Abstract
Flaviviruses include the most prevalent and medically challenging viruses. Persistent infection with flaviviruses of epithelial cells and hepatocytes that do not undergo cell death is common. Here, we report that, in epithelial cells, up-regulation of autophagy following flavivirus infection markedly enhances virus replication and that one flavivirus gene, NS4A, uniquely determines the up-regulation of autophagy. Dengue-2 and Modoc (a murine flavivirus) kill primary murine macrophages but protect epithelial cells and fibroblasts against death provoked by several insults. The flavivirus-induced protection derives from the up-regulation of autophagy, as up-regulation of autophagy by starvation or inactivation of mammalian target of rapamycin also protects the cells against insult, whereas inhibition of autophagy via inactivation of PI3K nullifies the protection conferred by flavivirus. Inhibition of autophagy also limits replication of both Dengue-2 and Modoc virus in epithelial cells. Expression of flavivirus NS4A is sufficient to induce PI3K-dependent autophagy and to protect cells against death; expression of other viral genes, including NS2A and NS4B, fails to protect cells against several stressors. Flavivirus NS4A protein induces autophagy in epithelial cells and thus protects them from death during infection. As autophagy is vital to flavivirus replication in these cells, NS4A is therefore also identified as a critical determinant of flavivirus replication.
Keywords: Apoptosis, Autophagy, Flaviviruses, Phosphatidylinositol 3-Kinase, Viral Replication, Virus, Dengue-2, Modoc, NS4A
Introduction
Over 2.5 billion people live in areas at high risk for Dengue and related flavivirus infections (1). Dengue hemorrhagic fever, a severe complication present in ∼5% of cases, claims more lives annually than all other hemorrhagic fevers combined (2); much of the damage is caused by death of infected cells. The fate of infected cells depends on cell type. Although flavivirus induces apoptosis of neurons and macrophages, infected hepatocytes and epithelial cells do not die. We find that flavivirus up-regulates autophagy in MDCK3 renal epithelial cells and other cell types and subsequently protects them. We further identify nonstructural protein NS4A of both Dengue-2 and Modoc viruses as the sole viral mediator of autophagy up-regulation and protection against death. Up-regulation of autophagosomes by either live virus infection or NS4A expression depends on PI3K and is important for replication of flavivirus in renal epithelial cells.
Flaviviruses often persist in liver and kidney following the acute phase of infection, suggesting that infected cells evade destruction by the host immune response, likely by flavivirus-induced up-regulation of autophagy in these cells. In vitro infection of hepatocytes and fibroblasts leads to up-regulation in autophagy and protection against death (3, 4). More relevant, Dengue-2 viruses replicate within the hepatocyte autophagosomes (5), and inhibition of autophagy attenuates virus replication (3). We extend these findings by identifying the nonstructural viral protein NS4A as the virus-encoded protein that up-regulates autophagy and thus protects the host cell against death, providing a well protected host cell for long term replication of virus.
The flavivirus genome contains 10 genes encoding an ∼11-kDa polyprotein precursor that binds to the ER membrane after translation at the rough ER as follows: three structural proteins (capsid (core) protein, pre-membrane protein, and envelope glycoprotein) and seven nonstructural genes (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) that mediate viral replication, assembly, and evasion of the host immune system. The active proteins are cleaved from the ER-bound viral polyprotein by cellular proteases signalase and furin and viral protease NS3/2B. NS3 is both a viral protease (with required cofactor NS2B (6)) and a viral ATP-dependent helicase (7, 8); NS5 is an RNA-dependent RNA polymerase and methyltransferase (9) responsible for virus genome replication, whereas NS1 is possibly a part of the viral replication complex (10). The small hydrophobic flavivirus proteins (NS2A, NS4A, and NS4B) remain the most poorly characterized. NS2A is required for the assembly of new flavivirus virions; NS4A associates with the virus replication complex and induces ER membrane rearrangements (discussed below), and NS4B is an antagonist of interferon. Flavivirus NS4A is an ∼16-kDa membrane-associated protein consisting of four transmembrane helices and an N-terminal cytosolic region. Once the viral genome is translated in the ER, NS4A is cleaved in the cytoplasm by viral NS2B-3 protease complex. The transmembrane domain of NS4A is cleaved by host signalase in the ER lumen (11). Mature NS4A then promotes membrane rearrangements by inducing curvature of the ER membrane (12–14). NS4A also colocalizes with double-stranded RNA. NS4A and NS1 must interact for viral RNA synthesis to be efficient, suggesting that NS4A is the docking site for the replication complex (12, 15). NS4A also associates with host cell polypyrimidine tract-binding protein to indirectly bind double-stranded RNA (16), stabilizing double-stranded viral genomic intermediates.
Although it is clear that NS4A is involved in virus replication, the mechanisms are uncertain. In this study, we identify flavivirus NS4A as the sole mediator of flavivirus-induced protection against cell death, induced by NS4A-mediated up-regulation of autophagy signaling. This up-regulation of autophagosomes can be induced by either whole virus or NS4A, depends on PI3K, and is important for flavivirus replication in renal epithelial cells.
EXPERIMENTAL PROCEDURES
Cell and Virus Culture and Treatment
MDCK, 293T, Vero, HeLa, and Swiss Webster primary macrophage cells were maintained in Dulbecco's minimum essential media (DMEM) with 10% FBS, 1% penicillin/streptomycin at 37 °C under a 5% CO2 atmosphere. C636 mosquito cells were maintained in Eagle's minimal essential media (10% FBS, 1 mm sodium pyruvate (S8636, Sigma), 1% nonessential amino acids (M7145, Sigma), 2 mm l-glutamine (25030-081, Invitrogen), 25 units/ml Fungizone (15290-018, Invitrogen), and 50 units/ml penicillin/streptomycin) at 28 °C, 5% CO2. Primary murine macrophages were obtained by peritoneal collection from pathogen-free Swiss Webster mice. Prior to all infections, cells were seeded and allowed to attach overnight in maintenance media. Cells were washed with 1× phosphate-buffered saline (PBS) before infecting at an m.o.i. = 10 (unless otherwise stated). Virus stocks were diluted with ice-cold flavivirus diluting media (1× PBS containing 0.75% bovine serum albumin fraction V, pH 8.0). Following application of virus dilutions, cells were incubated for 1.5 h at 37 °C (28 °C for C636), 5% CO2. Cells were then washed once with 1× PBS, covered with maintenance media, and incubated at 37 °C (28 °C for C636), 5% CO2 until data collection. Individual Dengue-2 genes (DEN2 E, C, prM, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) were expressed in mammalian cells from a pCAGGS-HA plasmid vector (kindly provided by Adolfo Garcia-Sastre, Mount Sinai Medical School, New York), and individual Modoc genes (NS1, NS2A, NS4A, and NS4B) were expressed in mammalian cells from a TOPO plasmid vector (gift of Dr Stephane Boissinot, Queens College, New York). Cells were plated to ∼85% confluency and allowed to attach overnight in penicillin/streptomycin-free cell type-appropriate culture media supplemented with 10% FBS. Cells were transfected using the Lipofectamine 2000 system (52887, Invitrogen) according to the manufacturer's protocol. Post-transfection, cells were incubated at 37 °C for at least 16 h before collection or treatment to allow plasmid expression.
Dengue-2 virus was generously provided by Dr. Garcia-Sastre, and Modoc virus (strain M544) was obtained from the American Type Tissue Collection (ATTC). For expansion of Dengue-2 stocks, subconfluent C636 mosquito cells were infected with virus and incubated at 28 °C for 6 days. For expansion of Modoc virus stocks, the virus was applied to subconfluent Vero cells and incubated at 37 °C for 7 days. In both cases, the culture media of the infected plates were agitated and collected, then mixed 2:1 with flavivirus freeze media (0.75% bovine serum albumin fraction V in 0.12 m NaCl, 0.05 m H3BO3, pH 9.0), and stored at −80 °C. Viral load of Dengue and Modoc stock solutions was then determined by plaque assay as described by Davis and Hardy (17).
Assessment of Cell Viability
Cells were infected and exposed to toxins at 24 hpi (hours post-infection), incubated for an additional 24 h at 37 °C, 5%CO2, collected by trypsin digestion, and stained with 0.4% trypan blue in 1× PBS. We have previously shown a direct correlation between trypan blue exclusion and other viability assays (18). Live (white) and dead (blue) cells, were counted on a hemocytometer, with cell viability expressed as percent dead cells above mock. Camptothecin (CPT) (C9911, Sigma) was applied at 50–100 μm final concentration, staurosporine (STS) (S5921, Sigma) at 20 μm, and cycloheximide (CHX) (01811, Sigma) at 150 μm. Influenza A was applied to cells at m.o.i. = 5, and the cells were incubated for 1 h at room temperature, washed once with 1× PBS, and covered with maintenance media. When appropriate, autophagy was inhibited using wortmannin, which inhibits both class I and class III PI3Ks, and 3-methyladenine (3MA), which specifically inhibits class III PI3K signaling. Autophagy was up-regulated by starvation in subculture media lacking FBS for 24 h prior to and following infection or rapamycin treatment for 1 h prior to and following infection. In all cases, cells were incubated with toxin for 24 h prior to collection.
Transmission Electron Microscopy
Reagents were purchased from Electron Microscopy Sciences (Fort Washington, PA). MDCK cells were plated at a density of 1.5 × 106, infected with influenza virus at an of m.o.i. of 5 with or without treatment. After 24 hpi, cells were fixed in 2.5% glutaraldehyde in 0.2 m cacodylate buffer, pH 7.4, for 20 min at room temperature on the plate. Cells were then scraped off and centrifuged in conical tubes at 1,200 × g for 5 min. After washing in cacodylate buffer, cells were post-fixed with 1% osmium tetroxide in the same buffer for 1 h at room temperature and washed again. Cells were then dehydrated through ascending ethanol concentrations (50–100%) for 10 min each and embedded in Agar 100 epoxy resin. Sections were counterstained with lead citrate and uranyl acetate. High resolution images were taken with a Philips 208 electron microscope.
Western Blot, Immunocytochemistry, and Cytochemistry
Cells were infected and treated as described above. At 48 hpi, cells were scraped and washed with ice-cold 1× PBS before whole lysate proteins were collected in RIPA buffer and quantified using the Bio-Rad protein assay and an Ultrospec III spectrophotometer (GE Healthcare). Western blot analysis was performed by SDS-PAGE as described by Lin et al. (18), using primary antibodies as follows: 1:500 anti-LC3II (L7543, Sigma) and 1:1000 anti-β-tubulin (sc9104, Santa Cruz Biotechnology) as a loading control. Positive signals were detected using ECL (RPN2132, GE Healthcare) and visualized using hyperfilm ECL photoradiographic film (28906835, GE Healthcare).
For immunocytochemical and cytochemical analysis, cells were seeded onto flame-sterilized glass coverslips, allowed to attach overnight, and infected and treated as above. At 48 hpi, cells were washed with 1× PBS and fixed with fresh, ice-cold 3% paraformaldehyde for 10 min, washed once, and stored in 1× PBS in the dark at 4 °C. Cells were then stained with 1:50 mouse anti-Flavi E antibody (Di-4G2-15) and 1:2,000 phalloidin-TRITC (P1951, Sigma). 1:500 anti-mouse IgG-AlexaFluor 488 (A11008, Invitrogen) was used as the secondary antibody. Nuclear localization was determined by staining with 4′,6-diamidino-2-phenylindole (DAPI) (D8417, Sigma). Cells were embedded by Gel Mount (M-01, Biomeda). LC3-GFP-expressing cells were embedded immediately following fixation. Cells were observed by confocal microscopy (Leica, Germany).
Assessment of Viral Replication
Cell culture media samples of Modoc- and Dengue-2-infected cells were collected at various times, and total viral RNA was extracted using a viral RNA extraction kit (52904; Qiagen), following the manufacturer's protocol. cDNA was then generated from extracted viral RNA using a Superscript III first strand synthesis kit (18080-400, Invitrogen), following the manufacturer's protocol. 1 μg of cDNA was amplified by qRT-PCR in 20-μl reactions using a LightCycler FastStart DNA Master SYBR Green 1 kit (03515869001, Roche Applied Science), with primers for Modoc (forward, GATTCAGGATGGCCCAAGAATC, and reverse, AGTAGGAAGGTGGACAGAATGA) and Dengue-2 (forward, TTAGAGGAGACCCCTCCC, and reverse, TCTCCTCTAACCTCTAGTCC), using a LightCycler 2.0 real time PCR machine (Roche Applied Science). Approximate viral titer (pfu/ml) was determined by quantifying viral RNA in serial dilutions of stock virus at a known concentration and generating a standard curve for comparison analysis. Relative RNA release in each cell type was compared with mock − infected cells and presented as (infected − mock).
For confirmation of qRT-PCR data, cell culture supernatant was also taken for replication analysis by plaque assay. Confluent baby hamster kidney cells in 12-well plates were infected with serial dilutions of supernatant for 2 h at 37 °C prior to overlay with plaquing media (45%, Eagle's minimal essential media; 5%, FBS; 50%, 2% low melting point-agarose) and incubation at 37 °C for 4 days. Agar was then removed, and cells were stained with crystal violet solution. Plaques were counted and virus titer was determined. Each sample was run in triplicate, and error bars indicate 1 S.D.
Assessment of Flavivirus-induced Autophagy
To examine flavivirus-induced autophagy, LC3 localization during infection was determined as described by Kabeya et al. (19). Briefly, cells were plated onto heat-sterilized glass coverslips in 35-mm plates, and LC3-GFP was transiently expressed by transfection with a C2-LC3-GFP construct (provided by Guido Kroemer, Institut Gustave-Roussy, Villejuif, France), using Lipofectamine 2000, and incubated for 16 h to allow LC3-GFP expression. Cells were then infected as described above. At 24 hpi, cells were fixed with ice-cold 3.5% paraformaldehyde (04042, Fisher) for 10 min and rinsed once with 1× PBS. Cells were then embedded by Gel Mount (M-01, Biomed) and observed by confocal microscopy. Generation of a punctate GFP expression pattern is indicative of LC3 translocation and autophagosome formation. Transfection efficiency of at least 50% was confirmed using fluorescence microscopy (data not shown). Mock-infected cells were also analyzed to ensure that LC3-GFP expression alone did not cause autophagy. Autophagy up-regulation was also examined as described above in HeLa cells stably expressing LC3-GFP.
RESULTS
Modoc Virus Does Not Kill Infected Epithelial Cells or Fibroblasts
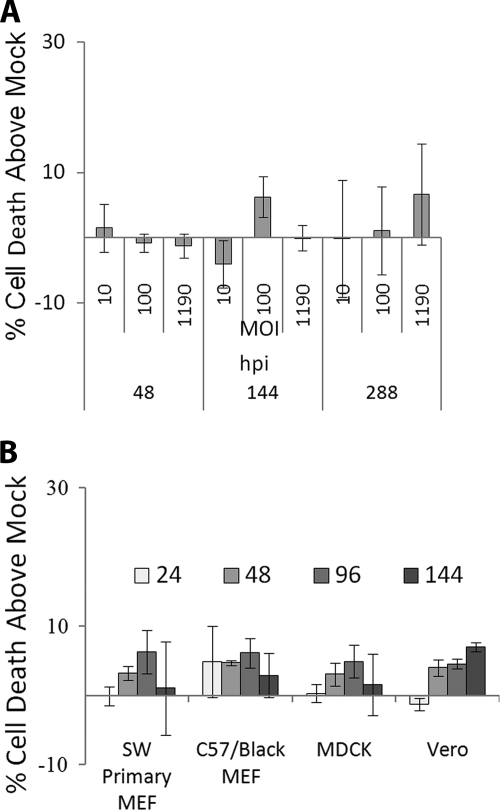
The impact of flavivirus on cell fate is specific to cell type; macrophages and neurons die following infection but hepatocytes and fibroblasts survive. Several authors have noted persistent flavivirus infection of the renal epithelium (20–24, 26), indicating that renal epithelial cells also survive infection. To understand the particular virus-host interactions involved in renal flavivirus infection, we chose MDCK renal epithelial cells as our main model for in vitro study. As Modoc virus has been proposed as a research model for flavivirus infection in vivo, we first characterized Modoc-induced cell death of MDCK renal epithelial cells. As shown in Fig. 1A, Modoc virus does not kill MDCK cells (<10% death, even after 288 hpi using undiluted stock virus (m.o.i. = 1190)).
FIGURE 1.
Flavivirus does not kill epithelial cells or fibroblasts. A, MDCK renal epithelial cells infected with Modoc virus at various m.o.i. show no significant death when analyzed by trypan blue exclusion, even very late following infection with high m.o.i. B, no significant death is observed following infection with Modoc virus at m.o.i. = 10 in several other cell types as late as 144 hpi. Because background cell death varied by cell type and condition, living and dead cells were counted, and percent cell death was calculated as % cell death above mock infection. Each assay was run in triplicate.
We next characterized the ability of Modoc virus to kill several other types of cells (Swiss Webster primary mouse embryonic fibroblasts (MEFs), C57Black primary MEFs, Vero, and MDCK cells) and assayed for death at 24, 48, 96, and 144 hpi to establish that this effect was not specific to MDCK cells. As shown in Fig. 1B, no significant levels of death (<10%) are observed in any of the cell types tested as late as 144 hpi at 10 m.o.i., demonstrating that Modoc virus does not kill epithelial cells or fibroblasts in vitro.
Modoc Virus and Dengue Virus Infect and Replicate in MDCK Renal Epithelial Cells
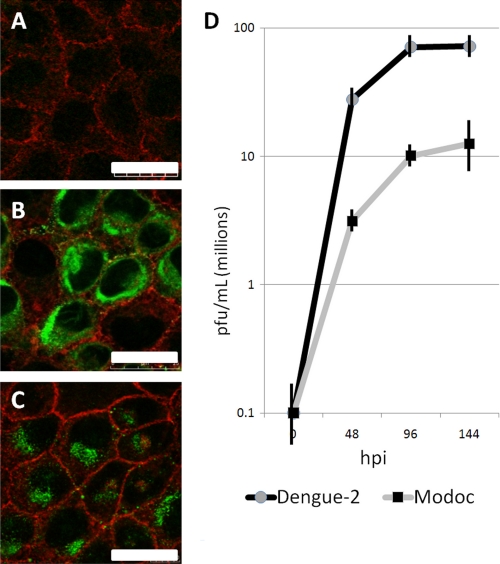
To ensure that MDCK cells support Dengue-2 and Modoc virus infection and replication, we evaluated by immunocytochemistry and qRT-PCR, the capability of each virus to infect and replicate in MDCK cells. For immunocytochemistry, cells were fixed at 48 hpi and stained for flavivirus E protein with antibody Di-4G2-4-15 (with AlexaFluor 488) and with phalloidin-TRITC to visualize cell boundaries and were observed with a confocal microscope. At 48 hpi, mock − infected cells show no positive staining for flavivirus protein (Fig. 2A). Both Dengue-2 (Fig. 2B) and Modoc (Fig. 2C) viruses are abundant in vesicles primarily within the perinuclear region of infected cells, a staining pattern consistent with previous findings of localization in the endoplasmic reticulum and Golgi bodies of infected cells (27). Following infection with Dengue-2 or Modoc virus, over half the cells within the infected populations are positive for flavivirus E protein (60 and 54%, respectively, data not shown), confirming that these viruses are capable of infecting and replicating in MDCK cells.
FIGURE 2.
Flaviviruses establish a productive infection in renal epithelial cells. Dengue-2 and Modoc virus-infected MDCK cells were fixed at 48 hpi and stained with phalloidin-TRITC (red, actin) and antibody Di-4G2-15 (green) flavivirus Envelope protein. A, mock − infected cells lack positive staining for virus protein. Massive flavivirus Envelope protein accumulation in cytoplasmic vesicles within the ER-rich perinuclear region of both Dengue-2 virus (B) and Modoc virus (C)- infected cells demonstrates the establishment of flavivirus translation and assembly. D, qRT-PCR of infected cell supernatant demonstrates rapid replication and release of virus.
To quantify flavivirus replication in MDCK cells, supernatant samples from Modoc- and Dengue-2-infected cells were collected at 48-h intervals until 144 hpi. Viral RNA was extracted from each sample and subjected to qRT-PCR. For both Dengue-2 and Modoc viruses, primers for the NS4A gene were used, and qRT-PCR was performed as described under “Experimental Procedures.” As shown in Fig. 2D, robust replication of both Dengue-2 and Modoc virus is evident by 48 hpi. A maximum virus titer of 107 pfu/ml is reached by Modoc virus at 144 hpi, whereas the maximum titer generated by Dengue-2 at 144 hpi is 108 pfu/ml. These data confirm that MDCK cells support in vitro infection and replication of both Dengue-2 and Modoc viruses and provide a basis for the use of MDCK.
Dengue-2 and Modoc Viruses Kill Macrophages while Protecting Epithelial Cells and Fibroblasts
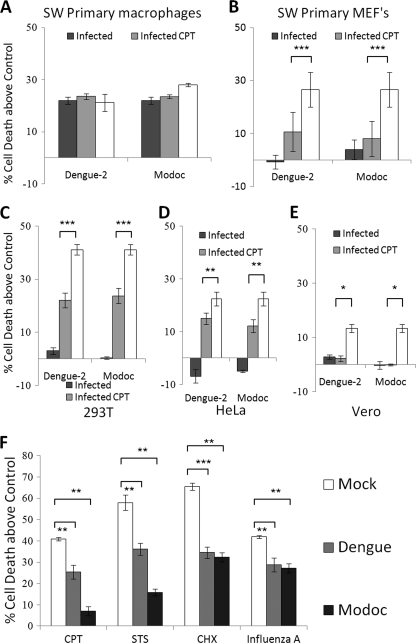
As killing of macrophages by flavivirus has been reported previously (23), we verified in our laboratory the ability of Modoc or Dengue-2 virus to kill or protect macrophages. We obtained primary macrophages by peritoneal perfusion of adult Swiss Webster mice. As shown in Fig. 3A, infection of macrophages with either Dengue-2 or Modoc virus results in ∼22% cell killing (p < 0.001 for both), roughly equal to that caused by exposure to 75 μm CPT. Conversely. Dengue-2 or Modoc virus do not kill primary Swiss Webster mouse embryonic fibroblasts (obtained from embryonic day 10.5 embryos). In fact, Dengue-2 infection caused a 59% decline and Modoc a 70% decline in cell death caused by CPT (Fig. 3B) (p < 0.0001, p = 0.0005, respectively), demonstrating that infection with either virus protects these cells. We also tested several other cells for killing or protection after flavivirus infection as follows: two human cell lines (293T (human kidney fibroblasts) and HeLa (human carcinoma)) and a primate cell line (Vero (green monkey kidney fibroblast)). As shown in Fig. 3C, infection with either Dengue-2 or Modoc virus does not kill 293T cells. Instead, Dengue-2 infection in 293T cells results in a 46% decline in death induced by CPT (p < 0.001), whereas Modoc infection leads to a 42% decline (p < 0.001). Likewise, neither virus kills HeLa cells. In HeLa cells (Fig. 3D), Dengue-2 infection leads to a 32% decline in CPT-induced cell death (p = 0.001), and Modoc infection causes a 45% decline (p = 0.001). Infection in Vero cells (Fig. 3E) by Dengue-2 leads to a 92% decline in cell death caused by CPT (p = 0.04), whereas Modoc infection in these cells causes an 85% decline in death by CPT (p = 0.03). These results demonstrate that cell fate following flavivirus infection depends on cell type and indicate that pro-survival signaling is up-regulated following infection in certain cell types.
FIGURE 3.
Flavivirus infection confers resistance to death in renal epithelial cells. In all panels, white (clear) bars represent death in cells exposed to cell death inducer alone, without flavivirus. A, both Dengue-2 and Modoc virus infections kill Swiss Webster primary macrophages with levels of death roughly equal that of 70 μm CPT-treated cells. B, no cell killing is observed after Dengue-2 or Modoc infection in Swiss Webster MEFs. Instead, these cells are significantly protected against CPT-induced death. 293T human kidney cells (C), HeLa human carcinoma cells (D), and Vero monkey kidney cells (E) are also protected against death by CPT. F, Dengue-2- and Modoc-infected MDCK cells demonstrate a significant reduction in cell killing compared with mock when exposed to four biochemically unique death stimuli, including DNA damage (CPT), kinase inhibition (STS), protein synthesis inhibition (CHX), and cytopathic viral infection (influenza A). In this and subsequent figures, * = p < 0.05; ** = p < 0.01, and *** = p < 0.001 for the bracketed comparisons.
Dengue-2 and Modoc Infection in MDCK Renal Epithelial Cells Protects against Death Induced by Several Stimuli
The fact that Modoc virus does not kill epithelial cells or fibroblasts from several mammalian species, even at high m.o.i. and late hpi, suggests that the virus can activate pro-survival signaling. We therefore tested whether Dengue-2 or Modoc virus could protect cells against agents that would normally activate apoptosis. Cells were infected with either Dengue-2 or Modoc virus and incubated at 37 °C until 24 hpi, at which time cells were exposed to toxin and held a further 24 h prior to collection at 48 hpi and trypan blue exclusion assay.
We analyzed whether Dengue-2 or Modoc could protect MDCK cells against CPT (70 μm), STS (20 μm), CHX (150 μm), and influenza A WSN/33 virus infection (m.o.i. = 5). As shown in Fig. 3F, both Dengue-2 and Modoc viruses protect MDCK cells against death by all insults examined. Preinfection with Dengue-2 virus leads to a 39% decline in death induced by CPT (p < 0.001), a 40% decline in that induced by STS (p < 0.002), a 46% decline in death caused by CHX (p < 0.0001), and a 31% decline in that caused by influenza A (p = 0.002) at 48 hpi when deaths are compared with mock − infected cells. Preinfection with Modoc virus results in an 83% decline in CPT-induced cell death (p < 0.002), a 72% decline in STS-induced death (p < 0.001), a 51% decline in CHX-induced cell death (p = 0.004), and a 36% decline in influenza A-induced cell death (p = 0.002) at 48 hpi. Thus, infection with either Dengue-2 or Modoc virus is sufficient to protect MDCK cells against death induced by a variety of insults. The generalized ability of infection to protect against such a wide range of insults suggests activation of a powerful and broad cell process.
Flavivirus Infection Induces PI3K-dependent Autophagy in MDCK Epithelial Cells
Up-regulation of autophagy often leads to PI3K-dependent pro-survival signaling (28–31). We confirmed that both Modoc and Dengue-2 viruses induce autophagy during infection. To this end, we examined LC3-GFP translocation following infection with Dengue-2 and Modoc virus in both HeLa and MDCK cells. LC3 remains in a cytosolic inactive distribution until activated during autophagy up-regulation. This cytosolic distribution results in a diffuse LC3-GFP expression pattern. Upon activation, LC3 is cleaved into LC3-I and LC3-II. LC3-II then translocates to and accumulates at autophagosomal membranes, resulting in a punctate GFP distribution.
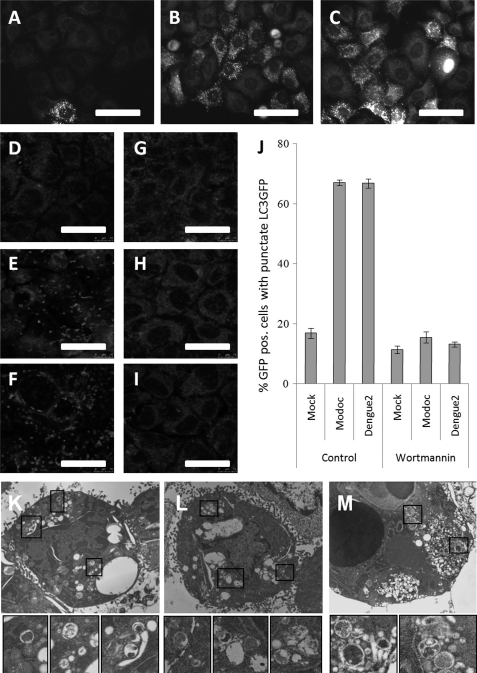
We first infected HeLa cells stably expressing LC3-GFP with either virus and fixed them at 24 hpi for observation by confocal microscopy. Mock-infected HeLa cells show little LC3-GFP punctation, indicating low levels of autophagy typical of healthy cells (Fig. 4A). Infection with either Dengue-2 (Fig. 4B) or Modoc (Fig. 4C) virus induces a substantial increase in the number of cells containing punctate LC3-GFP, indicating an up-regulation of autophagy following infection with either virus.
FIGURE 4.
Flavivirus infection leads to PI3K-dependent autophagy. A, cytoplasmic distribution of LC3-GFP observed in mock − infected HeLa cells indicates a low level of autophagy typical of healthy cells. However, following infection of HeLa cells with either Dengue-2 virus (B) or Modoc virus (C), LC3-GFP shifts to a punctate pattern, indicating up-regulation of autophagy and accumulation of LC3-GFP at autophagosomal membranes. D, mock − infected MDCK cells transiently expressing LC3-GFP display little LC3-GFP punctation, indicating a low level of autophagy. Both Dengue-2 (E) and Modoc virus-infected MDCK (F) cells display LC3-GFP punctation, indicating up-regulation of autophagy following infection. G, inhibition of PI3K by exposure to wortmannin has no effect on LC3-GFP punctation in mock − infected cells but abolishes LC3-GFP punctation induced by both Dengue-2 virus (H) and Modoc virus (I) infections, indicating that flavivirus-induced autophagy is PI3K-dependent. J, cells were scored for LC3-GFP punctation and expressed as the percent cells with punctate LC3-GFP. A limited amount of LC3-GFP punctation is seen in mock control cells, whereas infection by either Dengue-2 or Modoc virus increases punctation dramatically (to ∼70%), indicating flavivirus-induced up-regulation of autophagy. PI3K inhibition by wortmannin completely eliminates both Modoc and Dengue-2 virus-induced up-regulation of autophagy. K, boxes, mock − infected MDCK cells display double-membrane autophagosomes typical of housekeeping functions in healthy cells. Both Dengue-2 (L, boxes) and Modoc (M, boxes) virus-infected MDCK cells show an increase in double-membrane autophagosomes, demonstrating autophagy up-regulation following infection in these cells.
To confirm these results in our MDCK system, we next transfected MDCK cells with a C2-LC3-GFP plasmid and infected with either virus. At 48 hpi, cells were fixed and mounted for observation by confocal microscopy. At 48 hpi mock − infected cells display only limited LC3-GFP punctation, indicating little activation of autophagy (Fig. 4D), whereas both Modoc (Fig. 4E) and Dengue-2 (Fig. 4F) infection produce pronounced LC3-GFP punctation, indicating up-regulation of autophagosome formation. Although only 17% of mock − infected cells display punctate LC3-GFP (indicating normal housekeeping activity), nearly 70% of both Modoc and Dengue-2 virus-infected cells display LC3-GFP punctation (Fig. 4J), demonstrating that both viruses induce autophagy. Autophagosome formation is completely abolished following treatment with the PI3K inhibitor wortmannin in both Modoc- (Fig. 4H) and Dengue-2 (Fig. 4I)-infected cells, but it is not altered in mock − infected cells (Fig. 4G). PI3K inhibition by this method reduces the number of infected cells that display punctate LC3-GFP to 17%, similar to mock levels of GFP punctation (Fig. 4J). These results support a model by which PI3K-dependent autophagy results from infection in both Modoc and Dengue-2 viruses and are consistent with previous findings using Dengue-2 virus (3).
Absolute experimental confirmation of autophagy up-regulation has historically relied on the observation of increased numbers of double-membrane autophagosomes by electron microscopy. We therefore fixed and collected both Dengue-2 and Modoc virus-infected cells at 48 hpi for electron microscopy. Analysis of subcellular morphology of mock − infected cells reveals double-membrane vacuoles reminiscent of classic autophagosomes (Fig. 4K, boxes), representing normal housekeeping functions typical of healthy cells. Both Dengue-2 (Fig. 4L, boxes) and Modoc (Fig. 4M, boxes) virus-infected cells display an increase in the number of double-membrane autophagosomes, demonstrating an up-regulation of autophagy following flavivirus infection.
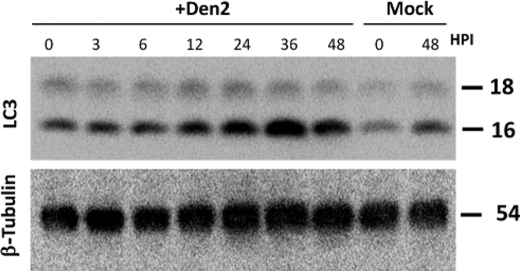
To complete our analysis, we collected whole-protein lysates from Dengue-2-infected cells at 0, 3, 6, 12, 24, 36, and 48 hpi, and we probed these lysates for cleaved LC3 by Western blot. The appearance of LC3-II is often used as an indicator of active autophagy although the amount seen in cells may also increase if clearance is slowed (28). Minimal LC3-II is seen in mock − infected cells at 0 and 48 hpi, reflecting normal housekeeping autophagy (Fig. 5A). However, in Dengue-2-infected cells, observed LC3-II increases steadily until 36 hpi. The decline in cleaved LC3 by 48 hpi potentially derives from completion of the autophagic cycle but, because these cells were in late-stage infection, the issue is more complex and was not pursued further here. β-Tubulin was used as a loading control (Fig. 5B). The total LC3 protein, as judged by Western blot, at least doubles in response to Dengue-2 or NS4A (supplemental Fig. S1). Our results from microscopy and Western blot suggest that autophagy is up-regulated in MDCK cells following both Dengue-2 and Modoc virus infection.
FIGURE 5.
Dengue-2 virus induces LC3 cleavage. Upper panel, analysis of whole-protein lysates of Dengue-2-infected cells at 0, 3, 6, 12, 24, 36, and 48 hpi demonstrates a steady increase in LC3 cleavage to 36 hpi, indicating Dengue-2-induced autophagy in infected cells. The decline in LC3 cleavage at 48 hpi suggests complete LC3 turnover at late hpi. Lower panel, β-tubulin was used as a loading control.
PI3K-dependent Autophagy Is Required for Protection Induced by Flavivirus Infection
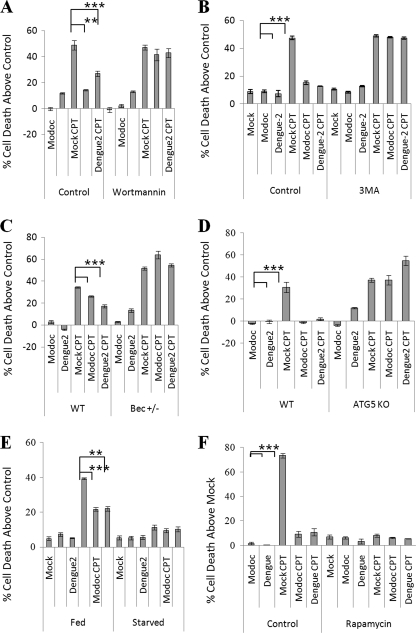
Dengue-2 and Modoc virus both affect survival of MDCK and several other cells in a similar manner, suggesting similarities in the biochemistry of protection. Flavivirus infection triggers up-regulation of autophagy (3, 5), and autophagy is known to protect against insults that trigger the activation of apoptosis (30–33). We therefore hypothesized that the protection observed following infection derives from activation of autophagy. We sought to identify the effect of key autophagy-related proteins on flavivirus-induced protection in epithelial cells. Wortmannin inhibits class I and III PI3K activity, subsequently inhibiting autophagy. Wortmannin treatment does little to cell survival in infected or uninfected cells, whereas CPT treatment kills 49% of mock − infected cells above control within 24 h of exposure (Fig. 6A). However, infection with either Dengue-2 or Modoc virus provides protection against death, with Dengue-2 infection causing a 76% decline (p = 0.0004) and Modoc infection causing a 98% decline (p = 0.0016) in CPT-induced death. Inhibition of PI3K by wortmannin treatment eliminates the protective response invoked by Modoc and Dengue-2 virus infection, allowing CPT-induced death to reach 50% above control in infected populations, killing these cells as effectively as uninfected cells.
FIGURE 6.
Inhibition of autophagy abolishes flavivirus-induced protection against death. A, nonspecific PI3K inhibition by wortmannin eliminates protection by both Dengue-2 and Modoc virus in MDCK renal epithelial cells. B, inhibition of class III PI3K activity by 3MA also eliminates protection against death in MDCK cells, indicating class III PI3K involvement in flavivirus-induced protection. C, inhibition of autophagy by Beclin-1 knockdown eliminates protection by both Dengue-2 and Modoc virus against CPT-induced death. D, inhibition of autophagy by Atg5 knock-out also eliminates protection by both Dengue-2 and Modoc virus. E, up-regulation of autophagy by starvation prior to infection allows no significant death by CPT treatment with or without infection by either virus. F, up-regulation of autophagy signaling by mammalian target of rapamycin inactivation using rapamycin treatment allowed no significant death by high concentration of CPT with or without infection by either virus.
As wortmannin inhibits both class I and class III PI3K proteins, we next inhibited specifically class III PI3K proteins during infection using the class III PI3K inhibitor 3MA, and we repeated the assay. As shown in Fig. 6B, Modoc and Dengue-2 infection and 3MA treatment alone do not kill cells, and levels of death for each are comparable with mock − infected cells at ∼10%. As in previous experiments, CPT treatment induces ∼50% death in mock − infected cells, whereas preinfection with either Dengue-2 or Modoc virus leads to a 70% reduction in death induced by CPT when compared with mock − infected, CPT-treated cells (p = 0.0004 and p = 0.0007, respectively). However, treatment with 3MA leads to the total abolition of both Dengue-2 and Modoc virus-induced protection against CPT-induced death. The data indicate that class III PI3K activity is a regulatory factor for protection against cell death and suggest more specifically that autophagy signaling may be responsible for the ability of the virus to trigger a response leading to protection.
Autophagy-related Proteins Beclin-1 and Atg5 Are Required for Flavivirus-induced Protection
These results demonstrate that class III PI3K signaling is required for Modoc- and Dengue-2-induced protection. Thus, PI3K protects against cell death, suggesting that the ability of the virus to trigger autophagy leads to protection. To confirm this, we characterized Modoc- and Dengue-2-induced protection in two different autophagy-deficient cell lines, Beclin+/− and Atg5−/− MEFs.
Beclin (also called Atg6) interacts with PI3Ks to initiate autophagosome formation. We examined CPT-induced death after flavivirus infection in Beclin knockdown cells, which are heterozygous for the Beclin gene and have a reduced capability to form autophagosomes. WT 4-1 and Beclin+/− cells were infected with Modoc or Dengue-2 virus and subsequently treated at 24 hpi with CPT. At 48 hpi, cells were collected, and percent cell death was analyzed by trypan blue exclusion. As shown in Fig. 6C, CPT kills 34% above control of treated wild-type cells, whereas both viruses were able to protect against cell death with Dengue-2 and Modoc viruses generating a 50% (p < 0.0001) and 24% (p = 0.0002) decline in cell death when compared, respectively, to mock. In mock − infected Beclin+/− cells, CPT kills more effectively, resulting in 52% cell death above control. Moreover, in these cells, virus-induced protection was eliminated, with Dengue-2- and Modoc-infected cells actually exhibiting more cell death after CPT treatment (p < 0.0001, p = 0.0001, respectively) when compared with infected CPT-treated wild-type cells. These figures were essentially equivalent to the amount of cell death provoked by CPT in the absence of virus. Thus, Beclin activation, a key event in the autophagy response, is essential to the protection afforded by Modoc and Dengue-2 virus.
We also analyzed the effect of Atg5 knock-out in mouse embryonic fibroblasts. Atg5 is an autophagy-related protein that acts downstream of Beclin-1 and is essential for formation of the isolation membrane. Wild-type and Atg5 KO MEFs were infected with either Dengue-2 or Modoc virus, incubated for 24 h, treated with CPT for an additional 24 h, and collected for analysis of trypan blue exclusion. As shown in Fig. 6D, CPT kills 31% above control of wild-type cells, whereas infection with either virus protects against death. Dengue-2 and Modoc infection both totally block CPT-induced cell death in wild-type cells (p < 0.001 and p = 0.001, respectively). In mock − infected Atg5 KO cells, CPT induces 37% cell death above control. Both Dengue-2 and Modoc infections fail completely to protect Atg5 KO cells against CPT, with CPT-induced death reaching levels similar to those seen in mock infections. Thus flavivirus-induced protection depends on Atg5. These results are consistent with those for Beclin+/−, suggesting a role for autophagy in flavivirus-induced protection against death.
Up-regulation of Autophagy Prior to Flavivirus Infection Does Not Enhance the Protective Effect
To determine whether flavivirus-induced protection results from the infection or derives from infection-induced autophagy, we examined flavivirus-induced protection while up-regulating autophagy. Prior to infection, autophagy was up-regulated by starvation or by addition of rapamycin (which inhibits mammalian target of rapamycin therefore stimulating autophagy). As shown in Fig. 6E, we found that starvation alone has no effect on cell survival and can protect cells from death induced by CPT. This protection remains the same in starved cells infected with Dengue-2 or Modoc virus. Thus autophagy, not infection itself, is responsible for Modoc- and Dengue-2-induced protection against death.
Up-regulation of autophagy using rapamycin treatment yielded similar results. For this experiment, we increased the concentration of CPT to 100 μm, to ensure that the protection we see following infection with either Dengue-2 or Modoc virus is genuine. As shown in Fig. 6F, mock − infected cells without rapamycin exhibited 74% cell death after 24 h of CPT. Dengue-2- and Modoc-preinfected cells each showed an ∼85% decline in death (p < 0.0001 for both) when compared with mock − infected CPT-exposed cells. (Slightly different conditions produced a modestly higher kill rate, but the pattern was consistent in all experiments.) Rapamycin protected all cells, mock − infected or infected with Modoc or Dengue-2, against CPT-induced death, each yielding only ∼8% cell death. Because of Modoc and Dengue-2 infection, rapamycin and starvation can each elicit autophagy and protection against CPT-induced death, the mechanism by which autophagy is up-regulated does not determine the level of protection. The data suggest that the protection elicited by these two viruses depends on the ability of the host cell to activate autophagy.
Autophagy Enhances Dengue-2 and Modoc Virus Replication
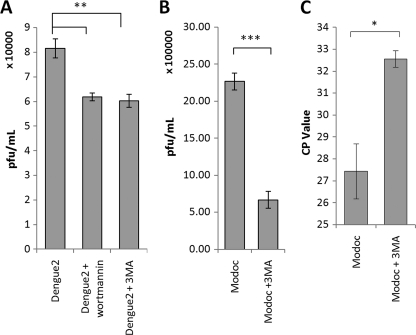
Others have demonstrated that autophagy up-regulation is important to Dengue-2 replication in hepatocytes (3) and that components of the virus replication complex colocalize with autophagosomal membranes (5). As reported above, a similar autophagy response is activated in epithelial cells during flavivirus infection. We therefore evaluated flavivirus replication by plaque assay in MDCK cells while inhibiting autophagy with wortmannin and 3MA. Each sample was run in triplicate, and error bars indicate ±1 S.D. For plaque assay results, virus titer is expressed as plaque-forming units per ml. As shown in Fig. 7A, Dengue-2 replication is apparent in control cells, generating 8.16 × 104 pfu/ml at 96 hpi. Inhibition of autophagy by 3MA or wortmannin treatment each reduces Dengue-2 viral titer to 6 × 104 pfu/ml at 96 hpi, a 27% reduction in viral replication (p = 0.001).
FIGURE 7.
Inhibition of autophagy reduces extracellular flavivirus titer. A, plaque assay of Dengue-2 virus replication in MDCK renal epithelial cells with and without inhibitors of autophagy by wortmannin or 3MA demonstrates a role for autophagy in Dengue-2 replication. Inhibition of autophagy by either wortmannin or 3MA reduces viral titer by 27% at 96 hpi. B, plaque assay analysis of Modoc virus replication in MDCK cells with and without 3MA reveals a similar role for autophagy in Modoc replication. Inhibition of autophagy by 3MA leads to a 71% decline in Modoc virus replication in MDCK cells at 96 hpi. C, RT-PCR quantification of extracellular Modoc viral RNA at 96 hpi also demonstrates that autophagy inhibition by 3MA reduces replication. Note that CP value indicates number of rounds of replication to measurability, and thus lower initial levels result in higher CP values.
Plaque assay of Modoc virus replication during inhibition of autophagy by 3MA revealed that Modoc virus is also dependent upon PI3K-mediated autophagy for optimum replication. Modoc virus replication is evident in control cells (Fig. 7B), generating 2.3 × 106 pfu/ml at 96 hpi. Inhibition of autophagy by 3MA treatment reduces Modoc viral titer to 6.7 × 105 pfu/ml by 96 hpi, a 72% reduction in viral replication (p = 0.02).
We also analyzed Modoc virus replication by RT-PCR. Autophagy was inhibited by 3MA, and virus RNA was extracted from each supernatant sample at 96 hpi and subjected to qRT-PCR analysis. As shown in Fig. 7C, total Modoc virus RNA in the supernatant of autophagy-inhibited cells is significantly reduced compared with control at 96 hpi (p = 0.02), indicating that replication of Modoc virus is enhanced by autophagy, a result similar to those using Dengue-2 virus. (CP value represents the number of cycles before a transcript is recognized. Thus higher numbers reflect lower initial value of RNA. Here the difference is ∼2−5 or a reduction to ∼3% of control.)
These results provide substantial evidence that autophagy enhances both Dengue-2 and Modoc virus replication in renal epithelial cells. The distant relationship of these two viruses also suggests that up-regulation of autophagy is a general characteristic of flavivirus infection in epithelial cells.
Protection against Cell Death Is Mediated by Flavivirus NS4A
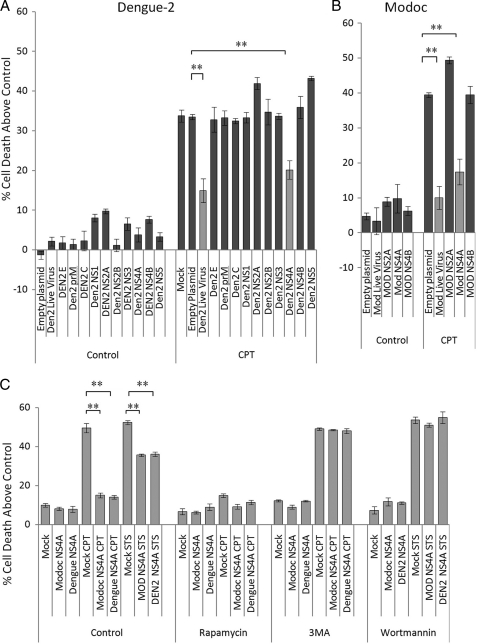
Having established links among flavivirus-induced protection, autophagy and flavivirus replication during infection with both Dengue-2 and Modoc viruses, we sought to identify key viral proteins triggering these effects. We transfected MDCK cells with one of each of the 10 viral genes of Dengue-2 virus and the NS2A, NS4A, and NS4B genes (the small hydrophobic proteins) of Modoc virus, and we allowed 20 h for protein expression. We confirmed expression of transfected plasmids by Western blot (supplemental Fig. S2). We then assayed for protection against cell death induced by CPT as above. Live virus infection was done in tandem as a control for comparison. As in the previous experiments, CPT kills 45% of control cells, and Dengue-2 and Modoc viruses both protect against CPT-induced death (Fig. 8, A and B, respectively), causing a 40 and 45% decline in CPT-caused death, respectively (p = 0.002; p = 0.003). Empty TOPO plasmid does not protect cells against CPT, allowing 47% of cells to die, similar to mock infections. Expression of C, prM, E, NS1, NS2B, NS3, NS4B, and NS5 genes from Dengue-2 and NS4B from Modoc virus does not protect cells against CPT, each protein allowing ∼48% CPT-induced death, similar to control mock − infected cells. Expression of either Dengue-2 or Modoc NS2A slightly increases cell death (p = 0.0002; p = 0.006, respectively). A similar increase in death was seen in CPT-treated NS2A (from both Dengue-2 (p = 0.02) and Modoc virus (p = 0.0001)) expressing cells. However, the situation is different for NS4A. NS4A from either Modoc or Dengue-2 is the only virus protein that protects in a manner similar to that of the whole virus. Dengue-2 NS4A expression decreases CPT toxicity by 35% (p = 0.002), whereas Modoc NS4A expression decreases it by 56% (p = 0.004) when compared with empty plasmid controls. These results demonstrate that NS4A expressed from either Dengue-2 or Modoc effectively mimics the protection afforded with either live virus.
FIGURE 8.
Expression of flavivirus NS4A alone leads to protection similar to that provided by infection with live flavivirus. A, individual expression of any of the 10 Dengue-2 structural and nonstructural genes does not lead to significant death in MDCK renal epithelial cells. Dengue-2 NS4A is the only gene that protects against cell killing by CPT, similar to live virus infection. NS2A and NS5 each allow slightly more death by CPT than mock − infected cells. Individual expression of empty plasmid vector, the structural protein capsid or core (C), premembrane (prM), and envelope (E) proteins. NS1, NS2B, NS3, or NS4B has no effect on CPT-induced death. B, infection with live Modoc virus infection or individual expression of Modoc NS4A also protects cells against CPT-induced death. Expression of NS2A, NS4A, or NS4B does not lead to significant cell killing in MDCK cells (<10%). Modoc NS4A expression protects in a manner similar to live virus infection. Modoc NS2A expression allowed slightly more killing than in mock − infected cells. Expression of the empty plasmid vector or Modoc NS4B had no effect on CPT-induced killing. C, NS4A expression from either Dengue-2 or Modoc virus protects against a DNA-damaging agent (CPT) and a kinase inhibitor (STS). Up-regulation of autophagy by rapamycin treatment protected against death by CPT regardless of expression of NS4A from either virus. Inhibition of autophagy by 3MA treatment abolishes Dengue-2 and Modoc NS4A-induced protection. Inhibition of autophagy by wortmannin also eliminates protection against STS-induced cell death induced by NS4A from either virus.
NS4A-induced protection is not specific to CPT-induced death and protects the cells from cell death induced by STS as well. As shown in Fig. 8C, CPT kills 57% of MDCK cells and STS kills 52% of mock − infected cells. Expression of empty plasmid control had no effect on either CPT- or STS-induced death, allowing 55 and 53% cell death, respectively. Expression of NS4A from either virus reduces CPT-caused death by 70% (Dengue-2 p = 0.0006; Modoc p = 0.0004) and STS-caused death by 30% (Dengue-2 p = 0.0012; Modoc p = 0.0012). Inducing autophagy with rapamycin protected cells regardless of the presence or absence of NS4A from either virus, allowing almost no CPT-induced death. As with live Modoc and Dengue-2 virus infections, inhibiting autophagy using either wortmannin or 3MA eliminated NS4A expression-induced protection against death by CPT and STS, allowing amounts of death similar to that of mock infection, at ∼50% in each case. These results demonstrate that expression of either Modoc or Dengue-2 NS4A is sufficient to protect the cells and indicate that NS4A-induced autophagy is responsible for NS4A-induced protection against death.
NS4A Expression Induces LC3 Cleavage and Translocation
As reported above, infection with either Modoc or Dengue-2 live virus stimulates autophagy, which protects infected cells against many toxins. We therefore determined whether NS4A expression also induces autophagy. We examined LC3 cleavage by Western blot in NS4A-expressing cells. Whole-cell lysates of both live virus-infected and NS4A-transfected cells were obtained, and Western blot analysis was performed using a LC3-specific primary antibody. As shown in Fig. 9A, mock − infected cells exhibit low levels of LC3 cleavage at 48 hpi. In contrast, both live Modoc- and Dengue-2-infected cells, as well as Dengue-2 and Modoc NS4A-expressing cells show up-regulation of LC3 cleavage by Western blot. β-Tubulin was used as a loading control (Fig. 9B). These results indicate that NS4A expression from either virus is sufficient to induce autophagy and suggest that NS4A-induced autophagy is responsible for conferring protection against death.
FIGURE 9.
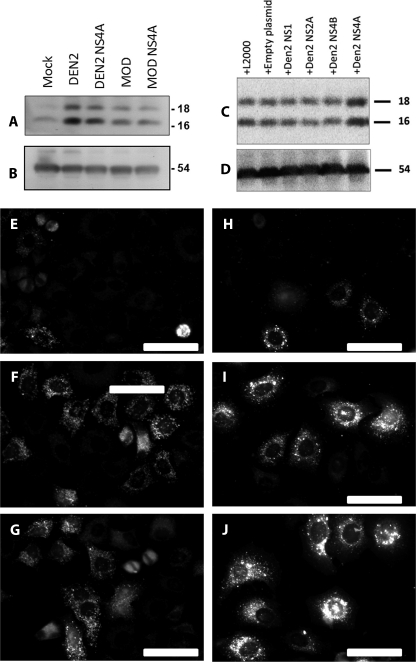
Flavivirus NS4A-induced protection in epithelial cells is dependent upon autophagy. A, Western blotting using a specific antibody for cleaved LC3II reveals an increase in LC3II cleavage following both live Dengue-2 and Modoc infections, as well as after individual expression of NS4A from either virus. B, β-tubulin was used as a loading control. C, whole-protein lysates were collected from MDCK cells transiently expressing Dengue-2 nonstructural proteins NS1, NS2A, NS4A, and NS4B at 24 h post-transfection, as well as cells transfected with empty plasmid and cells exposed to the transfection agent as controls. All conditions stimulate a moderate amount of LC3 cleavage, likely because of stress occurring during our transfection protocol as indicated by LC3 cleavage after exposure to the transfection agent alone. NS4A expression leads to a substantial increase in LC3 cleavage compared with controls and expression of other Dengue-2 nonstructural genes, indicating NS4A expression alone is uniquely sufficient to up-regulate autophagy in MDCK cells. D, β-tubulin was used as a loading control. E, mock − infected HeLa cells stably expressing LC3-GFP exhibit little LC3-GFP punctation, whereas Dengue-2 (F) and Modoc (G) virus infections induce LC3-GFP punctation following infection, indicating autophagy up-regulation. H, expression of empty plasmid vector control in MDCK cells transiently expressing LC3-GFP does not induce LC3-GFP punctation, whereas expression of Dengue-2 NS4A (I) or Modoc NS4A (J) both induce LC3-GFP punctation, indicating autophagy up-regulation following NS4A expression.
To confirm that NS4A alone is responsible for LC3 cleavage and autophagy in these cells, we collected whole-protein lysates from cells transiently individually expressing NS4A or control proteins NS1, NS2A, NS4B, or empty plasmid backbone. As shown in Fig. 9C, each condition caused a small amount of LC3 cleavage, likely due to cellular stress induced by our transfection protocol as indicated by the LC3 cleavage seen in cells treated with the transfection agent (Lipofectamine 2000) but lacking plasmid (+L2000). However, LC3 cleavage is markedly increased in NS4A-expressing cells, indicating that among the proteins examined, NS4A activity alone holds the ability to induce LC3 cleavage in MDCK cells. β-Tubulin was used as a loading control (Fig. 9D). Together with the results above, our data indicate that NS4A activity is unique among the flavivirus proteins in its ability to induce autophagy and cell protection during flavivirus infection.
We also examined LC3 translocation in HeLa cells stably expressing LC3-GFP following both live virus infection and NS4A expression. As shown in Fig. 9E, mock − infected cells display limited (housekeeping) LC3-GFP punctation. Consistent with our previous results, both Dengue-2 (Fig. 9F) and Modoc (Fig. 9G) viruses induce LC3-GFP punctation in a large subset of infected cells, indicating an up-regulation in autophagosome formation. Expression of empty plasmid vector induces little LC3-GFP punctation (Fig. 9H). Consistent with our results above, expression of both Dengue-2 NS4A (Fig. 9I) and Modoc NS4A (Fig. 9J) induces LC3-GFP punctation similar to live virus infection. Our results demonstrate that NS4A from both Modoc and Dengue-2 viruses is sufficient to induce autophagy and indicate that NS4A-induced autophagy is responsible for flavivirus-induced protection against death.
DISCUSSION
Both Dengue-2 and Modoc viruses naturally infect the kidneys, liver, spleen, lungs, salivary glands, several types of immune cells, and the central nervous system (35–37) of the host. These tissues represent many cell types, including neurons, macrophages, monocytes and lymphocytes, endothelial and epithelial cells, hepatocytes, and fibroblasts (36, 38, 39).
Flaviviruses cause apoptosis in neurons and macrophages (25, 40–44), resulting in much of the pathology associated with in vivo infection. Dengue-2 replicates to a very high titer in these cells (which are the primary sites of virus replication during acute infection) before the cells die. As infected macrophages cross epithelia within the body (including the hepatic and renal tubular epithelium), they transmit the virus to surrounding epithelial cells and fibroblasts. In contrast to neurons and macrophages, these latter cells do not die following infection. Instead they survive and often become persistently infected (21, 22, 29). Consistent with these results, we show that in vitro infection of macrophages with either Dengue-2 or Modoc virus results in cell killing similar to that of CPT at 48 hpi, whereas infection with either virus fails to kill renal epithelial cells and fibroblasts (MDCK, 293T, Vero, SW primary MEF, and C57/Bl primary MEF) even when the cells are infected at extremely high titer (m.o.i. = ∼1190), and the cells are evaluated after 12 days (288 hpi, the duration of the experiment).
One explanation for the lack of death in these cells is that our experimental in vitro protocol did not allow sufficient virus uptake and/or replication. We therefore assessed the infectivity and replication of Dengue-2 and Modoc viruses in MDCK cells using immunocytochemistry and RT-PCR, and we found that both viruses infect and replicate to high titer in MDCK cells. Therefore, although renal epithelial cells and fibroblasts are susceptible to flavivirus infection and allow efficient virus replication, infection does not kill the cells. This report therefore provides further evidence that cell fate following flavivirus infection is specific to cell type, as both Dengue-2 and Modoc viruses kill macrophages while sparing renal epithelial cells and fibroblasts.
Dengue-2 infection induces autophagy (3), and the autophagy is important for flavivirus replication in infected hepatocytes (5). Consistent with these results, we find that infection of both MDCK renal epithelial cells and HeLa cells with either Dengue-2 or Modoc virus induces PI3K-dependent autophagy, and chemical inhibition of autophagy by either wortmannin or 3MA in MDCK cells leads to a 27% reduction of extracellular virus titer. These results demonstrate that autophagosome formation enhances flavivirus replication in epithelial cells, similar to previously published results using hepatocytes (3, 5). As the liver and kidneys are both prominent sites of virus replication during persistent flavivirus infections, it is tempting to speculate that the cell type-specific activation of autophagy in these cells plays a major role in the initial establishment of persistent infection in vivo.
A common consequence of autophagy signaling is the up-regulation of pro-survival signaling and protection from death (30–32). To evaluate the link between flavivirus-induced autophagy and protection against death following infection, we assessed the protection offered by flavivirus infection following inhibition of autophagy by both chemical and genetic means. Inhibition of autophagy by wortmannin or 3MA treatment eliminates any protective effect that Dengue-2 or Modoc virus confers on infected cells. Inhibition of autophagosome formation by Beclin-1 knockdown or Atg5 knock-out also removes the protection against death conferred by infection with either virus. These results demonstrate that flavivirus-induced protection is dependent upon up-regulation of autophagy following infection. When autophagy is up-regulated prior to infection by either starvation or rapamycin treatment, cells are protected irrespective of flavivirus infection, providing evidence that it is specifically autophagy, and not other virus-mediated events, that provides protection against death following infection.
By expressing each virus gene individually, we determined that NS4A from either Dengue-2 or Modoc virus is the only flavivirus protein sufficient to protect against death by CPT. As with live virus infection, NS4A expression up-regulates autophagy and induces broad spectrum protection, shielding cells against both CPT- and STS-induced death. Expressing NS4A protects cells against CPT nearly as well as infecting the cells with live virus. Inhibition of PI3K signaling by wortmannin and 3MA eliminates autophagy-induced protection, whereas inducing autophagy by rapamycin confers protection regardless of NS4A expression. As with whole virus infection, we also find that of several Dengue-2 nonstructural genes tested, NS4A expression alone induces PI3K-dependent autophagy and subsequent protection against death. We conclude that flavivirus NS4A expression during whole virus infection leads to up-regulation of autophagy. NS4A-induced autophagy subsequently protects against death during infection. We do not yet know how the NS4A protein regulates autophagy. As autophagosomes figure prominently in flavivirus replication within these cells, flavivirus NS4A is thus identified as a potential target for development of antiviral drugs. We are currently exploring the specific biochemical virus-host interactions that govern NS4A-induced up-regulation of autophagy following flavivirus infection and searching for clues as to the mechanism(s) behind its cell-type specificity.
Supplementary Material
Acknowledgments
We thank Dr. Adolfo Garcia-Sastre of Mt. Sinai Medical School, New York, for providing the Dengue-2 virus as well as the Dengue-2 gene constructs. We thank Dr. Stephane Boissinot of City University of New York Queens College for the Modoc gene constructs and Dr. Guido Kroemer of Institut Gustave-Roussy, Villejuif, France, for the LC3-GFP construct. We also thank Narges Zali, Pierre Co, Alain Goldman, and Michelle Sahli for technical support. Dr. Areti Tsiola of the Queens College Core Facility for Imaging, Molecular and Cellular Biology, provided technical expertise. We especially thank Dr. Richard Lockshin for critical review of this manuscript prior to submission.
This work was supported, in whole or in part, by National Institutes of Health MARC U-STAR Grant 2T34GM070387-03 (to Z. Z.). This work was also supported by Professional Staff Congress-City University of New York (PSC-CUNY) award (to Z. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- MDCK
- Madin-Darby canine kidney cell
- m.o.i.
- multiplicity of infection
- 3-MA
- 3-methyladenine
- MEF
- mouse embryonic fibroblast
- CPT
- camptothecin
- STS
- staurosporine
- CHX
- cycloheximide
- TRITC
- tetramethylrhodamine isothiocyanate
- qRT
- quantitative RT
- pfu
- plaque-forming unit
- ER
- endoplasmic reticulum
- hpi
- hours post-infection.
REFERENCES
- 1. Halstead S. B. (2007) Lancet 370, 1644–1652 [DOI] [PubMed] [Google Scholar]
- 2. Morens D. M. (2009) Pediatr. Infect. Dis. J. 28, 635–636 [DOI] [PubMed] [Google Scholar]
- 3. Lee Y. R., Lei H. Y., Liu M. T., Wang J. R., Chen S. H., Jiang-Shieh Y. F., Lin Y. S., Yeh T. M., Liu C. C., Liu H. S. (2008) Virology 374, 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee C. J., Liao C. L., Lin Y. L. (2005) J. Virol. 79, 8388–8399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Panyasrivanit M., Khakpoor A., Wikan N., Smith D. R. (2009) J. Gen. Virol. 90, 448–456 [DOI] [PubMed] [Google Scholar]
- 6. Falgout B., Pethel M., Zhang Y. M., Lai C. J. (1991) J. Virol. 65, 2467–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li H., Clum S., You S., Ebner K. E., Padmanabhan R. (1999) J. Virol. 73, 3108–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takegami T., Sakamuro D., Furukawa T. (1995) Virus Genes 9, 105–112 [DOI] [PubMed] [Google Scholar]
- 9. Zhou Y., Ray D., Zhao Y., Dong H., Ren S., Li Z., Guo Y., Bernard K. A., Shi P. Y., Li H. (2007) J. Virol. 81, 3891–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lindenbach B. D., Rice C. M. (1999) J. Virol. 73, 4611–4621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin C., Amberg S. M., Chambers T. J., Rice C. M. (1993) J. Virol. 67, 2327–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miller S., Kastner S., Krijnse-Locker J., Bühler S., Bartenschlager R. (2007) J. Biol. Chem. 282, 8873–8882 [DOI] [PubMed] [Google Scholar]
- 13. Mackenzie J. M., Khromykh A. A., Jones M. K., Westaway E. G. (1998) Virology 245, 203–215 [DOI] [PubMed] [Google Scholar]
- 14. Roosendaal J., Westaway E. G., Khromykh A., Mackenzie J. M. (2006) J. Virol. 80, 4623–4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bartenschlager R., Lohmann V., Wilkinson T., Koch J. O. (1995) J. Virol. 69, 7519–7528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jiang L., Yao H., Duan X., Lu X., Liu Y. (2009) Biochem. Biophys. Res. Commun. 385, 187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davis J., Hardy J. (1973) J. Appl. Microbiol. 26, 344–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin L., Ye Y., Zakeri Z. (2006) Cell Death Differ. 13, 141–150 [DOI] [PubMed] [Google Scholar]
- 19. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tesh R. B., Siirin M., Guzman H., Travassos da Rosa A. P., Wu X., Duan T., Lei H., Nunes M. R., Xiao S. Y. (2005) J. Infect. Dis. 192, 287–295 [DOI] [PubMed] [Google Scholar]
- 21. Tonry J. H., Xiao S. Y., Siirin M., Chen H., da Rosa A. P., Tesh R. B. (2005) Am. J. Trop. Med. Hyg. 72, 320–324 [PubMed] [Google Scholar]
- 22. Siirin M. T., Duan T., Lei H., Guzman H., da Rosa A. P., Watts D. M., Xiao S. Y., Tesh R. B. (2007) Am. J. Trop. Med. Hyg. 76, 299–306 [PubMed] [Google Scholar]
- 23. Pogodina V. V., Frolova M. P., Malenko G. V., Fokina G. I., Levina L. S., Mamonenko L. L., Koreshkova G. V., Ralf N. M. (1981) Acta Virol. 25, 337–343 [PubMed] [Google Scholar]
- 24. Gritsun T. S., Frolova T. V., Zhankov A. I., Armesto M., Turner S. L., Frolova M. P., Pogodina V. V., Lashkevich V. A., Gould E. A. (2003) J. Virol. 77, 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gulati L., Chaturvedi U. C., Mathur A. (1982) Br. J. Exp. Pathol. 63, 194–202 [PMC free article] [PubMed] [Google Scholar]
- 26. Davis J. W., Hardy J. L. (1974) Infect. Immun. 10, 328–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cardiff R. D., Russ S. B., Brandt W. E., Russell P. K. (1973) Infect. Immun. 7, 809–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tanida I., Waguri S. (2010) Methods Mol. Biol. 648, 193–214 [DOI] [PubMed] [Google Scholar]
- 29. Murray K., Walker C., Herrington E., Lewis J. A., McCormick J., Beasley D. W., Tesh R. B., Fisher-Hoch S. (2010) J. Infect. Dis. 201, 2–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kroemer G., Jäättelä M. (2005) Nat. Rev. Cancer 5, 886–897 [DOI] [PubMed] [Google Scholar]
- 31. Bröker L. E., Kruyt F. A., Giaccone G. (2005) Clin. Cancer Res. 11, 3155–3162 [DOI] [PubMed] [Google Scholar]
- 32. Ogata M., Hino S., Saito A., Morikawa K., Kondo S., Kanemoto S., Murakami T., Taniguchi M., Tanii I., Yoshinaga K., Shiosaka S., Hammarback J. A., Urano F., Imaizumi K. (2006) Mol. Cell. Biol. 26, 9220–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blommaart E. F., Krause U., Schellens J. P., Vreeling-Sindelárová H., Meijer A. J. (1997) Eur. J. Biochem. 243, 240–246 [DOI] [PubMed] [Google Scholar]
- 34. Deleted in proof.
- 35. Lum L. C., Lam S. K., Choy Y. S., George R., Harun F. (1996) Am. J. Trop. Med. Hyg. 54, 256–259 [DOI] [PubMed] [Google Scholar]
- 36. Jessie K., Fong M. Y., Devi S., Lam S. K., Wong K. T. (2004) J. Infect. Dis. 189, 1411–1418 [DOI] [PubMed] [Google Scholar]
- 37. Davis J. W., Hardy J. L., Reeves W. C. (1974) Infect. Immun. 10, 1362–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leyssen P., Van Lommel A., Drosten C., Schmitz H., De Clercq E., Neyts J. (2001) Virology 279, 27–37 [DOI] [PubMed] [Google Scholar]
- 39. Solomon T. (2004) N. Engl. J. Med. 351, 370–388 [DOI] [PubMed] [Google Scholar]
- 40. Desprès P., Flamand M., Ceccaldi P. E., Deubel V. (1996) J. Virol. 70, 4090–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Desprès P., Frenkiel M. P., Ceccaldi P. E., Duarte, Dos Santos C., Deubel V. (1998) J. Virol. 72, 823–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jan J. T., Chen B. H., Ma S. H., Liu C. I., Tsai H. P., Wu H. C., Jiang S. Y., Yang K. D., Shaio M. F. (2000) J. Virol. 74, 8680–8691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Espina L. M., Valero N. J., Hernández J. M., Mosquera J. A. (2003) Am. J. Trop. Med. Hyg. 68, 48–53 [PubMed] [Google Scholar]
- 44. Halstead S. (1989) Rev. Infect. Dis. 11, 830–839 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.