Abstract
Primary cilia are found on many epithelial cell types, including renal tubular epithelial cells, where they participate in flow sensing. Disruption of cilia function has been linked to the pathogenesis of polycystic kidney disease. We demonstrated previously that the exocyst, a highly conserved eight-protein membrane trafficking complex, localizes to primary cilia of renal tubular epithelial cells, is required for ciliogenesis, biochemically and genetically interacts with polycystin-2 (the protein product of the polycystic kidney disease 2 gene), and, when disrupted, results in MAPK pathway activation both in vitro and in vivo. The small GTPase Cdc42 is a candidate for regulation of the exocyst at the primary cilium. Here, we demonstrate that Cdc42 biochemically interacts with Sec10, a crucial component of the exocyst complex, and that Cdc42 colocalizes with Sec10 at the primary cilium. Expression of dominant negative Cdc42 and shRNA-mediated knockdown of both Cdc42 and Tuba, a Cdc42 guanine nucleotide exchange factor, inhibit ciliogenesis in Madin-Darby canine kidney cells. Furthermore, exocyst Sec8 and polycystin-2 no longer localize to primary cilia or the ciliary region following Cdc42 and Tuba knockdown. We also show that Sec10 directly interacts with Par6, a member of the Par complex that itself directly interacts with Cdc42. Finally, we show that Cdc42 knockdown results in activation of the MAPK pathway, something observed in cells with dysfunctional primary cilia. These data support a model in which Cdc42 localizes the exocyst to the primary cilium, whereupon the exocyst then targets and docks vesicles carrying proteins necessary for ciliogenesis.
Keywords: Cdc42, Centriole, Kidney, MAP Kinases (MAPKs), Protein Targeting, Cilia, Exocyst
Introduction
Cilia are thin, rod-like organelles found on the surface of many eukaryotic cells, with complex functions in signaling, cell differentiation, and growth control. Cilia extend outward from the basal body, a cellular organelle related to the centriole. In kidney cells, a single primary cilium projects from the basal body, is non-motile, and exhibits an axoneme microtubule pattern of 9 + 0. This is in contrast to motile cilia that exhibit a typical 9 + 2 axoneme microtubule pattern of organization. In epithelia containing numerous motile cilia, cilia have been observed to have a propulsive function (1), whereas primary cilia are thought to have a mechanosensory function, with calcium acting as an intracellular second messenger (2).
In the mammalian kidney, primary cilia have been observed on cells in the parietal layer of Bowman's capsule, the proximal tubule, the distal tubule, and in the principal but not intercalated cells of the collecting duct (3). Importantly, the primary cilium of the kidney has been implicated in the pathogenesis of polycystic kidney disease (PKD)2. Multiple gene products that, when mutated, result in the development of PKD, including polycystin-1 and polycystin-2, have been localized to and are crucial for the function of renal primary cilia (as reviewed in Ref 2). What is not yet clear, however, is how membrane proteins such as the polycystins are targeted and delivered to cilia in these cells.
We previously showed that the exocyst complex localizes to the primary cilium (4), is essential for ciliogenesis (5), and genetically and biochemically interacts with polycystin-2 (6). The exocyst is a highly conserved 750-kDa complex comprised of Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, and Exo84 that was originally identified as regulating polarized exocytosis in Saccharomyces cerevisiae (7). Mammalian homologs of all eight exocyst proteins have been identified (8). Sec10 is a central component of the eight-protein exocyst complex, and knockdown of Sec10 but not Sec8 or Exo70 prevented ciliogenesis, whereas overexpression of Sec10 increased ciliogenesis in Madin-Darby canine kidney (MDCK) cells (5). Sec10 overexpression also resulted in an increase in cyst and tubule morphogenesis when MDCK cells were grown in a collagen matrix to the cyst stage and induced to tubulate with hepatocyte growth factor (9). Finally, we recently showed that Sec10 overexpression both protects MDCK cells from and enhances recovery following hydrogen peroxide-induced injury (10). On the basis of these data, we hypothesize that the exocyst plays a central role in the regulation of ciliary protein trafficking and ciliogenesis, although the molecular interactions that regulate the exocyst in ciliary function remain undiscovered.
A possible mechanism to target the exocyst to primary cilia is the Par complex, which includes the small GTPase Cdc42. We previously found that the exocyst coimmunoprecipitated and colocalized with Par3 (5), a main component of the Par complex that consists of Par3, Par6, atypical protein kinase C, and Cdc42 (11, 12). In addition to their studied function at cell-cell contacts, the Par complex has been immunolocalized to primary cilia and has been shown to be necessary for ciliogenesis (13, 14). It is known that the exocyst is regulated by multiple Rho and Rab family GTPases (reviewed in 15), which, like the exocyst, play central roles in cell polarization, morphogenesis, membrane trafficking, cell growth, and development (16, 17). This includes studies in yeast that revealed that Cdc42 regulated polarized exocytosis via interactions with the exocyst (18). Using inducible MDCK cell lines that express constitutively active or dominant negative forms of Cdc42 (19, 20), we established that Cdc42 is centrally involved in cystogenesis and tubulogenesis (21). Open questions remain as to if and how Cdc42 might participate in ciliogenesis and cooperate with the exocyst in ciliary membrane trafficking.
Here we show that Cdc42 colocalizes and interacts with exocyst Sec10, and that Cdc42 is necessary for primary ciliogenesis in that Cdc42 dominant negative expression, Cdc42 shRNA knockdown, and Tuba, a GEF for Cdc42, shRNA knockdown all result in inhibition of ciliogenesis. Exocyst Sec8 and polycystin-2 no longer localize to the primary cilium or the ciliary region following Cdc42 and Tuba knockdown. Sec10 directly binds to Par6, as others have shown that Cdc42 also does (22, 23), and knockdown of Sec10 (6) and Cdc42 increase MAPK activation. Thus, we identify Cdc42 GTPase as an upstream regulator of exocyst-mediated ciliogenesis.
MATERIALS AND METHODS
Cell Culture
Low passage type II MDCK cells were obtained from Dr. K. Mostov (University of California San Francisco, San Francisco, CA) and used between passages 3–10. These cells were originally cloned by Daniel Louvard at The European Molecular Biology Laboratory (EMBL) and came to Keith Mostov via Karl Matlin. The dominant negative form of Cdc42-myc was made using a well described single amino acid substitution (Cdc42N17) (19, 20). The tetracycline-repressible stable MDCK cell line expressing dominant negative Cdc42 was a gift from W. James Nelson and Tzuu-Shuh Jou and was used as we described previously (21). Cells were grown in modified Eagle's minimal essential medium containing Eagle's balanced salt solution and glutamine supplemented with 5% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin on plastic culture dishes. Some cells were grown on Transwell 0.45 μm polycarbonate filter units (Corning Life Sciences, Lowell, MA). The culture medium was changed daily. Doxycycline was used at 20 ng/ml of medium to inhibit expression of mutant Cdc42 protein.
shRNA Oligos
The shRNA sequences for Cdc42 and Tuba were cloned into the p199 cloning vector and then into a lentiviral delivery system by the Macara Laboratory and were generously sent to us (24). These were used for infection of MDCK cells. The p199 vector encodes GFP, which allowed us to identify and separate the infected cells using fluorescence-activated cell sorting (FACS Vantage S.E., BD Biosciences) as we described previously (5).
GST Pull-down
Purified GST and GST-Sec10 was generated as described previously (6). Briefly, full-length human Sec10 cDNA was cloned in-frame into the plasmid pGEX-4T-1 (Amersham Biosciences), and transformed into the DE3 strain of Escherichia coli (Stratagene, La Jolla, CA). GST fusion protein expression was induced by adding isopropyl-1-thio-β-d-galactopyranoside to growing cultures, and recombinant proteins were purified with glutathione-Sepharose (Amersham Biosciences) following bacterial cell lysis. For pull-down experiments, lysates from MDCK cells overexpressing Cdc42 or proteins generated from full-length cDNAs with myc epitopes using a rabbit reticulocyte lysate in vitro transcription and translation kit (Promega, Madison, WI) were incubated overnight with Sec10-GST or GST only bound to glutathione-Sepharose. Pull-downs were washed extensively and resuspended in Laemmli buffer, and bound Cdc42, Par3, Par6, Sec8, p53, and GAPDH were assayed for by Western blotting.
Western Blot Analysis
Cells grown on plastic and on Transwell filters were harvested in radioimmune precipitation assay buffer (Sigma-Aldrich) containing a proteinase inhibitor mixture (Sigma-Aldrich), and the lysates were centrifuged at 14,000 rpm for 20 min at 4 °C. Supernatants were collected, and protein concentration was determined using the BCA protein assay (Thermo Scientific, Waltham, MA). The protein samples were separated on 4–12% SDS-PAGE gels (Invitrogen) and then transferred to an Immobilon membrane (Millipore Corp., Bedford, MA). The membranes were blocked with 5% nonfat dry milk in PBS containing 0.1% Tween 20 and incubated with primary antibodies overnight at 4 °C. Antibodies used in this study include monoclonal anti-myc (9E10, a gift from Dr. Keith Mostov), monoclonal anti-Sec8 (Assay Designs, Ann Arbor, MI), monoclonal anti-GAPDH (G8795, Sigma), monoclonal anti-CDC42 (B-8, Santa Cruz Biotechnology, Santa Cruz, CA), polyclonal anti-Tuba (a gift of Dr. Pietro De Camilli), polyclonal anti-polycystin-2 (a gift from the Johns Hopkins Research and Clinical Core Center), polyclonal anti-phospho-ERK1/2 (#9101, Cell Signaling Technology, Danvers, MA), and polyclonal anti-total ERK1(/2) (sc-94, Santa Cruz Biotechnology). After washing with PBS containing Tween 20, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Finally, the membranes were exposed to a Western blotting chemiluminescence reagent (Pierce) and developed on x-ray film. For measuring band intensity, the films were scanned and analyzed by Kodak 1D software (Kodak, Rochester, NY).
Immunofluorescence Staining
To investigate ciliogenesis by immunofluorescence, tetracycline-repressible stable MDCK cells expressing dominant negative Cdc42 were grown on Transwell filters in the presence and absence of doxycycline for 14 days. The cells were then fixed with 4% paraformaldehyde for 20 min on ice, permeabilized for 15 min at 37 °C with 0.025% saponin in phosphate-buffered saline containing 0.7% fish skin gelatin (PFS buffer), and incubated with primary antibody against acetylated α-tubulin (T5192, Sigma), which labels primary cilia, overnight at 4 °C and secondary antibody for 1 h at 37 °C. Additional antibodies used for immunofluorescence were monoclonal anti-Sec8 (Assay Designs) and polyclonal polycystin-2 (a gift from the Johns Hopkins Research and Clinical Core Center). The cells were counterstained with DAPI (for staining of cell nuclei) and mounted with mounting medium (Vectashield).
Electron Microscopy
Cells grown on Transwell filters for 14 days were fixed in a solution containing 2% glutaraldehyde, 0.8% paraformaldehyde, and 0.1 m cacodylate. The fixed cells were rinsed with 100 mm of cacodylate buffer, dehydrated through a graded ethanol series, washed with hexamethyldisilazane (Electron Microscopy Sciences), dried for 5 min at 60 °C, coated with platinum, and analyzed on a scanning electron microscopy machine (XL20 S.E., Phillips, Inc.).
Statistics
Student's t test was used to determine the significance of the degree of ciliogenesis impairment in tetracycline-repressible MDCK cells expressing dominant negative Cdc42, containing shRNA targeting Cdc42, and containing shRNA targeting Tuba.
RESULTS
Exocyst Sec10 Interacts and Colocalizes with Cdc42
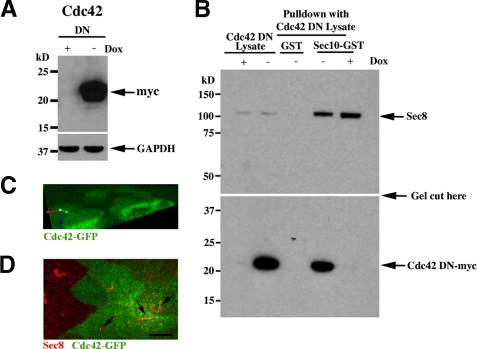
Our previous data showed that Cdc42 was centrally involved in cystogenesis and tubulogenesis and likely acted through the exocyst complex (21). Therefore, we proceeded to test for a biochemical interaction between the central exocyst component Sec10, which is essential for ciliogenesis (5), and Cdc42. We used GST pull-down assays, as relatively larger amounts of binding proteins can be obtained from the affinity column. Stable MDCK cell transfects inducibly expressing a dominant negative form of Cdc42 were grown in the presence or absence of doxycycline, a tetracycline derivative. In the absence of doxycycline, the Cdc42 dominant negative protein was expressed (Fig. 1A). A Sec10-GST fusion protein was purified on glutathione-Sepharose and used as an affinity matrix for the purification of specific binding proteins from tetracycline-repressible Cdc42-myc MDCK cell lysates. Dominant negative Cdc42-myc interacted with the exocyst in vitro, i.e. was in the pull-down fraction (Fig. 1B, bottom panel). As a positive control, the glutathione-Sepharose-immobilized Sec10-GST pulled down exocyst Sec8 from the lysate (Fig. 1B, top panel). In the presence of doxycycline (+ dox), no Cdc42 was expressed and no binding was detected with the anti-myc antibody. As an additional negative control, bead-immobilized GST alone was used, and no Cdc42 protein was detected in the pull-down fraction.
FIGURE 1.
Exocyst Sec10 Interacts and Colocalizes with Cdc42. A, Western blot analysis of MDCK cells expressing dominant negative (DN) Cdc42, with a myc epitope tag, were grown in the presence or absence of doxycycline (Dox), a tetracycline derivative. In the absence of doxycycline, dominant negative Cdc42 was expressed. B, Sec10-GST fusion protein, generated and purified from E. coli., was incubated with MDCK cell lysate from Cdc42-myc DN mutant cells. After extensive washing, bound proteins were analyzed by Western blot analysis. DN Cdc42-myc was found in the pull-down fraction and, therefore, is a Sec10 binding partner. As a positive control for the Cdc42 DN lysates, Sec8 is shown to bind to Sec10-GST (top panel). As a negative control, GST alone was used, and no Cdc42 mutant protein was pulled down, even in the absence of doxycycline. The top and bottom panels are from the same gel, and the exposure for the top and bottom halves of the gel was the same. C, MDCK cells stably expressing Cdc42-GFP cells were grown on Transwell filters, and apical staining was seen as described previously (25). D, the Cdc42-GFP MDCK cells were fixed and stained with antibody against endogenous exocyst Sec8. Colocalization of Sec8 and Cdc42 is seen (arrows). Scale bar = 5 μm..
MDCK cells expressing Cdc42-GFP were obtained from Keith Mostov and colleagues (25) and grown on Transwell filters. Similar to what they previously reported, apical expression of Cdc42-GFP was seen (Fig. 1C). Costaining with antibody to exocyst Sec8 showed colocalization at the primary cilium (Fig. 1D).
Dominant Negative Cdc42 Inhibits Ciliogenesis
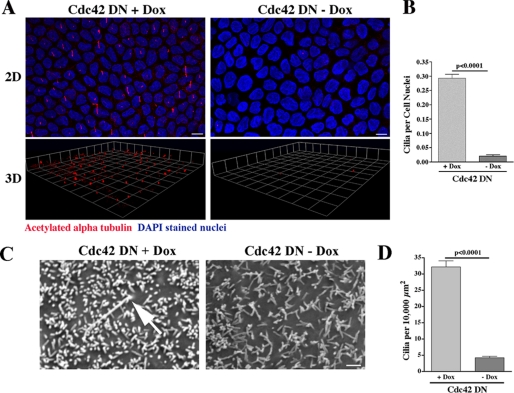
MDCK cells containing a stably transfected dominant negative Cdc42 were grown on Transwell filters in the presence or absence of doxycycline, which represses the expression of the dominant negative Cdc42 transgene. Using an antibody against acetylated α-tubulin, which identifies primary cilia, ciliogenesis was examined by immunofluorescence staining and confocal microscopy combined with evaluation of three-dimensional reconstruction of the stacked series. Ciliogenesis was virtually completely inhibited when the Cdc42 dominant negative protein was expressed following withdrawal of doxycycline (Fig. 2A). Quantification of ciliogenesis was performed using a ratio of cilia to cell nuclei, and the decreased ciliogenesis observed in the MDCK cells expressing Cdc42 dominant negative protein (minus doxycycline) was highly significant (p < 0.0001, Fig. 2B).
FIGURE 2.
Dominant Negative Expression of Cdc42 Inhibits Ciliogenesis. A, MDCK cells expressing dominant negative (DN) Cdc42 in the presence and absence of doxycycline were grown on Transwell filters for 14 days. Using an antibody against acetylated α-tubulin (red), ciliogenesis was examined by confocal microscopy combined with three-dimensional (3D) reconstruction of the stacked series. Ciliogenesis was virtually completely inhibited when Cdc42 DN protein was expressed (in the absence of doxycycline (Dox)). DAPI (blue) is a nuclear stain. DAPI staining is at a different level in the cell than staining for acetylated α-tubulin but is included in the merged figure to delineate individual cells and allow for statistical analysis. Scale bar = 5 μm. B, quantification of ciliogenesis was performed using a ratio of cilia to cell nuclei. Significantly fewer cilia were seen in the MDCK cells expressing Cdc42 DN protein (-Dox). C, Cdc42 DN cells were grown on Transwell filters in the presence (+) and absence (-) of doxycycline for 14 days. The cells were fixed in glutaraldehyde, and SE microscopy was performed (Phillips XL20). Confirming the results in A, primary cilia were rarely seen when Cdc42 DN protein was expressed (-Dox). Scale bar = 1.0 μm. D, quantification of ciliogenesis was performed by counting the number of cilia per surface area, as individual cells could not be identified. Significantly fewer cilia were seen in the MDCK cells expressing Cdc42 DN protein (-Dox)..
To determine the Cdc42 requirement for ciliogenesis by another method, we again grew Cdc42 dominant negative cells on Transwell filters in the presence and absence of doxycycline and then analyzed the fixed cells by scanning electron (SE) microscopy. Microvilli were readily apparent in the presence of dominant negative Cdc42 expression. Primary cilia were rarely present when the Cdc42 dominant negative protein was expressed (minus doxycycline) but were counted in levels comparable with normal MDCK cells (5) when dominant negative Cdc42 expression was repressed by doxycycline (Fig. 2C). Cilia were counted in identical cell surface areas in multiple microscopy images, and when compared between cells grown with and without doxycycline, the results were highly statistically significant and similar to the immunofluorescence results (p < 0.0001, Fig. 2D).
shRNA-mediated Cdc42 and Tuba Knockdown Inhibits Ciliogenesis
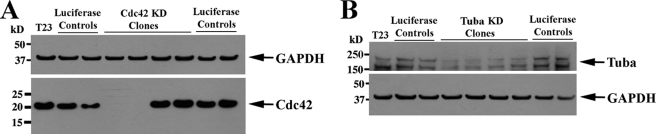
The dominant negative mutant of Cdc42 is thought to work by sequestering the upstream Rho guanine nucleotide exchange factors (GEFs). However, there are more than 70 mammalian GEFs, many of which are known to regulate multiple Rho GTPases (26–28). Thus, overexpression of dominant negative Cdc42 can block endogenous Cdc42 activity while at the same time may impact on multiple GEF functions and affect the activities of multiple Rho GTPases (24, 28–30). Therefore, we decided to perform a more definitive experiment and use shRNA to inhibit expression of Cdc42 and Tuba, a GEF of Cdc42 that was recently shown to be required for polarized spindle orientation during epithelial cyst formation (24). To generate stable cell lines with knockdown of Cdc42 and Tuba, we used a lentiviral vector delivery system containing shRNA sequences targeting Cdc42 and Tuba that had been designed and verified in MDCK cells by Qin et al. (24). The vectors encoding Cdc42 and Tuba shRNA also encoded GFP, which allowed us to identify and clone single infected cells using FACS. By Western blot analysis, there was no detectable Cdc42 protein following shRNA-mediated knockdown (Fig. 3A). Tuba was also significantly knocked down at the protein level but was still present at detectable levels by Western blotting (Fig. 3B).
FIGURE 3.
Cdc42 and Tuba were significantly knocked down using shRNA. Lentivirus-encoding shRNA targeting Cdc42 (A) and Tuba (B) were used to infect MDCK cells (24). The lentiviral construct contained a GFP, which allowed us to use FACS to generate single cell clones. Stable knockdown cell lines were then grown, and Western blotting was performed to determine efficacy of knockdown. By Western blotting, there was complete knockdown of Cdc42 (A) and 70–80% knockdown of Tuba (B). Western blotting of GAPDH is shown to demonstrate equal protein loading in the gel lanes.
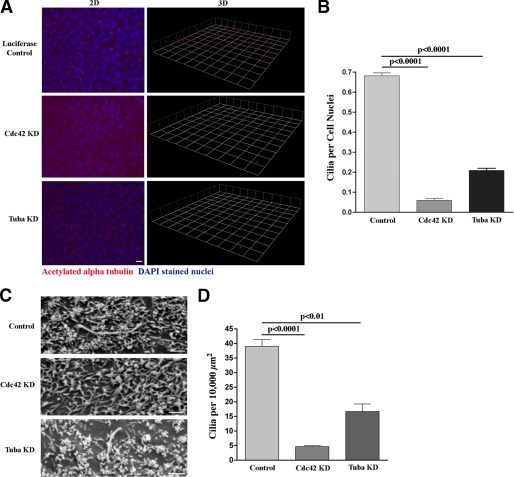
Using MDCK cells with stable knockdown of Cdc42, we found an almost complete absence of primary cilia by both immunofluorescence (p < 0.0001, Fig. 4, A and B) and SE (p < 0.0001, Fig. 4, C and D). Using the MDCK cells with stable knockdown of Tuba, we also found a highly significant decrease in ciliogenesis, although somewhat less pronounced than that following knockdown of Cdc42 (p < 0.0001 by immunofluorescence, Fig. 4, A and B, and p < 0.01 by SE, Fig. 4, C and D).
FIGURE 4.
Knockdown of Cdc42 and Tuba inhibit ciliogenesis. A, MDCK cells containing stable shRNA-mediated knockdown of Cdc42 and Tuba were grown on Transwell filters for 14 days. Using an antibody against acetylated α-tubulin (red), ciliogenesis was examined by confocal microscopy combined with three-dimensional (3D) reconstruction of the stacked series. Ciliogenesis was virtually completely inhibited when Cdc42 was knocked down and significantly inhibited following Tuba knockdown. Again, please note that DAPI staining (blue) is at a different level in the cell than staining for acetylated α-tubulin but is included in the merged figure to delineate individual cells and allow for statistical analysis. Scale bar = 5 μm. B, quantification of ciliogenesis was performed using a ratio of cilia to cell nuclei. Significantly fewer cilia were seen in the MDCK cells following Cdc42 and Tuba knockdown. C, the same Cdc42 and Tuba knockdown MDCK cells were grown on Transwell filters for 14 days, fixed in glutaraldehyde, and then SE microscopy was performed (Phillips XL20). Confirming the results in (A), primary cilia were almost never seen following Cdc42 knockdown and were rarely seen following Tuba knockdown. Scale bar = 1.0 μm. D, quantification of ciliogenesis was again performed by counting the number of cilia per surface area, as individual cells could not be identified. Significantly fewer cilia were seen in the MDCK cells following Cdc42 and Tuba knockdown..
We showed previously that the exocyst localized to the primary cilium in MDCK cells (5). In the Cdc42 and Tuba knockdown MDCK cells, the exocyst (detected by monitoring Sec8, which serves as a marker for the holo complex (31)) no longer localized at the primary cilium or ciliary region (p < 0.001, supplemental Fig. 1, A and B). Following Cdc42 and Tuba knockdown, polycystin-2 also no longer localized at the primary cilium or ciliary region (supplemental Fig. 2).
The Exocyst Directly Interacts with Par6
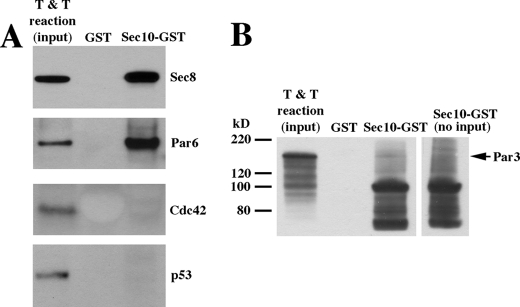
We had shown previously that several exocyst components coimmunoprecipitated with Par3 (5), a member of the Par complex along with Cdc42, Par6, and atypical protein kinase C. We wanted to determine whether the biochemical interaction between Sec10 and Cdc42 was direct or possibly occurred following exocyst binding to another Par complex component. Using in vitro transcription and translation of Cdc42, Par3, and Par6 as inputs for GST pull-downs, we found that Par6 directly bound Sec10 (Fig. 5A), whereas Cdc42 and Par3 did not directly bind Sec10 (Fig. 5, A and B). We used in vitro transcription and translation of Sec8 and p53 as positive and negative controls, respectively (Fig. 5A). Cdc42 has been shown to bind directly to Par6 (22, 23), suggesting that Par6 acts as a scaffolding to bring the exocyst and Cdc42 into close proximity at the primary cilium.
FIGURE 5.
The Exocyst Directly Interacts with Par6. A, GST pull-downs using in vitro transcription and translation reactions (T & T) as input revealed that Par6 directly bound Sec10, whereas Cdc42 did not. We had previously shown by coimmunoprecipitation that the exocyst interacted with the Par complex (5), but we had not yet identified which member of the Par complex was directly binding to the exocyst. Sec10-GST pull-down of Sec8 is shown as a positive control, and p53, which is not known to bind the exocyst, is shown as a negative control. B, using in vitro transcription and translation reaction as input for a GST pull-down, the Par3-Sec10 interaction we previously showed by coimmunoprecipitation (5) is not detectable, indicating that the Par3-Sec10 interaction is indirect. The GST-Sec10 protein, at ∼100 kDa, gives some background on Western blot analyses, so we have included a control lane (right lane) of GST-Sec10 with no input protein for comparison.
Cdc42 Knockdown Activates the MAPK Pathway
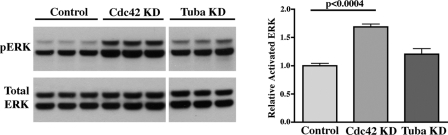
We recently showed that Sec10 knockdown, both in vitro and in vivo, led to phosphorylation (activation) of ERK, the final step in the MAPK pathway (6). Following shRNA-mediated knockdown of Cdc42, MAPK activation was also significantly increased (p < 0.0004, Fig. 6, A and B). MAPK activation in Tuba knockdown cells was also increased but did not reach statistical significance (p = 0.12, Fig. 6, A and B).
FIGURE 6.
Cdc42 Knockdown Activates the MAPK Pathway. A, control, Cdc42 knockdown, and Tuba knockdown MDCK cells were grown until confluence, and Western blotting was performed on cell lysate using antibodies against active (phosphorylated) ERK (pERK) and total ERK. Each sample was grown and analyzed in triplicate. All lanes shown were from the same blot with the same exposure. However, the Tuba lanes were separated from the other lanes, which is denoted by the space. B, measurement of the intensity of the bands showed that Cdc42 knockdown led to a significant increase in MAPK activation (p < 0.0004). MAPK activation in Tuba knockdown cells, although increased, did not reach statistical significance (p = 0.12).
DISCUSSION
In this study, we report four principal findings. First, we show, by two different methods, dominant negative expression and shRNA knockdown, that the small GTPase Cdc42 is necessary for primary ciliogenesis in renal tubule epithelial cells. Additionally, shRNA-mediated knockdown of Tuba, a Cdc42 GEF (24), also inhibits ciliogenesis in these cells. Second, we show that Cdc42 biochemically interacts with the exocyst complex, colocalizes with the exocyst at the primary cilium, and appears to be necessary for localization of the exocyst and exocyst-interacting proteins such as polycystin-2 at the primary cilium. Third, we demonstrate that Sec10 directly binds to Par6, a known Cdc42 interacting protein (22, 23) that may act as a bridging molecule for Cdc42 and the exocyst at the primary cilium. Finally, knockdown of Cdc42, like knockdown of Sec10 and polycystin-2 (6), results in MAPK pathway activation, as has been observed in autosomal dominant PKD mutant cells (32, 33). Therefore, we provide direct biochemical and mechanistic links between Cdc42 and key components with known roles in ciliogenesis.
Using MDCK cell lines that express dominant negative forms of Cdc42 (19, 20), we previously showed involvement of Cdc42, likely acting through the exocyst complex, in cystogenesis and tubulogenesis (21). In addition, a recent proteomics study identified Cdc42 as being present in the photoreceptor cilium of retinal pigmented epithelium (34). Here, we used shRNA to specifically knockdown Cdc42 and Tuba expression in MDCK cells. After selecting single clones with high percentages of knockdown, we discovered a similar inhibition of ciliogenesis. This is consistent with previous reports showing that Cdc42 regulates the exocyst in yeast (18) and is part of the Par complex (11, 12).
We showed previously that members of the exocyst, including Sec8, Sec10, and Exo70, coimmunoprecipitated with Par3 from MDCK cell lysate. A separate study also demonstrated that members of the exocyst complex coimmunoprecipitated with Par3 and atypical protein kinase C in rat cortical neurons (35). Here we show, using GST pull-downs of in vitro transcription and translation reactions, that the direct binding partner of Sec10 is actually Par6, another member of the Par complex. The Par complex is highly conserved in nature and, like the exocyst, has been associated with sites of cell-cell contact in polarized epithelial cells (as reviewed in Ref. 36). Recent findings have also localized Par3, Par6, and atypical protein kinase C to primary cilia. These proteins seem to be important in ciliogenesis, perhaps through their interactions with critical ciliary proteins such as KIF3A and Crumbs3a (13, 14). This is the first report to show that Cdc42 and Tuba, a Cdc42 GEF, also contribute to ciliogenesis in epithelial cells.
A key question then becomes how Cdc42 is activated during ciliogenesis, given the widespread localization over the apical surface observed by us and others (25). The model we favor is that one or more localized GEFs produce Cdc42-GTP activity at or near the primary cilium. Tuba, a Cdc42 GEF, was recently shown to be concentrated subapically, where the primary cilium forms (24). Knockdown of Tuba in our study inhibited ciliogenesis, although not to the same degree as knockdown of Cdc42. This may be due to incomplete knockdown of Tuba in our MDCK cells (Fig. 3B), or, alternatively, there may be one or more other GEFs that activate Cdc42 at the primary cilium. In fact, in a recent screen for modulators of ciliogenesis, 7784 therapeutically relevant genes across the human genome were tested using high-throughput siRNA (37). In that screen, intersectin 2, another Cdc42 GEF, was shown to be a positive regulator of ciliogenesis. Importantly, intersectin 2 was recently localized to the centrosome/basal body, which is at the base of the primary cilium, in MDCK cells (38). In the high-throughput siRNA screen by Kim et al., Cdc42 was listed as a rejected gene on the basis of “siRNA toxicity,” and the effect of Cdc42 knockdown on ciliogenesis was therefore not determined (37).
PKD2, encoding polycystin-2, is one of two genes which, when mutated, cause autosomal dominant polycystic kidney disease (2). We recently showed that the exocyst interacted biochemically and genetically with polycystin-2, and when Sec10 was knocked down during zebrafish development using antisense morpholinos, the embryos phenocopied zebrafish with polycystin-2 knockdown (6). Here we show that Sec8 and polycystin-2 no longer localize to the primary cilium or the ciliary region following Cdc42 and Tuba knockdown. The link between primary cilia and MAPK pathway signaling is thought to occur by calcium influx through polycystin-2, following bending of the primary cilia during normal urine fluid flow (39). Calcium acts as a second messenger to suppress growth in renal epithelial cells, and in cells with defective or absent cilia, such as Cdc42 knockdown cells, phosphorylated (active) ERK, the final step in the MAPK pathway, is consistently elevated. Similar to the reasons we proposed for Tuba knockdown not inhibiting ciliogenesis to the same degree as was seen with Cdc42 knockdown, there are several possibilities for why Tuba knockdown did not increase ERK activation. The first possibility is that there was incomplete knockdown of Tuba in our MDCK cells (Fig. 3B) and, had the Tuba knockdown been more robust, we would have seen an increase in phosphorylated ERK. Second, one or more other GEFs (such as intersectin 2 (37, 38)) could activate Cdc42 at the primary cilium, and we would also need to knock down the other GEFs to see an increase in phosphorylated ERK. Finally, we cannot rule out the possibility that although both Cdc42 and Tuba knockdown inhibit ciliogenesis, the two events may not be related. Interestingly, in one mouse model of polycystic kidney disease, prevention of ERK activation was shown to prevent abnormal cyst formation (40), whereas in other mouse models, although active ERK was increased, reversal did not prevent abnormal cystogenesis (41).
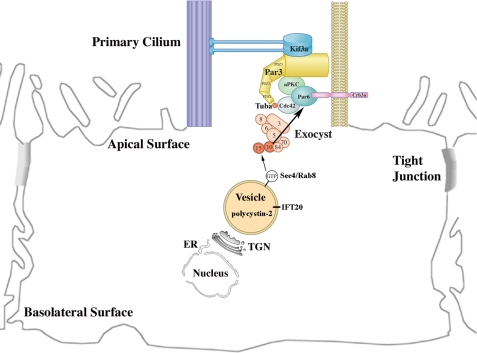
Together, these findings provide the basis for a model in which the exocyst complex is localized to the primary cilium by Cdc42, is stabilized at the primary cilium by binding to the Par complex through Par6, and then targets and docks vesicles carrying proteins necessary for ciliogenesis, such as polycystin-2 (Fig. 7). Given the importance of the primary cilium in many “ciliopathies,” including polycystic kidney disease, identifying the mechanisms of ciliary assembly governed by the exocyst could reveal novel therapeutic targets.
FIGURE 7.
Model for the Delivery of Ciliary Proteins. Our data support a model in which the exocyst complex is targeted to the primary cilium by Cdc42 and is then stabilized by binding to the Par complex via Par6. Once the exocyst complex is stabilized at the primary cilium, it then targets and docks vesicles carrying ciliary proteins, such as polycystin-2, by interacting with Rab8 found on the vesicles..
Supplementary Material
Acknowledgments
We thank the University of Pennsylvania Biomedical Imaging Core Facility of the Cancer Center for providing imaging services and the Johns Hopkins Research and Clinical Core Center for providing anti-polycystin-2 antibody.
This work was supported, in whole or in part, by National Institutes of Health Grants DK069909 (to J. H. L.) and K01DK087852 (to B. F.). This work was also supported by a Veterans Affairs Merit Award (to J. H. L.), a Satellite Healthcare Norman S. Coplon extramural research grant (to J. H. L.), and a University of Pennsylvania Translational Medicine Institute pilot grant (to J. H. L).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- PKD
- polycystic kidney disease
- MDCK
- Madin-Darby canine kidney
- SE
- scanning electron
- GEF
- guanine nucleotide exchange factor.
REFERENCES
- 1. Fawcett D. W., Porter K. R. (1954) J. Morphol. 94, 221–281 [Google Scholar]
- 2. Smyth B. J., Snyder R., Balkovetz D. F., Lipschutz J. H. (2003) Int. Rev. Cytol. 231, 51–89 [DOI] [PubMed] [Google Scholar]
- 3. Webber W. A., Lee J. (1975) Anat. Rec., 182, 339–343 [DOI] [PubMed] [Google Scholar]
- 4. Rogers K. K., Wilson P. D., Snyder R. W., Zhang X., Guo W., Burrow C. R., Lipschutz J. H. (2004) Biochem. Biophys. Res. Commun. 319, 138–143 [DOI] [PubMed] [Google Scholar]
- 5. Zuo X., Guo W., Lipschutz J. H. (2009) Mol. Biol. Cell 20, 2522–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fogelgren B., Lin S. Y., Zuo X., Jaffe K. M., Park K. M., Reichert R., Bell P. D., Burdine R. D., Lipschutz J. H. (2011) PLoS Genet. 7, 100/361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Novick P., Field C., Schekman R. (1980) Cell 21, 205–215 [DOI] [PubMed] [Google Scholar]
- 8. Hsu S. C., Ting A. E., Hazuka C. D., Davanger S., Kenny J. W., Kee Y., Scheller R. H. (1996) Neuron 17, 1209–1219 [DOI] [PubMed] [Google Scholar]
- 9. Lipschutz J. H., Guo W., O'Brien L. E., Nguyen Y. H., Novick P., Mostov K. E. (2000) Mol. Biol. Cell 11, 4259–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Park K. M., Fogelgren B., Zuo X., Kim J., Chung D. C., Lipschutz J. H. (2010) Am. J. Physiol. Renal Physiol. 298, F818–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Joberty G., Petersen C., Gao L., Macara I. G. (2000) Nat. Cell Biol. 2, 531–539 [DOI] [PubMed] [Google Scholar]
- 12. Lin D., Edwards A. S., Fawcett J. P., Mbamalu G., Scott J. D., Pawson T. (2000) Nat. Cell Biol. 2, 540–547 [DOI] [PubMed] [Google Scholar]
- 13. Sfakianos J., Togawa A., Maday S., Hull M., Pypaert M., Cantley L., Toomre D., Mellman I. J. (2007) J. Cell Biol. 179, 1133–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fan S., Hurd T. W., Liu C. J., Straight S. W., Weimbs T., Hurd E. A., Domino S. E., Margolis B. (2004) Curr. Biol. 14, 1451–1461 [DOI] [PubMed] [Google Scholar]
- 15. Lipschutz J. H., Mostov K. E. (2002) Curr. Biol. 12, R212-R214 [DOI] [PubMed] [Google Scholar]
- 16. Hall A. (1998) Science 279, 509–514 [DOI] [PubMed] [Google Scholar]
- 17. Van Aelst L., D'Souza-Schorey C. (1997) Genes Dev. 11, 2295–2322 [DOI] [PubMed] [Google Scholar]
- 18. Zhang X., Bi E., Novick P., Du L., Kozminski K. G., Lipschutz J. H., Guo W. J. (2001) J. Biol. Chem. 276, 46745–46750 [DOI] [PubMed] [Google Scholar]
- 19. Jou T. S., Nelson W. J. J. (1998) J. Cell Biol. 142, 85–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jou T. S., Schneeberger E. E., Nelson W. J. (1998) J. Cell Biol. 142, 101–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rogers K. K., Jou T. S., Guo W., Lipschutz J. H. (2003) Kidney Int. 63, 1632–1644 [DOI] [PubMed] [Google Scholar]
- 22. Atwood S. X., Chabu C., Penkert R. R., Doe C. Q., Prehoda K. E. J. (2007) J. Cell Sci. 120, 3200–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yamanaka T., Horikoshi Y., Suzuki A., Sugiyama Y., Kitamura K., Maniwa R., Nagai Y., Yamashita A., Hirose T., Ishikawa H., Ohno S. (2001) Genes Cells 6, 721–731 [DOI] [PubMed] [Google Scholar]
- 24. Qin Y., Meisen W. H., Hao Y., Macara I. G. J. (2010) J. Cell Biol. 189, 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martin-Belmonte F., Gassama A., Datta A., Yu W., Rescher U., Gerke V., Mostov K. (2007) Cell 128, 383–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bustelo X. R., Sauzeau V., Berenjeno I. M. (2007) BioEssays 29, 356–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Etienne-Manneville S., Hall A. (2002) Nature 420, 629–635 [DOI] [PubMed] [Google Scholar]
- 28. Melendez J., Grogg M., Zheng Y. J. (2010) J. Biol. Chem. 286, 2375–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schwartz M.A., Meredith J. E., Kiosses W. B. (1998) Oncogene 17, 625–629 [DOI] [PubMed] [Google Scholar]
- 30. Woo S., Gomez T. M. (2006) J. Neurosci. 26, 1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grindstaff K. K., Yeaman C., Anandasabapathy N., Hsu S. C., Rodriguez-Boulan E., Scheller R. H., Nelson W. J. (1998) Cell 93, 731–740 [DOI] [PubMed] [Google Scholar]
- 32. Yamaguchi T., Nagao S., Wallace D. P., Belibi F. A., Cowley B. D., Pelling J. C., Grantham J. J. (2003) Kidney Int. 63, 1983–1994 [DOI] [PubMed] [Google Scholar]
- 33. Yamaguchi T., Pelling J. C., Ramaswamy N. T., Eppler J. W., Wallace D. P., Nagao S., Rome L. A., Sullivan L. P., Grantham J. J. (2000) Kidney Int. 57, 1460–1471 [DOI] [PubMed] [Google Scholar]
- 34. Liu Q., Tan G., Levenkova N., Li T., Pugh E. N., Jr., Rux J. J., Speicher D. W., Pierce E. A. (2007) Mol. Cell. Proteomics 6, 1299–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lalli G. J. (2009) J. Cell Sci. 122, 1499–1506 [DOI] [PubMed] [Google Scholar]
- 36. Anderson J. M., Van Itallie C. M., Fanning A. S. (2004) Curr. Opin. Cell Biol. 16, 140–145 [DOI] [PubMed] [Google Scholar]
- 37. Kim J., Lee J. E., Heynen-Genel S., Suyama E., Ono K., Lee K., Ideker T., Aza-Blanc P., Gleeson J. G. (2010) Nature 464, 1048–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rodriguez-Fraticelli A. E., Vergarajauregui S., Eastburn D. J., Datta A., Alonso M. A., Mostov K., Martin-Belmonte F. J. (2010) J. Cell Biol. 189, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Torres V. E., Harris P. C. (2006) Nat. Clin. Pract. Nephrol. 2, 40–55; quiz 55 [DOI] [PubMed] [Google Scholar]
- 40. Omori S., Hida M., Fujita H., Takahashi H., Tanimura S., Kohno M., Awazu M. J. (2006) J. Am. Soc. Nephrol. 17, 1604–1614 [DOI] [PubMed] [Google Scholar]
- 41. Shibazaki S., Yu Z., Nishio S., Tian X., Thomson R. B., Mitobe M., Louvi A., Velazquez H., Ishibe S., Cantley L. G., Igarashi P., Somlo S. (2008) Hum. Mol. Genet. 17, 1505–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.