essential for chlamydial acquisition of host glycerophospholipids. J Biol Chem. 2004;279:9409–9416. doi: 10.1074/jbc.M312008200. [DOI] [PubMed] [Google Scholar]
- Subtil A, Delevoye C, Balana ME, Tastevin L, Perrinet S, Dautry-Varsat A. A directed screen for chlamydial proteins secreted by a type III mechanism identifies a translocated protein and numerous other new candidates. Mol Microbiol. 2005;56:1636–1647. doi: 10.1111/j.1365-2958.2005.04647.x. [DOI] [PubMed] [Google Scholar]
- Unal CM, Schaar V, Riesbeck K. Bacterial outer membrane vesicles in disease and preventive medicine. Semin Immunopathol. 2010 doi: 10.1007/s00281-010-0231-y. [DOI] [PubMed] [Google Scholar]
- Valdivia RH. Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr Opin Microbiol. 2008;11:53–59. doi: 10.1016/j.mib.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Vandahl BB, Stensballe A, Roepstorff P, Christiansen G, Birkelund S. Secretion of Cpn0796 from Chlamydia pneumoniae into the host cell cytoplasm by an autotransporter mechanism. Cell Microbiol. 2005;7:825–836. doi: 10.1111/j.1462-5822.2005.00514.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhang Y, Lu C, Lei L, Yu P, Zhong G. A Genome-Wide Profiling of the Humoral Immune Response to Chlamydia trachomatis Infection Reveals Vaccine Candidate Antigens Expressed in Humans. J Immunol. 2010;185:1670–1680. doi: 10.4049/jimmunol.1001240. [DOI] [PubMed] [Google Scholar]
- Zhong G, Reis e Sousa C, Germain RN. Production, specificity, and functionality of monoclonal antibodies to specific peptide-major histocompatibility complex class II complexes formed by processing of exogenous protein. Proc Natl Acad Sci U S A. 1997;94:13856–13861. doi: 10.1073/pnas.94.25.13856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G, Fan T, Liu L. Chlamydia inhibits interferon gamma-inducible major histocompatibility complex class II expression by degradation of upstream stimulatory factor 1. J Exp Med. 1999;189:1931–1938. doi: 10.1084/jem.189.12.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G, Liu L, Fan T, Fan P, Ji H. Degradation of transcription factor RFX5 during the inhibition of both constitutive and interferon gamma-inducible major histocompatibility complex class I expression in chlamydia-infected cells. J Exp Med. 2000;191:1525–1534. doi: 10.1084/jem.191.9.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G, Fan P, Ji H, Dong F, Huang Y. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J Exp Med. 2001;193:935–942. doi: 10.1084/jem.193.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G. Killing me softly: chlamydial use of proteolysis for evading host defenses. Trends Microbiol. 2009;17:467–474. doi: 10.1016/j.tim.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Weininger M, Pirbhai M, Dong F, Zhong G. Inhibition of staurosporine-induced activation of the proapoptotic multidomain Bcl-2 proteins Bax and Bak by three invasive chlamydial species. J Infect. 2006;53:408–414. doi: 10.1016/j.jinf.2005.12.028. [DOI] [PubMed] [Google Scholar]