Abstract
GPER is a Gs-coupled seven-transmembrane receptor that has been linked to specific estrogen binding and signaling activities that are manifested by plasma membrane-associated enzymes. However, in many cell types, GPER is predominately localized to the endoplasmic reticulum (ER), and only minor amounts of receptor are detectable at the cell surface, an observation that has caused controversy regarding its role as a plasma membrane estrogen receptor. Here, we show that GPER constitutively buds intracellularly into EEA-1+ endosomes from clathrin-coated pits. Nonvisual arrestins-2/-3 do not co-localize with GPER, and expression of arrestin-2 dominant-negative mutants lacking clathrin- or β-adaptin interaction sites fails to block GPER internalization suggesting that arrestins are not involved in GPER endocytosis. Like β1AR, which recycles to the plasma membrane, GPER co-traffics with transferrin+, Rab11+ recycling endosomes. However, endocytosed GPER does not recycle to the cell surface, but instead returns to the trans-Golgi network (TGN) and does not re-enter the ER. GPER is ubiquitinated at the cell surface, exhibits a short half-life (t½ <1 h), and is protected from degradation by the proteasome inhibitor, MG132. Disruption of the TGN by brefeldin A induces the accumulation of endocytosed GPER in Rab11+ perinuclear endosomes and prevents GPER degradation. Our results provide an explanation as to why GPER is not readily detected on the cell surface in some cell types and further suggest that TGN serves as the checkpoint for degradation of endocytosed GPER.
Keywords: Endocytosis, G-protein-coupled Receptors (GPCRs), Lysosomes, Trafficking, Ubiquitination, GPER, GPR30, trans-Golgi Network
Introduction
Seven-transmembrane receptors (7TMRs)2 comprise the largest class of plasma membrane receptors (> 800 members), and they initiate intracellular signaling by binding extracellular ligands and employ heterotrimeric G-proteins (1). Upon binding their cognate ligands, activated G-protein-coupled receptors (GPCRs) undergo a series of adaptive changes that effectively prevent excessive receptor signaling. This process, termed receptor desensitization is typically associated with receptor phosphorylation by G-protein-coupled receptor kinases and the recruitment of β-arrestins, which not only facilitate receptor uncoupling from G-proteins, but promote recruitment of activated receptors into clathrin-coated pits via interaction of arrestin with clathrin and the clathrin adaptor, AP2. Subsequent recruitment of dynamin, or other small GTPases, to the receptor-enriched, clathrin-coated pits results in the scission of newly formed intracellular vesicles or endosomes (2, 3). Alternatively, GPCRs are also known to cluster in a clathrin-independent manner in caveolae prior to entry into endosomes formed by either dynamin-dependent or -independent mechanisms (4–7).
Once internalized, GPCRs face distinct trafficking destinations: either they recycle to the cell surface and re-associate with their G-proteins (resensitization) or they are targeted to lysosomes for degradation resulting in complete termination of receptor signaling (down-modulation) (3, 8). A third fate has been described for endocytosed GPCRs, with receptors accumulating in perinuclear compartments and exhibiting slow sorting times into either degradative and/or recycling pathways (8–10). The fate of GPCR sorting appears to be determined by the nature of the association of GPCRs with arrestin (10–15). In general, receptors that contain serine-threonine clusters at the C terminus tail are phosphorylated, form avid interactions with arrestin, and are subsequently degraded (11, 12) or retained in the perinuclear area (9, 10) or slowly dephosphorylated and recycled (12, 13). In contrast, receptors that contain phosphorylation sites but not serine-threonine clusters form transient interactions with arrestins and are rapidly dephosphorylated and recycled (13). Thus, internalization, recycling, and degradation of GPCRs collectively serve as a mechanism to mediate receptor desensitization, resensitization, and down-regulation, thereby, contributing to the spatiotemporal regulation of GPCR-promoted cell signaling.
A number of studies suggest that GPER functions as a unique estrogen receptor that promotes rapid estrogen action and is associated with female reproductive cancers (16–19). Both ectopic expression and RNA interference studies have shown that GPER is involved in high-affinity specific binding of [3H]17β-estradiol at the plasma membrane (20), activation of G-proteins (16, 20), adenylyl cyclase (17, 20, 21), mobilization of intracellular calcium stores (22), and a series of 17β-estradiol (E2)-induced functional activities, including activation of integrin α5β1 and subsequent fibronectin matrix assembly, and EGFR transactivation (16, 19). These results strongly suggest that GPER acts similarly to other 7TMRs as a cell surface receptor. Moreover, subcellular fractionation studies also support the plasma membrane as a site of GPER action, and stimulation of GPER-expressing HEK-293 cells with E2 induced sequestration of GPR30 into clathrin-coated vesicles (22). However, some controversy still exists with regard to GPER and its identification as a plasma membrane estrogen receptor (23–25). A major concern with the proposed function of GPER as a plasma membrane receptor is the fact that the protein largely concentrates in the endoplasmic reticulum (ER). Likewise, binding sites for synthetic estrogen fluorophores are not observed at the cell surface but are concentrated in the ER (26). GPER is predominately localized intracellularly in endometrial, ovarian, and breast cancer cells obtained in tissue biopsies (18, 27, 28). However, cell surface expression is not only observed upon forced overexpression of GPER in HEK-293 cells (22) but also in mammalian tissues, such as rat pyramidal neurons (29) and renal epithelia (30), suggesting that GPER is not restricted from the plasma membrane.
Here, we have further explored the endocytic fate of GPER in HEK-293 cells and compared its endocytic trafficking pattern to other GPCRs, which either recycle (β1-adrenergic receptor (β1AR)) or are sorted to lysosomes for proteolytic degradation (CXCR4). Our results show that, although GPER, CXCR4, and β1AR each enter cells via clathrin-dependent endocytosis, they ultimately have distinct endocytic fates. GPER endocytosis does not involve recruitment of arrestins. Contrary to the rapid recycling route employed by β1AR and rapid lysosomal degradation route of CXCR4, GPER accumulates in the trans-Golgi network (TGN) and suffers degradation through an ubiquitin-proteasome pathway.
EXPERIMENTAL PROCEDURES
Cell Lines and Culture Conditions
Human embryonal kidney 293 cells (HEK-293) were purchased from the American Tissue Culture Collection (Manassas, VA). HEK-293 cells that stably express hemagglutinin (HA)-tagged GPER (HA-GPER) or HA-β1-adrenergic receptor (HA-β1AR) were previously described (22). HEK-293 (HA-CXCR4) cells were generated in a similar manner by transfection and drug selection as described previously (22) using HA-CXCR4 plasmid DNA kindly provided by Jeffrey Benovic (Thomas Jefferson University). All cells were grown at 37 °C in phenol red-free DMEM/Ham's F-12 medium (Invitrogen) supplemented with 5% fetal bovine serum and 50 μg/ml gentamicin.
Antibodies
Monoclonal GPER antibody, 2F2, was generated as described previously (22). Fab fragments of mouse HA antibody (Covance) were produced by limited papain digestion and isolated as the non-bound fraction on protein A-immunoabsorbent beads using a commercial kit (Thermo Scientific). Other commercial antibodies included: rabbit anti-HA epitope monoclonal antibody (Cell Signaling, MA, at 1:1000), mouse anti-HA (Covance, at 1:200), mouse anti-EEA1 (IgG1, at 1:500), mouse anti-LAMP-1 (Santa Cruz Biotechnology, IgG1, at 1:40), sheep anti-TGN46 (AbD Serotec, 1:1000), mouse anti-Rab11 (1:200), mouse anti-rab-7 (Cell Signaling, 1:200), rabbit anti-calnexin (Stressgen Bioreagents, 1:200), rabbit anti-ubiquitin (Santa Cruz Biotechnology, at 1:200), goat anti-rabbit Alexa-Fluor 594 and anti-mouse Alexa-Fluor 488 (1:1000, Molecular Probes), donkey anti-sheep Alexa-Fluor 647 and 555 (1:1000, Molecular Probes), and donkey anti-rabbit Alexa-Fluor 555 and anti-mouse Alexa-Fluor 488 (1:1000) and 647 (1:500) (Molecular Probes).
Plasmids
Molecular clones for HA-CXCR4, arrestin-2-GFP, and arrestin-3-GFP were kindly provided by Jeffrey Benovic. Plasmids encoding amino acids 319–418 of bovine arrestin 2, or mutants of the same protein lacking the clathrin (ΔLIELD) or β-adaptin (F391A) interaction sites of bovine arrestin-2, were also provided by Dr. Benovic (31).
Endocytosis Analysis
Cells were seeded onto glass coverslips (0.3 × 106/35-mm dish) and washed with serum-free medium several times before use. When needed, cells were transiently transfected with cDNAs encoding arrestin-2/-3-GFP or arrestin-2 mutants using Lipofectamine (Invitrogen). Cells were chilled in cold medium and then incubated with rabbit or mouse anti-HA antibody/normal rabbit IgG alone or with both anti-HA antibody and Alexa Fluor 488-conjugated transferrin (50 μg/ml, Molecular Probes) in ice-cold, serum-free medium for 25 min. Cells were washed with cold serum-free medium to remove excess antibody or transferrin and then treated in medium prewarmed to 37 °C with vehicle or respective agonists and/or reagents (cycloheximide, 36 μm; MG132, 10 μm; brefeldin A (BFA), 2.5 μg/ml; chloroquine, 100 μm) and incubated at 37 °C for various times. Cells were fixed in 4% paraformaldehyde in PBS and then processed for immunofluorescence or ELISA analysis.
Immunofluorescence
Fixed cells were permeabilized in blocking buffer containing 0.1% Triton X-100, 3% BSA, and 3% normal goat or donkey serum in PBS (pH 7.4) or incubated in the blocking buffer without Triton X-100 for 20–30 min at room temperature. Cells were then incubated for 1 h in primary antibodies diluted in blocking buffer. Unbound primary antibody was removed by washing in PBS. Cells were incubated at room temperature in secondary antibodies for 1 h, washed in PBS, and mounted in anti-quench mounting medium with DAPI (Vector Laboratories, Inc., Burlingame, CA). Immunofluorescence images were visualized with an Eclipse 80i microscope (Nikon, Inc., Melville, NY) equipped with a Nikon Plan Fluor 100 × 0.5–1.3 oil iris with differential interference contrast and epifluorescence capabilities using a Qimaging Retiga 2000R digital camera and Nikon imaging software (NIS-Elements-BR 3.0), or with the Zeiss LSM 710 confocal laser scanning microscope with ZEN 2009 software (Zeiss). The figures were processed with brightness/contrast adjustment using Photoshop CS2 (Adobe).
ELISA Assay
Cells were seeded onto poly-l-lysine-coated 6-well plates (1 × 106/well) and prelabeled with rabbit anti-HA antibody as described above. After removal of excess antibody, cells were incubated with vehicle or E2 (1 nm) or Stromal cell-derived factor-1 (SDF-1, 25 ng/ml) for 0, 15, 30, and 60 min at 37 °C and then fixed in 4% paraformaldehyde. Fixed cells were permeabilized with 0.1% Triton X-100 in blocking buffer containing 3% BSA and 3% normal donkey serum in PBS for 20 min at room temperature. Cells fixed at the 0 time point (representing total surface receptors) were not permeabilized and were incubated only with the blocking buffer. After several rinses with PBS, the cells were incubated for 1 h at room temperature with HRP-conjugated donkey anti-rabbit IgG (1:8000) diluted in 3% BSA in PBS, and then incubated with the HRP substrate 3,3′,5,5′-tetramethylbenzidine (Sigma) for 20 min. The reaction was stopped by the addition of hydrochloric acid. The absorbance of the reaction product was then measured at 450 nm. The nonspecific background binding of antibody to cells was determined by omission of primary antibody.
Decay Rate and Ubiquitination Analysis
HA-GPER or HA-β1AR cells were prelabeled with HA antibody as described above. Cells were then directly harvested (0 min) or incubated at 37 °C with cycloheximide (36 μm) alone or together with chloroquine (100 μm) or MG132 (10 μm) or BFA (2.5 μg/ml) for various time periods. Cells were harvested and solubilized in radioimmune precipitation assay-buffered detergent supplemented with protease inhibitors as described previously (16). Insoluble proteins were clarified from detergent lysates by centrifugation at 12,000 rpm for 25 min. Samples were prepared for electrophoresis by mixing 20–30 μg of detergent-soluble protein in 2× reducing Laemmli sample buffer (Bio-Rad) for 10 min at room temperature prior to gel loading. The remainder of lysate was incubated with protein A-coupled agarose beads overnight at 4 °C. Immunoabsorbed proteins were pelleted by centrifugation at 1000 rpm for 2 min. Protein A beads containing surface-labeled receptor were washed six times with lysis buffer, and then captured proteins were eluted from the beads using 1× reducing Laemmli sample buffer for 10 min at room temperature.
Immunoblotting Analysis
Samples were resolved by 10% SDS-PAGE according to standard procedures. The same amount of proteins, determined using a BCA protein assay, was loaded into the gel. Equal protein loading was also verified by staining of the transferred nitrocellulose membrane using red reversible dye Ponceau S. After blocking with 5% nonfat milk in TBS-T buffer for 30 min, the transferred membrane was incubated overnight in primary antibody, diluted in 3% or 5% BSA in TBS-T buffer, at 4 °C. The membrane was washed three times, incubated for 1 h at room temperature with HRP-conjugated donkey anti-rabbit IgG (Pierce), and then treated with enhanced chemiluminescence substrate (SuperSignal, Pierce) and exposed on film (Kodak).
RESULTS
Internalization of Recombinant HA-GPER
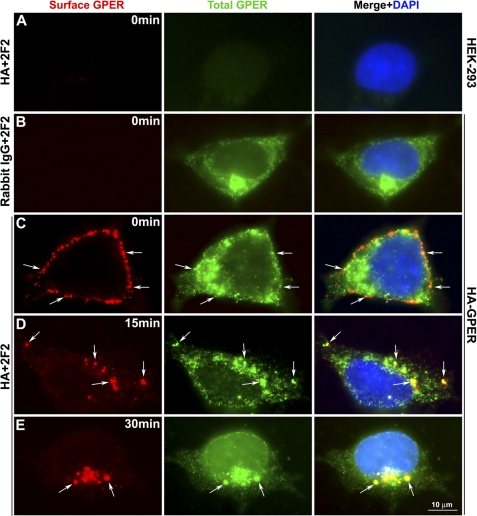
To investigate the molecular events that regulate GPER internalization, and to determine its endocytic fate, the widely adopted scheme of tracking cell surface receptors using antibodies specific for amino-terminally located epitope tags was employed (32). To this end, live HEK-293 cells stably expressing HA-GPER were surface-labeled at 4 °C with HA-specific rabbit antibodies. For controls, normal HEK-293 cells or HEK-293 cells expressing HA-GPER were surface-labeled with HA antibody or normal rabbit IgG, respectively. Cells were then warmed to 37 °C for various lengths of time in the presence of E2. After treatment, cells were fixed, permeabilized, and labeled with monoclonal antibody (mAb) 2F2 specific for the C terminus of GPER. Surface and intracellular receptors were distinguished using species-specific fluorescent antibodies using epifluorescence analysis. In control experiments, no immunoreactivity was observed in normal HEK-293 cells either at the surface or in the cytoplasm. Similarly, no immunoreactivity was observed at the surface of HA-GPER cells (Fig. 1, A and B). mAb 2F2 showed no immunoreactivity in permeabilized control HA-β1AR cells (data not shown). These results suggest no nonspecific binding in HA-GPER HEK-293 cells for both HA and 2F2 antibodies. Intracellular GPER was abundantly expressed and concentrated in a perinuclear pattern in permeabilized cells that were immunostained with mAb 2F2 (Fig. 1, B–E). Coincident staining of HA antibodies applied to live cells and mAb 2F2 applied post fixation and permeabilization was observed at the surface prior to warming (Fig. 1C) and intracellularly after warming (Fig. 1, D and E) suggesting that the recombinant HA-GPER protein was intact during trafficking. HA antibody-bound immunoreactivity was not intracellularly detected in the cells fixed at 0 min (Fig. 1C). This suggests that dissociation of HA antibodies from surface receptors and subsequent binding to intracellular GPER during staining procedures employed in this study did not occur. At early time points (<15 min) as represented in Fig. 1D, the bulk of HA-GPER was observed in vesicles near the inner face of the plasma membrane, whereas at later times (≥30 min) GPER concentrated in a large compartment in close proximity to the nucleus (Fig. 1E). These observations support the idea that intact HA-GPER protein is measured during GPER internalization.
FIGURE 1.
Endocytosis of recombinant HA-GPER. HEK-293 cells (A) or HEK-293 cells stably expressing HA-GPER (B–E) were prelabeled with rabbit HA antibody (A and C–E) or normal rabbit IgG (B) at 4 °C and then stimulated with E2 (1 nm) at 37 °C for 15 min (D) or 30 min (E). Fixed cells were permeabilized and then incubated with mouse mAb 2F2 specific for the carboxyl terminus of GPER. Surface and endocytosed GPERs were visualized with Alexa 594-conjugated goat anti-rabbit IgG (red), and total GPER by Alexa 488-conjugated anti-mouse IgG (green). Co-localization is shown in yellow (arrows) at the surface (C) or intracellularly (D and E) in the merged images. Nuclei were detected by DAPI (blue). Images are representatives from three independent experiments.
GPER Undergoes Constitutive Endocytosis
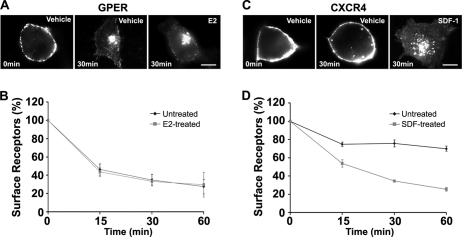
Many 7TMRs, including CXCR4 (33, 34), undergo both agonist-dependent and agonist-independent internalization (7, 35–40). Therefore, the capacity of GPER and CXCR4 to internalize in the absence of their cognate ligands was tested (Fig. 2). For these experiments, HA-GPER cells or HA-CXCR4 cells were prelabeled in the cold with HA antibodies and then cells were exposed to vehicle or their respective ligands, E2 or SDF-1. Cells were fixed at various time intervals and evaluated by immunofluorescence. The majority of surface-labeled GPER redistributed to the intracellular compartment with a similar endocytic fate concentrated in a perinuclear patch whether or not agonist was added (Fig. 2A). Quantitative ELISA assay further revealed that GPER disappeared from the cell surface in the absence of E2 at 15, 30, and 60 min (53.8 ± 6.0%, 65.2 ± 6.3%, and 72.6 ± 7.9%, respectively). Similar results were obtained at these time points in the presence of E2 (56.4 ± 4.9%, 66.7 ± 3.1%, and 70.4 ± 13.6%, respectively) (Fig. 2B). These findings indicate that GPER undergoes constitutive endocytosis. CXCR4 undergoes either constitutive or agonist-dependent endocytosis in different cell types (34, 41, 42). In our immunofluorescence analyses performed under similar receptor-labeling conditions, HA-CXCR4 exhibited constitutive endocytosis to a much lesser degree than GPER, with most receptors requiring SDF-1 for endocytosis (Fig. 2C). Quantitative ELISA assay showed that, although 46.4 ± 4.1%, 65.6 ± 1.46%, and 74.5 ± 2.0% of surface CXCR4 disappeared from the cell surface with SDF-1 stimulation at 15, 30, and 60 min, respectively, significantly less receptor exited the plasma membrane in the absence of SDF-1 (25.3 ± 2.3%, 24.3 ± 3.8%, and 30.1 ± 2.8%) at these respective time points (Fig. 2D).
FIGURE 2.
HA-GPER undergoes agonist-independent endocytosis. HA-GPER (A and B) or HA-CXCR4 cells (C and D) were prelabeled with rabbit HA antibody at 4 °C. After removing excess antibody, cells were treated with E2 (1 nm), or SDF-1 (25 ng/ml), or vehicle for various time intervals at 37 °C and then fixed in paraformaldehyde. Cells were then processed for immunostaining (A and C) or ELISA assay (B and D). Results shown represent the mean ± S.E. of three independent experiments.
The rate of GPER constitutive endocytosis is faster than that of CXCR4, as approximately half (53.8 ± 6.0%) of surface-labeled GPER disappeared by 15 min, whereas roughly one quarter (25.3 ± 2.3%) of surface-labeled CXCR4 entered cells in the absence of agonist at 15 min. After 15 min, surface GPER disappearance continued progressively although at a slower rate; however, CXCR4 constitutive endocytosis remained constant at a low level over 1 h. Contrary to the effect of E2 stimulation on GPER endocytosis, SDF-1 treatment of CXCR4 cells nearly tripled the amount of surface-labeled receptor that entered cells in the absence of agonist (Fig. 2, B and D). In contrast, as reported previously (2, 3) constitutive endocytosis of HA-β1AR was not observed in HEK-293 cells. To directly rule out the possibility that constitutive receptor endocytosis may be caused by cross-linking receptors by whole HA Ig, HA antibody Fab fragments were prepared and were used to prelabel HA-GPER cells. No significant difference was measured in HA-GPER endocytosis whether cells were prelabeled with monovalent or bivalent HA antibodies (data not shown).
Our previous study showed that GPER is sequestered in clathrin-coated vesicles in the presence of E2 in HEK-293 cells (22). It has been reported that some GPCRs undergo clathrin-independent constitutive internalization but switch to clathrin-dependent internalization with agonist stimulation (7). To examine if this is also the case for GPER, E2-treated and untreated cells were co-immunostained for clathrin. Our results demonstrate that GPER co-localized with clathrin both in the presence and absence of E2, indicating that GPER constitutive internalization is clathrin-dependent (supplemental Fig. S1).
GPER Endocytosis Occurs Independently of Arrestin-2/-3
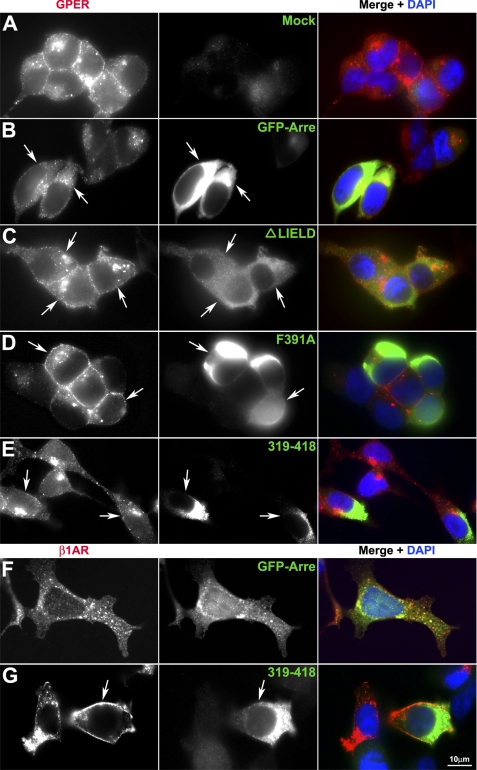
Non-visual arrestins (arrestin-2 and arrestin-3) function as adaptor proteins to recruit 7TMRs to clathrin-coated pits via interaction with receptors, clathrin, and clathrin-associated protein-2 complexes (2). To examine if arrestin-2 or -3 redistributes in response to agonist stimulation and forms detectable complexes with receptor, HA-GPER or HA-β1AR cells were transiently transfected with arrestin-2-GFP or arrestin-3-GFP and then stimulated with their respective agonists. Isoproterenol stimulation of HA-β1AR/arrestin-2-GFP cells caused rapid recruitment of arrestin-2 into vesicular bodies containing HA-β1AR (Fig. 3F). In contrast, E2 stimulation of HA-GPER/arrestin-2-GFP cells neither facilitated the receptor endocytosis as compared with mock-transfected cells nor induced the redistribution of arrestin-2 or its association with the receptor (Fig. 3, A and B). Similar results were obtained in HA-β1AR cells or HA-GPER cells that were transfected with arrestin-3-GFP (data not shown). These results indicate that non-visual arrestins do not appear to become activated during GPER endocytosis.
FIGURE 3.
GPER receptor fails to form complexes with arrestin-2. HA-GPER (A–E) or HA-β1AR (F and G) cells transiently transfected with cDNAs encoding GFP-tagged arrestin-2 (GFP-Arre) or indicated mutant arrestin-2 (green) were prelabeled with rabbit HA antibody and then stimulated with E2 (1 nm) or isoproterenol (10 μm) at 37 °C for 30 min. After removing excess antibody, cells were fixed and permeabilized. Surface or endocytosed receptors were visualized by Alexa 594-conjugated antibody (red). Arrows indicate transfected cells. Nuclei were detected by DAPI (blue). Images are representatives from three independent experiments.
Because HA-GPER cells express significantly less surface receptor than HA-β1AR cells, we considered the possibility that our inability to measure arrestin-2/-3 mobilization in HA-GPER cells may be a consequence of reduced receptor expression. To address this possibility, the influence of forced overexpression of dominant-negative arrestins on GPER internalization was evaluated. Overexpression of a mutant arrestin-2 peptide encoding amino acids 319–418 and containing the β-adaptin and clathrin binding sites, but not the receptor interaction domain (31, 43), negatively blocked HA-β1AR endocytosis (Fig. 3G) yet had no impact on GPER internalization (Fig. 3E). Similarly, arrestin-2 mutants lacking β-adaptin (F391A) or clathrin (ΔLIELD) interaction sites, respectively, negatively impacted HA-β1AR (data not shown) but not GPER internalization (Fig. 3, C and D) even though HA-GPER cells expressed less detectable surface receptor than HA-β1AR cells. Collectively, these results suggest that non-visual arrestins are not involved in GPER endocytosis.
Endocytosed GPER Enters Early and Recycling Endosomes
7TMRs that enter endosomal vesicles generally recycle back to the plasma membrane where they form functional receptors or are directed to lysosomes for degradation resulting in receptor down-regulation or termination of signaling (2). To define the fate of endocytosed GPER, the endocytic route of HA-GPER was followed using endosome-specific markers that distinguish early and recycling endosomes as well as late endosomes and lysosomes.
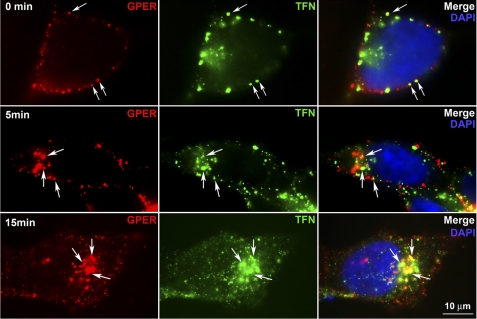
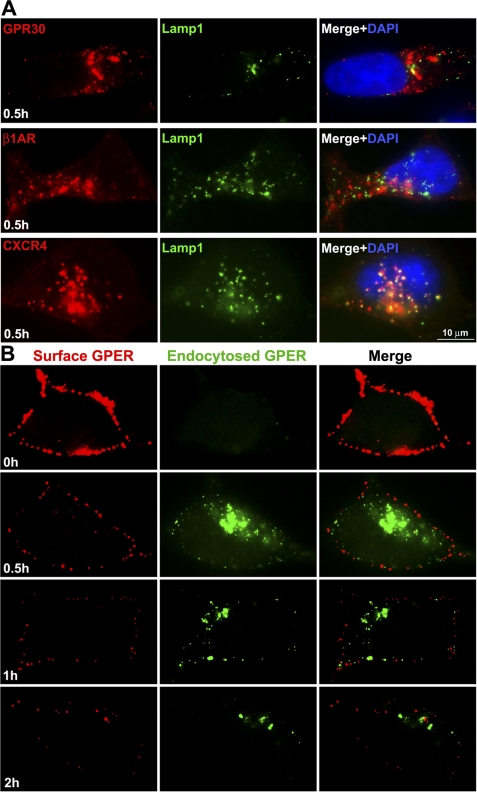
Early endosomal antigen-1 (EEA-1) and transferrin are widely employed as markers of early and recycling endosomes (33, 34, 44). Dual immunostaining for surface-labeled receptor and EEA-1 demonstrated that GPER, similar to β1AR and CXCR4, rapidly enters EEA-1+ endosomes (supplemental Fig. S2). Although each receptor type still remains associated with EEA-1, a significant difference was observed by 30 min between receptor types in the distribution pattern of receptor-positive, EEA-1+ endosomes with HA-GPER endosomes concentrating in a perinuclear fashion, whereas HA-β1AR and HA-CXCR4 endosomes were homogenously distributed throughout the cytosol. Dual labeling of surface GPER with HA-antibodies and transferrin receptors with Alexa 488-conjugated transferrin revealed the co-localization of GPER with transferrin receptors at the cell surface prior to warming and within the cytoplasm by 5 min post warming. After 15 min, the majority of GPER co-localized with transferrin around the perinuclear area. These results suggest that, like transferrin, the bulk of GPER enters recycling endosomes (Fig. 4).
FIGURE 4.
Endocytosed HA-GPER co-localizes with transferrin. HA-GPER cells were prelabeled with rabbit anti-HA antibody and Alexa 488-conjugated transferrin (green) at 4 °C and incubated at 37 °C for the indicated time points, fixed, and permeabilized. HA-GPER was detected with anti-rabbit Alexa 594 (red). Arrows indicate the co-localization of GPER and transferrin at the cell surface or in the cytoplasm (in yellow). Nuclei were detected with DAPI (blue). Images are representatives from three independent experiments.
GPER Does Not Traffic to Lysosomes or Recycle to the Plasma Membrane
To address whether GPER is sorted to a late endosome or lysosome for degradation, endocytosed GPER was co-immunostained with the late endosome marker, Rab7, or lysosome marker, lysosome-associated membrane protein-1 (LAMP-1). Like HA-β1AR, which recycles to the plasma membrane (43, 45), only a small portion of endocytosed GPER (<10%) co-stained with Rab7 (data not shown) or LAMP-1 (Fig. 5A), suggesting that the bulk of GPER does not enter a late endosome-lysosomal degradative pathway. In contrast, CXCR4, a well known example of a GPCR that suffers lysosomal degradation, was strongly positive in either Rab7 endosomes or LAMP-1-labeled lysosomes (Fig. 5A).
FIGURE 5.
HA-GPER neither accumulates in the lysosome nor recycles to the cell surface. A, HA-GPER or HA-β1AR or HA-CXCR4 cells were prelabeled with rabbit anti-HA antibody at 4 °C and incubated with their respective ligands at 37 °C for 30 min. Fixed cells were permeabilized and immunostained for lysosomal membrane antigen-1 (LAMP-1). HA-GPER or LAMP-1 was detected with anti-rabbit Alexa Fluor 594 (red) or anti-mouse Alexa Fluor 488 (green), respectively. Co-localization of the receptors and LAMP-1 was demonstrated in the merged images (yellow). Nuclei were detected with DAPI (blue). B, the levels of surface-labeled GPER decreased progressively over time. Cells were surface-labeled, incubated with 17β-E2 for the indicated time points, and then fixed. Cells with surface-labeled GPER were incubated with goat anti-rabbit Alexa 594 (surface-labeled receptor, red), and then permeabilized and incubated with goat anti-rabbit Alexa 488 (endocytosed receptor, green) after permeabilization. Results are representatives from four independent experiments.
To further examine the fate of endocytosed GPER, surface and endocytosed GPER were monitored over time by sequentially applying anti-rabbit Alexa 594 (prior to permeabilization) and anti-rabbit Alexa 488 (post-permeabilization) (Fig. 5B). This differential-labeling strategy discriminates surface GPER (red) versus endocytosed (green) GPER. We found that both surface and endocytosed receptors decreased progressively over 2 h (Fig. 5B). These results are consistent with the finding from the ELISA assay in Fig. 2 showing that surface pre-labeled GPER disappeared increasingly over time (∼1 h). Together, these findings suggest that, unlike β1AR, the bulk of GPER does not recycle to the cell surface or enter the lysosome for degradation and, instead, is driven to an alternative degradative pathway.
GPER Accumulates in the TGN
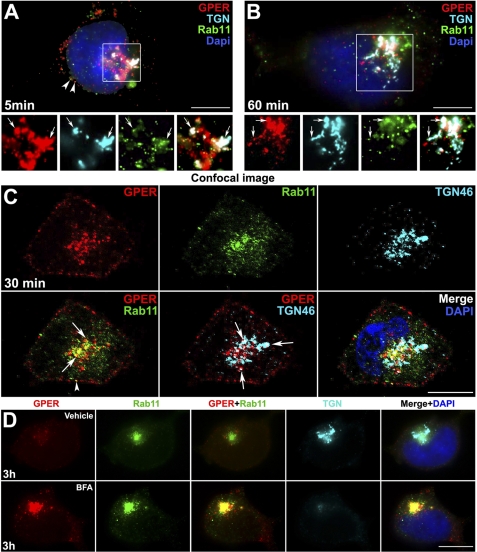
The perinuclear compartment is not a commonly described endocytic destination for GPCRs, however, retrograde transport of other cell surface receptors, such as vasopressin type 2A receptor (10, 15) and somatostatin type 2A receptor (14, 46), from the plasma membrane toward the nucleus has been reported. Moreover, the TGN is a well known regulatory checkpoint for newly synthesized GPCRs during their biogenesis and export to the plasma membrane (8). Therefore, we tested the hypothesis that endocytosed GPER might return to the TGN from recycling endosomes. Indeed, GPER was found to co-localize with TGN-46, a marker for the TGN, as early as 5 min (Fig. 6A) and continued to accumulate in the TGN over 1 h (Fig. 6B). Confirmation of this finding was obtained by evaluating the co-localization of GPER with TGN-46 as well as the small GTPase, Rab11, which has been linked to perinuclear recycling endosomes (47, 48). To ensure that the co-localization is not due to superimposition of separate labeling, images were also captured using confocal microscopes with 0.8-μm-thick optical sections (Fig. 6C). In experiments in which endocytosed receptor was co-stained with TGN-46 and Rab11, some Rab11 overlapped with TGN-46-labeled compartments where GPER was also localized, suggesting that GPER-containing recycling endosomes may fuse into the TGN. In contrast, only a minor portion of β1AR co-localized with TGN-46, whereas a large number of receptors co-distributed with Rab11 (data not shown). Similarly, CXCR4 did not co-localize with TGN-46 over the time of study (data not shown). These data suggest that GPER enters the TGN via a Rab11+ recycling endosome.
FIGURE 6.
HA-GPER accumulates in the TGN. A–C, epifluorescence (A and B) and confocal (C) microscope images show the co-localization of GPER with TGN46 and Rab11. HA-GPER cells were prelabeled with rabbit HA antibody (red) and incubated with E2 (1 nm) at 37 °C for 5 (A), 30 (C), and 60 min (B). Fixed and permeabilized cells were immunostained with sheep anti-TGN46 (cyan) and mouse anti-Rab11 antibodies (green). The individual channels of the boxed regions are shown in the bottom panel (A and B). D, endocytosed GPER was concentrated in Rab11-labeled endosome after brefeldin A (BFA) treatment. HA-GPER cells were surface-labeled with HA antibody and then incubated at 37 °C with E2 for 3 h in the presence of vehicle or the TGN-disrupting agent, BFA. Fixed and permeabilized cells were stained as in A–C. Nuclei were detected with DAPI (blue). Arrows indicate the co-localization of all three labels (white). Arrowheads indicate the co-localization of GPER and Rab11 (yellow). Scale bars, 10 μm. Results are representatives from six independent experiments.
To further assess whether GPER enters the TGN via Rab-11 endosomes, cells were treated with the TGN disrupter, BFA, or vehicle (Fig. 6D). By 3 h, most of the endocytosed GPER (∼95%) concentrated in a perinuclear patch that strongly co-labeled with Rab11 in a pattern in which the typical structure of the TGN was destroyed by BFA treatment, indicating an accumulation of GPER in Rab11-positive perinuclear recycling endosomes. Moreover, accumulation of a large amount of endocytosed GPER in the perinuclear area of BFA-treated cells remained for over 3 h as compared with untreated cells, implying that blockade of receptor trafficking from Rab11+ recycling endosomes to the TGN prevents receptor degradation and that the TGN is the checkpoint for determining GPER degradation. Less than 5% of vesicles were co-labeled with GPER and Rab11 at the cortex of the cells over 3 h, which suggests that only a few GPERs could recycle to the plasma membrane directly from the recycling endosome.
GPER Does Not Re-enter the ER
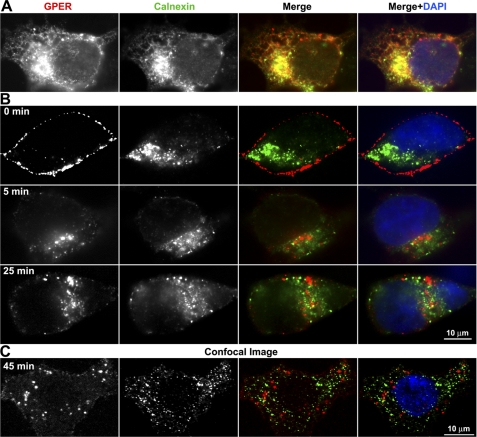
GPER is expressed at high levels in the ER of human breast cancer cells (26). Similarly, we observed that the bulk of recombinant HA-GPER is coassociated with calnexin (Fig. 7A), a finding consistent with the idea that the receptor accumulates in the ER. Experiments evaluating the ability of surface-labeled GPER to return to the ER revealed that little detectable GPER was found to co-localize with calnexin at 5, 25, and 45 min using both epifluorescence (Fig. 7B) and confocal microscopy (Fig. 7C), indicating that GPER selectively returns to the TGN, but does not reenter the ER.
FIGURE 7.
Endocytosed GPER does not return to the ER. A, the majority of recombinant HA-GPER localizes to the ER. HA-GPER cells were fixed, permeabilized, and immunostained for calnexin, a marker of ER. Co-localization is shown in yellow in the merged image. B and C, endocytosed GPER does not co-localize with calnexin using epifluorescence (B) or confocal microscopes (C). HA-GPER cells were surface-labeled with mouse monoclonal HA antibody and incubated with E2 at 37 °C for the indicated time intervals. Fixed cells were permeabilized and labeled with rabbit anti-calnexin. GPER and calnexin were visualized by anti-mouse Alexa 594 (red) and anti-rabbit Alexa 488 (green), respectively. Nuclei were detected with DAPI (blue). Results are representatives from three independent experiments.
Degradation of GPER via the Ubiquitin-Proteasomal Pathway
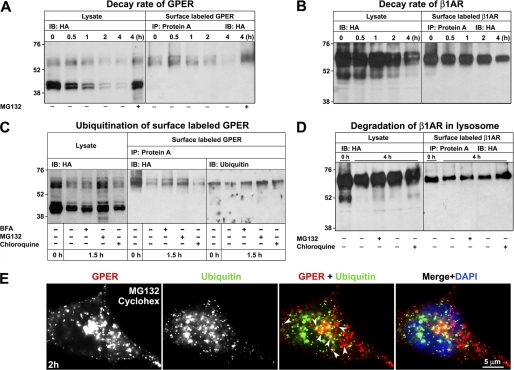
Accumulation of endocytosed GPER in BFA-treated cells (Fig. 6D) implies that GPER undergoes degradation post TGN sorting. To assess the decay rate, the half-life of total and surface GPER versus β1AR (Fig. 8, A and B) was measured in cells that were treated with the protein synthesis inhibitor, cycloheximide. To analyze surface receptor, lysates were prepared post rabbit HA antibody labeling of live cells at 4 °C, and receptor-antibody complexes were immunoabsorbed onto protein A-agarose beads. The immunoabsorbed proteins were then eluted and probed with HA-specific antibodies. Immunoblot analysis comparing detergent lysates prepared from whole cells versus surface-labeled cells identified surface versus intracellular forms of both HA-GPER and HA-β1AR. ∼10% of the total HA-GPER proteins were expressed on the cell surface. Both the major (43- and 45-kDa) and large (60-, 62-, and 67-kDa) forms of GPER were rapidly degraded with an estimated half-life of <30 min (Fig. 8A). In contrast, >95% of the total HA-β1AR was expressed on the cell surface, and the degradation of total and surface β1AR was slow with measured half-lives of >4 h. Collectively, these findings are consistent with the idea that HA-β1AR is recycled and does not rely upon receptor biogenesis to replenish surface receptors, whereas GPER is rapidly degraded.
FIGURE 8.
HA-GPER is degraded via an ubiquitin-proteasomal pathway. HA-GPER or HA-β1AR cells were prelabeled with HA antibody at 4 °C and incubated at 37 °C with chloroquine, MG132, or BFA in the presence of cycloheximide (36 μm) at the indicated time points. Whole cell lysates or prelabeled receptors immunoabsorbed with protein A-coupled beads were separated by 10% SDS-PAGE. A–D, whole cell lysates or surface-labeled receptors were immunoblotted with HA antibody. In C, surface-labeled receptors were immunoabsorbed on protein A as in A, B, and D, but elutes were divided into equivalent fractions and then either probed with HA or ubiquitin antibodies. E, endocytosed GPER co-localizes with ubiquitin. Surface-labeled HA-GPER cells were incubated at 37 °C with cycloheximide and MG132 for 2 h and then immunostained for ubiquitin. Co-localization is shown in yellow in the merged image (arrowheads). Results for all experiments shown here are representative from three independent experiments.
Our immunofluorescence data indicate that the bulk of endocytosed GPER neither enters a lysosomal degradative pathway nor recycles to the cell surface (Fig. 5), which suggests the existence of another major degradative pathway for GPER. We hypothesized that GPER might be degraded through a ubiquitin-proteasomal pathway. To test this hypothesis and to further assess the impact of the lysosome as a site of GPER degradation, surface and total GPER decay rates were measured in cells that had been treated either with the proteasome inhibitor, MG132, or the lysosomotrophic agent, chloroquine (Fig. 8C). Consistent with our finding that GPER does not accumulate in lysosomes (Fig. 5), chloroquine treatment had only a modest effect in blocking the degradation of total GPER and only had a slight influence on the degradation of surface-labeled receptors. In contrast, MG132 treatment almost completely blocked the degradation of total GPER suggesting, that the proteasome is a major site for the degradation of both intracellular and plasma membrane-derived GPER.
Probing immunoprecipitates prepared from surface-labeled protein with ubiquitin-specific antibodies revealed that the 62-kDa isoform of GPER was preferentially ubiquitinated and that 43- and 45-kDa bands of GPER were not easily detected. Moreover, ubiquitinated GPER was present at the plasma membrane prior to endocytosis and treatment with BFA or MG132 prevented further decay of the 62-kDa GPER isoform. Chloroquine treatment had little effect on the degradation of ubiquitinated GPER (Fig. 8C). No significant effect was measured upon total or surface-labeled HA-β1AR in cells that were treated with either MG132 or chloroquine, consistent with the current and previous findings that little β1AR was degraded over 4 h (Fig. 8B) (49). Co-immunostaining for ubiquitin and endocytosed GPER showed that endocytosed GPER co-localized with ubiquitin in the cells treated with cycloheximide and MG132 (Fig. 8E). Collectively, these data suggest that the TGN is an important regulatory checkpoint for GPER degradation via a ubiquitin-proteasome pathway.
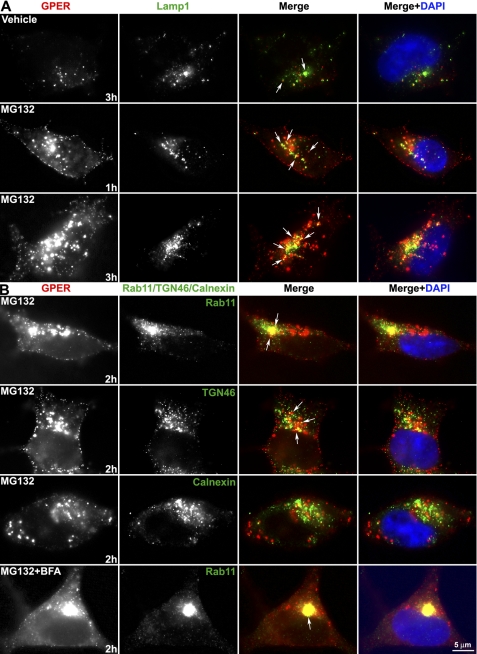
Consistent with this idea, immunofluorescence also showed that inhibition of proteasome activity with MG132 resulted in robust accumulation of endocytosed GPER relative to untreated cells that only contain relatively little endocytosed GPER over time (∼3 h) (Fig. 9A). Blockade of proteasomal proteolysis resulted in more endocytosed GPER co-localized with LAMP-1. Treatment with MG132 allows for a more thorough evaluation of the subcellular distribution of endocytosed GPER and reveals that many, but not all, endocytosed GPER is distributed in a perinuclear pattern with Rab11 or TGN-46, but not with calnexin (Fig. 9B). Combined treatment of MG132 with BFA revealed that endocytosed GPER is almost exclusively accumulated in Rab11 endosomes (Fig. 9B) and is not easily detected in other subcellular organelles, including the ER, or endosomes (data not shown).
FIGURE 9.
TGN is a regulatory checkpoint for GPER degradation. HA-GPER cells prelabeled with HA antibody at 4 °C were incubated at 37 °C for the indicated time intervals in the presence of E2 and MG132 or BFA or both. Fixed and permeabilized cells were immunostained for surface-labeled GPER and LAMP-1 (A) or Rab11/TGN46/calnexin (B). Co-localization (arrows) is shown in yellow in the merged images. Nuclei were detected with DAPI (blue). Results are representatives from four independent experiments.
Collectively, these data indicate that GPER is ubiquitinated at the plasma membrane, traffics to the TGN via Rab11, does not re-enter the ER, and suffers degradation via the 26 S proteasome.
DISCUSSION
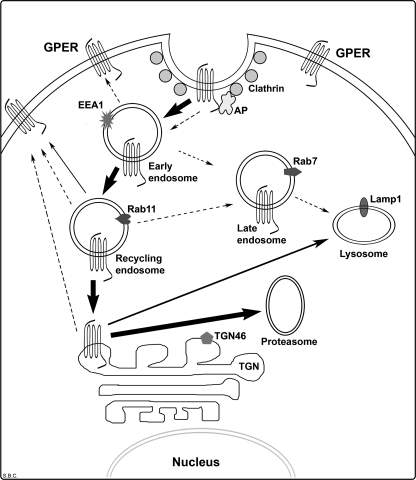
We have previously reported that ectopic expression of recombinant HA-GPER in HEK-293 cells results in the assembly of a functional receptor that is detected in the plasma membrane and promotes estradiol-dependent release of intracellular calcium and entry into cells via clathrin-coated vesicles (22). Here we show that the G-protein-coupled estrogen receptor, GPER, employs an endocytic trafficking mechanism that is unusual for members of the GPCR superfamily, resulting in its rapid down-modulation with a measured half-life of surface-derived GPER of <30 min. Salient features of our data as outlined in Fig. 10 include: (a) constitutive entry from the plasma membrane into clathrin-coated pits, EEA-1+ early endosomes, and transferrin+ and Rab11+ recycling endosomes; (b) a failure to form transient or stable interactions with arrestin-2 or -3; (c) perinuclear accumulation of endocytosed GPER in the TGN; (d) BFA-sensitive ubiquitination and degradation of endocytosed GPER in proteasomes; and (e) while GPER is commonly detected in the ER, endocytosed receptor does not re-enter this compartment. Collectively, these data indicate that the TGN is an important checkpoint for GPER endocytosis prior to degradation and may, in part, explain why GPER is commonly detected intracellularly.
FIGURE 10.
Schematic model of GPER internalization. GPER undergoes rapid constitutive endocytosis, a process involving recruitment of GPER into clathrin-coated pits in an arrestin-independent fashion. The majority (>90%) of endocytosed GPER traffics through the early and recycling endosomal pathway, accumulating in the trans-Golgi network (TGN), and suffering degradation through the ubiquitin-proteasome pathway. Only a minor portion (<10%) of GPER is degraded in the lysosome through the TGN and recycled to the plasma membrane through the recycling endosome. Dotted arrows indicate the endocytic pathways that have been documented for other GPCRs.
GPER Undergoes Constitutive Endocytosis by an Arrestin-independent Mechanism
HA-tagged GPER, β1AR, or CXCR4, stably expressed in HEK-293 cells, was labeled at the cell surface at 4 °C with HA-specific antibody, and the intracellular destination of antibody-labeled receptors was followed after removal of excessive, unbound antibody. The reliability of this method for measuring GPER trafficking was further addressed in experiments that showed that HA antibody-bound GPER co-stained in both the plasma membrane and perinuclear compartments with C-terminally bound GPER 2F2 monoclonal antibody suggesting that recombinant HA-GPER remained intact during its intracellular trafficking (Fig. 1). Our quantitative analysis reveals that, in contrast to β1AR and CXCR4, the majority of surface-labeled GPER (>50%) is endocytosed by 15 min without agonist stimulation, indicating the existence of rapid constitutive endocytosis for GPER. Constitutive endocytosis of GPER was also observed when monovalent Fab fragments of HA antibody were used to label surface receptor (data not shown), indicating that receptor cross-linking by bivalent antibody does not trigger GPER internalization (42). Moreover, antibody prelabeling did not alter agonist-dependent internalization of similarly engineered HA-β1AR or HA-CXCR4, suggesting that HA antibody binding does not activate HA-β1AR and HA-CXCR4 and subsequently triggers their endocytosis. Furthermore, because our endocytosis assays were conducted on cells that were extensively washed and maintained in serum-free media for prolonged periods of time (>24 h), it is unlikely that constitutive endocytosis of GPER was due to the presence of residual agonist. However, the experiments detailed here do not completely exclude the possibility that the binding of monovalent HA antibody triggered conformational activation of GPER and subsequently facilitated the endocytosis.
Constitutive activity, observed both in native and recombinant systems, is suggested to support the ability of a GPCR to adopt an active conformation in the absence of agonist (36). Constitutive activity of 7TMRs was first reported for the opioid receptor, and subsequently more than 60 7TMRs, coupled to Gs-, Gi-, and Gq-proteins, also have been found to exhibit constitutive internalization and constitutive activity (36). To date, the mechanism and physiological role for 7TMRs constitutive endocytosis is still elusive. Constitutive endocytosis may serve as a mechanism for controlling the number of functionally active 7TMRs on the cell surface and providing a tonic support for basal cell function, such as neuronal activity (34). Constitutive endocytosis may also play a role in the axonal targeting of CB1 receptors in cultured neurons (39). Intriguingly, many 7TMRs with naturally occurring mutations have been identified to display increased constitutive activity as compared with wild type. Increased constitutive activity is associated with various clinical phenotypes or diseases (36). We have previously shown that E2 treatment of (HA-GPER) HEK-293 cells results in HA-GPER-dependent mobilization of intracellular calcium (22), suggesting agonist-induced signaling is measured in these cells. We have also found that HEK-293 cells expressing HA-GPER display higher levels of pCREB in the nucleus than HEK-293 cells expressing HA-β1AR (unpublished data), indicating that GPER exhibits constitutive activity. During the preparation of the manuscript, Leeb-Lundberg and colleagues also observed the existence of constitutive endocytosis of FLAG-tagged GPER in MDCK and HEK-293 cells by directly applying FLAG antibody to the live cells at 37 °C instead of prelabeling with antibody at low temperature as used in our study, although no more detailed information about endocytic trafficking of the receptor was provided (50). Prior studies have shown that CXCR4 displays distinct degrees of constitutive endocytosis in different cell types (33, 34). Our current results show that CXCR4 exhibits predominant agonist-dependent internalization and a low level of constitutive endocytosis in HEK-293 cells (Fig. 2), which is consistent with previous findings in U-937 and CEM cells (33, 34).
Upon activation, 7TMRs are rapidly phosphorylated by GPCR kinases leading to subsequent recruitment of β-arrestins and receptor desensitization. Arrestins are adaptor proteins that link 7TMRs to clathrin, adaptor protein AP-2, and phosphoinositides; facilitate receptor uncoupling from G-proteins; and lead to the targeting of GPCRs into clathrin-coated pits and subsequent endocytosis (2). Our findings show that GPER constitutive endocytosis is clathrin-mediated (supplemental Fig. S1) but independent of arrestin (supplemental Fig. S2), suggesting that GPER constitutive endocytosis does not require GPCR kinase-mediated phosphorylation. In fact, many other 7TMRs have also been identified that do not require GPCR kinase and arrestin for entering clathrin-coated pits, such as thromboxane-A2β receptor (51), CXCR2 (52), and PAR1 (36). On the other hand, there have been studies reporting that GPCRs undergo endocytosis via recruitment into caveolae instead of clathrin-coated pits (6).
Intracellular Distribution Pattern of GPER
Multiple investigators have shown that GPER promotes rapid estrogen-dependent signals in both endogenous and exogenous settings, including stimulation of intracellular cAMP and the release of pro-HB-EGF from the plasma membrane (16, 17, 20–22, 50, 53, 54). Its biochemical actions, and the mechanisms employed to promote these signals, are familiar to GPCRs, because they are regulated by Gs- and Gβγ-proteins and manifested by enzymes that are positioned exoplasmically (MMP-3) (55) and integrally (adenylyl cyclase) (17) within the plasma membrane. These findings strongly suggest that the plasma membrane is the likely site of GPER action. However, GPER is difficult to detect in the plasma membrane and is commonly found to be concentrated intracellularly and predominately co-localized with the KDEL (Lys-Asp-Glu-Leu) ER retention marker (26). Use of Alexa dye-conjugated 17α-[4-aminomethylphenylethnyl]-estra-1,3,5(10)-triene 3,17β-diol as a means to measure the subcellular distribution of GPER binding sites has shown that detergent permeabilization is necessary, implying that GPER may function uniquely among 7TMRs as an intracellular receptor (26). This study further showed that expression of GFP-tagged GPR30 resulted in a diffuse intracellular staining pattern further supporting this concept. However, our current results and prior work have demonstrated that GPER can be detected in the plasma membrane (29, 50) and undergoes endocytosis (22, 50), although in this report and in our study, GPER mainly resides intracellularly (Fig. 1). In fact, this distribution pattern is not unique for GPER, because many other 7TMRs have been reported to display a similar intracellular distribution pattern, such as the type A cholecystokinin receptor (56), GABAA (35), CXCR4 (34), 5-HT1A (57), and CB1 receptors (37). It has been suggested that constitutive endocytosis of these receptors may account for their low or undetectable levels at the plasma membrane (34, 35, 56). Here, we propose that the deficit balance between a minor component of recycling receptors and rapid constitutive endocytosis as well as rapid degradation, may provide a partial explanation for the predominant intracellular localization observed for GPER. Accumulation of GPCRs in the ER and TGN is commonly measured during receptor biogenesis and export to the plasma membrane and has been associated with post-translational editing mechanisms, including proper receptor folding, the formation of intrachain disulfide bonds, glycosylation, and carbohydrate processing. Intriguingly, our current findings indicate that endocytosed GPER returns to the TGN but does not re-enter the ER either through early or recycling endosomes.
The TGN Serves as a Regulatory Checkpoint for Endocytosed GPER
Once entering the early endosome, GPCRs face two general endocytic trafficking fates. GPCRs are targeted to lysosomes for degradation via Rab7+ late endosomes, or Rab11+ recycling endosome-to-late endosome pathway, a mechanism, which leads to receptor down-modulation and termination of receptor signaling. Alternatively, GPCRs are either rapidly recycled to the plasma membrane directly via Rab4 and/or Rab5+ early endosomes (the “short cycle”) or slowly recycled to the plasma membrane via Rab11+recycling endosomes and/or the TGN (the “long cycle”), which results in receptor resensitization and signal recovery (reviewed in Refs. 3, 8). By dual and triple immunofluorescence, we show that the vast majority of endocytosed GPER co-localizes with EEA-1, transferrin, Rab11, and the TGN marker, TGN-46, but not with the later endosome marker, Rab7, or the resident lysosomal membrane marker, LAMP-1, or the ER marker, calnexin. Disruption of the TGN by BFA treatment results in the accumulation of endocytosed GPER in Rab11 vesicles. By monitoring the surface levels of prelabeled GPER, we found that surface GPER was decreased progressively over the time course. Together, these findings suggest that, in contrast to classic pathways of GPCR endocytic trafficking, the bulk of endocytosed GPER neither recycles back to the plasma membrane nor traffics to the late endosome and lysosome but, rather, traffics in a retrograde fashion through the early endosomes and Rab11+ perinuclear recycling endosomes to accumulate in the TGN. Therefore, the TGN serves as a regulatory checkpoint for GPER endocytic trafficking. Slow receptor recycling times and perinuclear accumulation of the vasopressin 2A receptor has also been noted, although the endosomal trafficking pattern, and the involvement of Rab-related proteins and/or the TGN, is unclear (10, 15).
Degradation of GPER via an Ubiquitin/Proteasome Pathway
Degradation of endocytosed GPCRs generally occurs in lysosomes via ubiquitin-dependent or -independent machinery, such as β2AR, CXCR4, and PAR1 (3). However, our immunofluorescence analysis demonstrates that only a minor portion of endocytosed GPER co-localizes with Rab-7+ late endosomes and LAMP-1-marked lysosomes, suggesting the existence of an alternative degradative pathway for GPER (Fig. 6). Indeed, Western blotting and immunofluorescence analyses revealed that endocytosed GPER underwent rapid degradation and that its degradation was mainly blocked by MG132, an inhibitor of proteasomal proteolysis, but only slightly blocked by the lysosomotrophic agent chloroquine. However, we did notice that treatment of cells with MG132 caused more receptors to co-localize with LAMP-1, implying that degradation of GPER occurs in lysosomes as a secondary target when proteasomal proteolysis is blocked. Further ubiquitination analysis clearly demonstrated that GPER is ubiquitinated at the plasma membrane prior to endocytosis and that MG132 treatment rescues ubiquitinated GPER after endocytosis. This result is supported by dual immunofluorescent staining showing the co-localization of endocytosed GPER with ubiquitin after MG132 treatment. Taken together, these findings suggest that endocytosed GPER undergoes ubiquitination and subsequent degradation via proteasomal proteolysis.
Intriguingly, BFA treatment rescued ubiquitinated endocytosed GPER in Western blotting and resulted in a larger number of receptors concentrating in Rab11+ perinuclear endosomes as compared with untreated cells even at late time points (e.g. 3 h) in immunofluorescence. This implies that disruption of the TGN prevents degradation of endocytosed GPER. Moreover, combined treatment of BFA and MG132 revealed that endocytosed GPER is almost exclusively concentrated in Rab11+ endosomes and is not easily detected in other subcellular sites (Figs. 6D and 9B), indicating an essential role for the TGN in GPER degradation. Collectively, these data suggest that the TGN may serve as the checkpoint for sorting of GPER mainly to the proteasomes and only to a lesser extent, to lysosomes for ubiquitin-dependent degradation. To our knowledge, somatostatin type 2A receptor is the first GPCR that has been reported in vivo to traffic to the TGN directly from early endosomes, through which the receptor is not directed to the lysosome for degradation, but slowly recycles to the cell surface for functional resensitization (46, 58). Our current results indicate that GPER is the first reported GPCR that is degraded through the recycling endosome-TGN-proteasomal degradative pathway after endocytosis.
During biogenesis, newly synthesized proteins are translocated into the lumen of the ER and scrutinized by ER quality control machinery before sorting into the vesicles of the secretory pathway. Misfolded or incompletely proteins are retained in the ER and retrotranslocated to the cytosolic side of ER membrane through the Sec61 translocon complex-formed channel and subsequently degraded in the 26 S proteasome via ubiquitination (59, 60). Several GPCRs have been reported to utilize this machinery for degradation, including A2a-adenosine receptor and δ-opioid receptor (61, 62). Interestingly, our data reveal that the 26 S proteasome is also the destination for endocytosed GPER through the TGN. Co-localization of endocytosed GPER with the ER marker, calnexin, was not observed even in the presence of the proteasome inhibitor, MG132 (Figs. 7 and 9B), indicating that endocytosed GPER does not re-enter the ER. The mechanism by which endocytosed GPER is translocated from the TGN to the proteasome for degradation is yet to be determined. However, other integral plasma membrane receptors, such as platelet-derived growth factor β-receptor and growth factor receptor, also undergo degradation in the proteasome after endocytosis, albeit whether the receptors are translocated from the TGN or not is unclear (63, 64).
Disengagement from G-proteins and endocytic trafficking of a given GPCR from the plasma membrane to the intracellular compartment is the key mechanism by which cells protect themselves from chronic stimulation by their cognate ligands. At present, it is unclear why endocytosed GPER is preferentially sorted to proteasomes instead of lysosomes, but the fact that this novel membrane estrogen receptor is linked to female reproductive cancers that are driven by chronic estrogen stimulation suggests that future research is necessary to address the molecular determinants encoded within GPER and/or its associated chaperone proteins that define its unique endocytic sorting mechanism.
Supplementary Material
Acknowledgment
We thank Hilary Magruder for proofreading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA119165-01A2 (to E. J. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- 7TMRs
- seven-transmembrane receptors
- GPR30/GPER
- G-protein-coupled estrogen receptors
- GPCR
- G-protein-coupled receptor
- TGN
- trans-Golgi network
- β1AR
- β1-adrenergic receptor
- CXCR4
- C-X-C chemokine receptor type 4
- PAR1
- Proteinase-activated receptor 1
- EEA-1
- early endosome antigen 1
- LAMP-1
- lysosomal-associated membrane protein 1
- ER
- endoplasmic reticulum
- SDF-1
- stromal cell-derived factor-1
- E2
- 17β-estradiol
- AP
- adaptor protein
- BFA
- brefeldin A
- HB-EGF
- heparin-bound epidermal growth factor
- MMP-3
- matrix metalloproteinase-3.
REFERENCES
- 1. Fredriksson R., Lagerström M. C., Lundin L. G., Schiöth H. B. (2003) Mol. Pharmacol. 63, 1256–1272 [DOI] [PubMed] [Google Scholar]
- 2. Marchese A., Chen C., Kim Y. M., Benovic J. L. (2003) Trends Biochem. Sci. 28, 369–376 [DOI] [PubMed] [Google Scholar]
- 3. Marchese A., Paing M. M., Temple B. R., Trejo J. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 601–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lamb M. E., De Weerd W. F., Leeb-Lundberg L. M. (2001) Biochem. J. 355, 741–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lavezzari G., Roche K. W. (2007) Neuropharmacology 52, 100–107 [DOI] [PubMed] [Google Scholar]
- 6. Hong Y. H., Kim J. Y., Lee J. H., Chae H. G., Jang S. S., Jeon J. H., Kim C. H., Kim J., Kim S. J. (2009) J. Neurochem. 111, 61–71 [DOI] [PubMed] [Google Scholar]
- 7. Scarselli M., Donaldson J. G. (2009) J. Biol. Chem. 284, 3577–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jean-Alphonse F., Hanyaloglu A. C. (2011) Mol. Cell. Endocrinol. 331, 205–214 [DOI] [PubMed] [Google Scholar]
- 9. Innamorati G., Sadeghi H., Birnbaumer M. (1999) J. Recept. Signal. Transduct Res. 19, 315–326 [DOI] [PubMed] [Google Scholar]
- 10. Innamorati G., Le Gouill C., Balamotis M., Birnbaumer M. (2001) J. Biol. Chem. 276, 13096–13103 [DOI] [PubMed] [Google Scholar]
- 11. Bremnes T., Paasche J. D., Mehlum A., Sandberg C., Bremnes B., Attramadal H. (2000) J. Biol. Chem. 275, 17596–17604 [DOI] [PubMed] [Google Scholar]
- 12. Oakley R. H., Laporte S. A., Holt J. A., Barak L. S., Caron M. G. (1999) J. Biol. Chem. 274, 32248–32257 [DOI] [PubMed] [Google Scholar]
- 13. Oakley R. H., Laporte S. A., Holt J. A., Barak L. S., Caron M. G. (2001) J. Biol. Chem. 276, 19452–19460 [DOI] [PubMed] [Google Scholar]
- 14. Tulipano G., Stumm R., Pfeiffer M., Kreienkamp H. J., Höllt V., Schulz S. (2004) J. Biol. Chem. 279, 21374–21382 [DOI] [PubMed] [Google Scholar]
- 15. Bouley R., Lin H. Y., Raychowdhury M. K., Marshansky V., Brown D., Ausiello D. A. (2005) Am. J. Physiol. Cell Physiol. 288, C1390–1401 [DOI] [PubMed] [Google Scholar]
- 16. Filardo E. J., Quinn J. A., Bland K. I., Frackelton A. R., Jr. (2000) Mol. Endocrinol. 14, 1649–1660 [DOI] [PubMed] [Google Scholar]
- 17. Filardo E. J., Quinn J. A., Frackelton A. R., Jr., Bland K. I. (2002) Mol. Endocrinol. 16, 70–84 [DOI] [PubMed] [Google Scholar]
- 18. Filardo E. J., Graeber C. T., Quinn J. A., Resnick M. B., Giri D., DeLellis R. A., Steinhoff M. M., Sabo E. (2006) Clin. Cancer Res. 12, 6359–6366 [DOI] [PubMed] [Google Scholar]
- 19. Quinn J. A., Graeber C. T., Frackelton A. R., Jr., Kim M., Schwarzbauer J. E., Filardo E. J. (2009) Mol. Endocrinol. 23, 1052–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thomas P., Pang Y., Filardo E. J., Dong J. (2005) Endocrinology 146, 624–632 [DOI] [PubMed] [Google Scholar]
- 21. Kanda N., Watanabe S. (2003) J. Invest. Dermatol. 121, 1500–1509 [DOI] [PubMed] [Google Scholar]
- 22. Filardo E. J., Quinn J., Pang Y., Graeber C., Shaw S., Dong J., Thomas P. (2007) Endocrinology 148, 3236–3245 [DOI] [PubMed] [Google Scholar]
- 23. Otto C., Rohde-Schulz B., Schwarz G., Fuchs I., Klewer M., Brittain D., Langer G., Bader B., Prelle K., Nubbemeyer R., Fritzemeier K. H. (2008) Endocrinology 149, 4846–4856 [DOI] [PubMed] [Google Scholar]
- 24. Levin E. R. (2009) Endocrinology 150, 1563–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kang L., Zhang X., Xie Y., Tu Y., Wang D., Liu Z., Wang Z. Y. (2010) Mol. Endocrinol. 24, 709–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Revankar C. M., Cimino D. F., Sklar L. A., Arterburn J. B., Prossnitz E. R. (2005) Science 307, 1625–1630 [DOI] [PubMed] [Google Scholar]
- 27. Smith H. O., Leslie K. K., Singh M., Qualls C. R., Revankar C. M., Joste N. E., Prossnitz E. R. (2007) Am. J. Obstet. Gynecol. 196, 386.e1–9; discussion 386.e9–11 [DOI] [PubMed] [Google Scholar]
- 28. Smith H. O., Arias-Pulido H., Kuo D. Y., Howard T., Qualls C. R., Lee S. J., Verschraegen C. F., Hathaway H. J., Joste N. E., Prossnitz E. R. (2009) Gynecol. Oncol. 114, 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Funakoshi T., Yanai A., Shinoda K., Kawano M. M., Mizukami Y. (2006) Biochem. Biophys. Res. Commun. 346, 904–910 [DOI] [PubMed] [Google Scholar]
- 30. Cheng S. B., Graeber C. T., Quinn J. A., Filardo E. J. (2011) Steroids, in press [DOI] [PubMed] [Google Scholar]
- 31. Kim Y. M., Benovic J. L. (2002) J. Biol. Chem. 277, 30760–30768 [DOI] [PubMed] [Google Scholar]
- 32. Benovic J. L. (2000) Regulation of G-Protein-Coupled Receptor Function and Expression, pp. 4–13, Wiley-Liss, Inc., New York [Google Scholar]
- 33. Tarasova N. I., Stauber R. H., Michejda C. J. (1998) J. Biol. Chem. 273, 15883–15886 [DOI] [PubMed] [Google Scholar]
- 34. Zhang Y., Foudi A., Geay J. F., Berthebaud M., Buet D., Jarrier P., Jalil A., Vainchenker W., Louache F. (2004) Stem Cells 22, 1015–1029 [DOI] [PubMed] [Google Scholar]
- 35. Connolly C. N., Uren J. M., Thomas P., Gorrie G. H., Gibson A., Smart T. G., Moss S. J. (1999) Mol. Cell Neurosci. 13, 259–271 [DOI] [PubMed] [Google Scholar]
- 36. Seifert R., Wenzel-Seifert K. (2002) Naunyn. Schmiedebergs Arch. Pharmacol. 366, 381–416 [DOI] [PubMed] [Google Scholar]
- 37. Leterrier C., Bonnard D., Carrel D., Rossier J., Lenkei Z. (2004) J. Biol. Chem. 279, 36013–36021 [DOI] [PubMed] [Google Scholar]
- 38. Paing M. M., Johnston C. A., Siderovski D. P., Trejo J. (2006) Mol. Cell. Biol. 26, 3231–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McDonald N. A., Henstridge C. M., Connolly C. N., Irving A. J. (2007) Mol. Pharmacol. 71, 976–984 [DOI] [PubMed] [Google Scholar]
- 40. Mohammad S., Baldini G., Granell S., Narducci P., Martelli A. M., Baldini G. (2007) J. Biol. Chem. 282, 4963–4974 [DOI] [PubMed] [Google Scholar]
- 41. Signoret N., Oldridge J., Pelchen-Matthews A., Klasse P. J., Tran T., Brass L. F., Rosenkilde M. M., Schwartz T. W., Holmes W., Dallas W., Luther M. A., Wells T. N., Hoxie J. A., Marsh M. (1997) J. Cell Biol. 139, 651–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Förster R., Kremmer E., Schubel A., Breitfeld D., Kleinschmidt A., Nerl C., Bernhardt G., Lipp M. (1998) J. Immunol. 160, 1522–1531 [PubMed] [Google Scholar]
- 43. Liang W., Curran P. K., Hoang Q., Moreland R. T., Fishman P. H. (2004) J. Cell Sci. 117, 723–734 [DOI] [PubMed] [Google Scholar]
- 44. Lok C. N., Loh T. T. (1998) Biol. Signals Recept. 7, 157–178 [DOI] [PubMed] [Google Scholar]
- 45. Gardner L. A., Delos Santos N. M., Matta S. G., Whitt M. A., Bahouth S. W. (2004) J. Biol. Chem. 279, 21135–21143 [DOI] [PubMed] [Google Scholar]
- 46. Lelouvier B., Tamagno G., Kaindl A. M., Roland A., Lelievre V., Le Verche V., Loudes C., Gressens P., Faivre-Baumann A., Lenkei Z., Dournaud P. (2008) J. Neurosci. 28, 4336–4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Trischler M., Stoorvogel W., Ullrich O. (1999) J. Cell Sci. 112, 4773–4783 [DOI] [PubMed] [Google Scholar]
- 48. Seachrist J. L., Ferguson S. S. (2003) Life Sci. 74, 225–235 [DOI] [PubMed] [Google Scholar]
- 49. Liang W., Fishman P. H. (2004) J. Biol. Chem. 279, 46882–46889 [DOI] [PubMed] [Google Scholar]
- 50. Sandén C., Broselid S., Cornmark L., Andersson K., Daszkiewicz-Nilsson J., Mårtensson U., Olde B., Leeb-Lundberg F. L. (2011) Mol. Pharmacol. 79, 400–410 [DOI] [PubMed] [Google Scholar]
- 51. Parent J. L., Labrecque P., Driss Rochdi M., Benovic J. L. (2001) J. Biol. Chem. 276, 7079–7085 [DOI] [PubMed] [Google Scholar]
- 52. Fan G. H., Yang W., Wang X. J., Qian Q., Richmond A. (2001) Biochemistry 40, 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vivacqua A., Bonofiglio D., Recchia A. G., Musti A. M., Picard D., Andò S., Maggiolini M. (2006) Mol. Endocrinol. 20, 631–646 [DOI] [PubMed] [Google Scholar]
- 54. Albanito L., Madeo A., Lappano R., Vivacqua A., Rago V., Carpino A., Oprea T. I., Prossnitz E. R., Musti A. M., Andò S., Maggiolini M. (2007) Cancer Res. 67, 1859–1866 [DOI] [PubMed] [Google Scholar]
- 55. Filardo E. J., Quinn J. A., Graeber C. T. (2003) in Identities of Membrane Steroid Receptor (Watson C. S. ed) pp. 139–146, New York, Kluwer Press [Google Scholar]
- 56. Tarasova N. I., Stauber R. H., Choi J. K., Hudson E. A., Czerwinski G., Miller J. L., Pavlakis G. N., Michejda C. J., Wank S. A. (1997) J. Biol. Chem. 272, 14817–14824 [DOI] [PubMed] [Google Scholar]
- 57. Xu H., Qin S., Carrasco G. A., Dai Y., Filardo E. J., Prossnitz E. R., Battaglia G., Doncarlos L. L., Muma N. A. (2009) Neuroscience 158, 1599–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Csaba Z., Lelouvier B., Viollet C., El Ghouzzi V., Toyama K., Videau C., Bernard V., Dournaud P. (2007) Traffic 8, 820–834 [DOI] [PubMed] [Google Scholar]
- 59. Römisch K. (1999) J. Cell Sci. 112, 4185–4191 [DOI] [PubMed] [Google Scholar]
- 60. Cross B. C., High S. (2009) J. Cell Sci. 122, 1768–1777 [DOI] [PubMed] [Google Scholar]
- 61. Milojevic T., Reiterer V., Stefan E., Korkhov V. M., Dorostkar M. M., Ducza E., Ogris E., Boehm S., Freissmuth M., Nanoff C. (2006) Mol. Pharmacol. 69, 1083–1094 [DOI] [PubMed] [Google Scholar]
- 62. Petaja-Repo U. E., Hogue M., Laperriere A., Bhalla S., Walker P., Bouvier M. (2001) J. Biol. Chem. 276, 4416–4423 [DOI] [PubMed] [Google Scholar]
- 63. Mori S., Tanaka K., Omura S., Saito Y. (1995) J. Biol. Chem. 270, 29447–29452 [DOI] [PubMed] [Google Scholar]
- 64. Lipkowitz S. (2003) Breast Cancer Res. 5, 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.