Abstract
A key component of excitation contraction (EC) coupling in skeletal muscle is the cytoplasmic linker (II-III loop) between the second and third transmembrane repeats of the α1S subunit of the dihydropyridine receptor (DHPR). The II-III loop has been previously examined in vitro using a linear II-III loop with unrestrained N- and C-terminal ends. To better reproduce the loop structure in its native environment (tethered to the DHPR transmembrane domains), we have joined the N and C termini using intein-mediated technology. Circular dichroism and NMR spectroscopy revealed a structural shift in the cyclized loop toward a protein with increased α-helical and β-strand structure in a region of the loop implicated in its in vitro function and also in a critical region for EC coupling. The affinity of binding of the II-III loop binding to the SPRY2 domain of the skeletal ryanodine receptor (RyR1) increased 4-fold, and its ability to activate RyR1 channels in lipid bilayers was enhanced 3-fold by cyclization. These functional changes were predicted consequences of the structural enhancement. We suggest that tethering the N and C termini stabilized secondary structural elements in the DHPR II-III loop and may reflect structural and dynamic characteristics of the loop that are inherent in EC coupling.
Keywords: Calcium Channels, Ion Channels, NMR, Protein-Protein Interactions, Skeletal Muscle, Intrinsically Unstructured Protein, Molecular Recognition, Protein Cyclization, Skeletal Dihydropyridine Receptor, Skeletal Ryanodine Receptor
Introduction
The release of Ca2+ ions from the sarcoplasmic reticulum of skeletal muscle fibers in response to depolarization of the surface membrane, excitation-contraction (EC)5 coupling, has three essential molecular components. These are the cytoplasmic linker between the second and third transmembrane repeats (the II-III loop) of the α1S subunit of the skeletal dihydropyridine receptor (skDHPR), the β1a subunit of the DHPR, and the skeletal muscle type 1 ryanodine receptor (RyR1). It is generally accepted that membrane depolarization leads to the movement of charged residues in the fourth membrane-spanning helix (S4 segment) of the α1S subunit and that this “charge movement” causes a conformational change in the II-III loop, which is physically transmitted to the RyR1 channel, to open the ion channel and allow Ca2+ ion flow (1). The role of the II-III loop in EC coupling has been extensively probed and confirmed over the past few decades using in vivo chimeric techniques (2–5), and the critical regions and residues involved in this process have been identified (3, 6). Physical interactions between the II-III loop and RyR1 are also seen in in vitro studies, which reveal an increase in the activity of RyR1 channels when they are exposed to the recombinant DHPR II-III loop (7–10). However, in vitro structure and function studies of the II-III loop have thus far been performed with loop constructs expressed as a linear protein with free or untethered N- and C- terminal ends. Although the ends of the untethered loop are in close proximity in the three-dimensional structure (11), the geometrical constraints are obviously very different from those experienced in the intact protein, where the N and C termini of the loop are “tethered” to the S6 helix of the second transmembrane repeat and the S1 helix of the third transmembrane repeat, respectively, both of which are inserted in the membrane of the transverse tubule extensions of the fiber surface.
To more closely mimic the tethered nature of the II-III loop N and C termini in the intact membrane-bound protein and the in vivo situation, we have used an intein-based method to link the N- and C- terminal ends of the loop. Intein-mediated protein splicing is a post-translational reaction in which a precursor protein undergoes self-catalyzed head-to-tail intramolecular rearrangement with the consequent exclusion of an internal protein domain and the formation of a mature intein protein (12, 13). Intein-mediated protein splicing has been used extensively in biotechnology in the purification of recombinant proteins, ligation of polypeptides, and the synthesis of protein microarrays, where it links the protein with a solid surface binding domain (14). Intein-mediated cyclization has also proved a useful tool for investigating the stability of native fold proteins (15).
In this study, we used the cyanobacterium Synechocystis sp. strain PCC6803 DnaB 429 amino acid intein protein to generate a split functional N- and C- terminal intein (16). The terminal inteins come together to form a functional protein splicing domain that was engineered to contain the DHPR II-III loop. We found that tethering the ends of the II-III loop increased α-helical and β-strand structure in particular regions of the loop involved in key protein-protein interactions and EC coupling. This structural change strikingly increased the affinity of the II-III loop for the SPRY2 domain and enhanced its ability to activate RyR1 channels in lipid bilayers.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of the Linear and Cyclic II-III Loop
The linear II-III loop was cloned, expressed, and purified as we have previously described (11). For the intein plasmid construction, the pNW1120 vector, which houses the Synechocystis sp. strain PCC6803 DnaB split intein gene, was used (kindly donated by Pavel Prosselkov from the Australian National University Research School of Chemistry). The vector carried nucleotides encoding a 9-amino acid residue linker (TRESGSIEF). The linker length was chosen to avoid conformation strain and interference with the structure of the cyclized protein (15, 17). The linker was used as the initiation point of intein-mediated protein cyclization via the serine and glutamate residues.
The vector was linearized using endonuclease digestion with 1 unit each of MluI (New England BioLabs) and EcoRI (New England BioLabs) at 37 °C for 1 h, in the presence of 1× NEB buffer 3 (New England BioLabs). The cleaved product was resolved on an agarose gel, visualized under UV light, immediately excised from the gel, and purified using the QIAQUICK gel extraction kit following the manufacturer's instructions (Qiagen).
As previously described, the expression vector (18) hosting the Oryctolagus cuniculus skDHPR II-III loop gene (9) was used as a template to amplify the skDHPR II-III loop cDNA and add an EcoRI site and His6 codons at the 5′-end and an MluI site at the 3′-end. The amplification was carried out with primers DHPRiHF (5′-GAGAATTCCACCATCACCATCACCATGAGGCGGAGAGCCTG-3′) and DHPRiR (5′-GTACGCGTCAGGACACGGACCTTATTG-3′) (GeneWorks) in a 20-μl reaction mix containing PCR buffer (Promega), 1.75 mm MgCl2, 0.2 mm dNTPs, 2 units of TaqDNA polymerase (Promega), 5 ng of DNA template, and 0.3 pmol of primers. PCR was carried out for 35 cycles in the PTC-200 DNA Engine (MJ Research). The 417-bp amplicon was subjected to agarose gel electrophoresis at 50 mA, excised from the gel, and purified with QIAQUICK gel extraction kit (Qiagen). After a 1-h digestion with MluI and EcoRI at 37 °C, in the presence of 1× NEB buffer 3 (New England BioLabs), the digested product was again purified by agarose gel electrophoresis as described above.
The purified fragment was ligated into the linearized pNW1120 vector in the presence of 1× ligation buffer (Promega) and 10 units of T4 DNA ligase (Promega). The ligation reaction was performed overnight at 4 °C, and the plasmid was transformed into Escherichia coli DH5α. The insertion and the orientation of the fragment into the vector were confirmed by MluI-EcoRI double digestion and ABI Prism DNA sequencing (Applied Biosystems). The pNW1120 vector containing the skeletal DHPR II-III loop gene (Fig. 1A) was then transformed into the E. coli BL21 for recombinant protein expression. The BL21 inoculation culture was incubated and agitated overnight in Luria Bertani (LB) broth supplemented with 100 μg/ml ampicillin at 30 °C and then transferred to a larger volume of LB broth supplemented with 100 μg/ml ampicilin, and agitation-incubation at 30 °C continued until A600 was between 0.8 and 1.0. Protein production was induced by rapidly increasing the incubation temperature to 42 °C, and incubation continued for another 4 h. Cultures were then harvested by centrifugation and stored at −20 °C. The protein was purified using a method similar to that described for the linear loop (11). For NMR samples, uniformly 15N-labeled and 13C/15N-labeled protein was produced by growing the expression strain in M9 salt media with 15NH4Cl and [13C]-glucose as the sole nitrogen and carbon source.
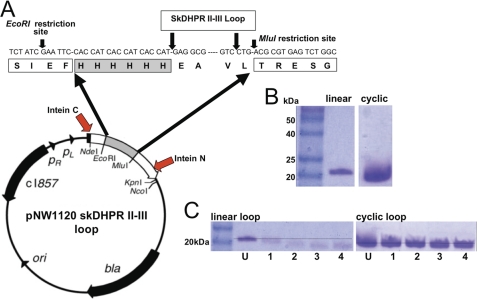
FIGURE 1.
Expression and characterization of the cyclic II-III loop. A, the pNW1120 skDHPR II-III loop expression vector with the His6 tag (gray box) and nine-residue linker (white box). The location of Intein N and Intein C in the vector is shown by the red arrows (modified from Ref. 17). B, Coomassie Blue-stained 12% BisTris-Cl polyacrylamide gels containing Invitrogen BenchMarkTM prestained protein ladders (Invitrogen) in the left lanes. The linear skDHPR II-III loop and the cyclic skDHPR II-III loop are in the right lanes. The dotted line passes through the 20-kDa marker. C, Coomassie Blue-stained 12% BisTris-Cl polyacrylamide gels of the linear (left gel) and cyclized (right gel) skDHPR II-III loop after a 4-h digestion with carboxypeptidase-Y. The first lane in the left gel shows the 20-kDa marker from the protein ladder. In each gel, lanes labeled U show the undigested protein, and lanes labeled 1–4 show aliquots taken after 1–4-h periods of digestion. The dotted lines indicate the position of the undigested protein.
Carboxypeptidase-Y Digestion
The purified protein was digested with carboxypeptidase-Y (Cp-Y) (Roche Applied Science) to confirm the cyclization. The sample was first dialyzed overnight against 100 mm sodium citrate buffer (BDH Chemicals, Australia), pH 6.5, at 4 °C and then incubated with Cp-Y (the Cp-Y/protein ratio was 1:100 (v/v)) at 25 °C for 4 h. Aliquots were taken at 1-h intervals and run on 12% BisTris-Cl denaturing PAGE at 200 V for ∼40 min.
Synthesis of Linker-His6 Tag Peptide
A 15-residue peptide (linker peptide) corresponding to the 9-residue linker and the His6 tag inserted upstream of the cyclized skDHPR II-III loop was synthesized by the Australian Cancer Research Foundation Biomolecular Resource Facility using an Applied Biosystems 430A protein synthesizer with purification using HPLC (Shimadzu) and mass spectroscopy (MALDI TOF/TOFTM model 4800, Applied Biosystems). Stock peptide solution (1 mm) was prepared in Milli-Q water (Millipore).
Nuclear Magnetic Resonance (NMR), Circular Dichroism (CD), and Spectrofluorimetry Experiments
All NMR experiments for the cyclic II-III loop were performed using methods previously employed for the linear II-III loop (11). The NMR sample was made up of 100 μm 15N/13C-labeled cyclic II-III loop in 50 mm phosphate, 200 mm KCl buffer at pH 6.5. DSS was added in one sample for internal reference for the chemical shift. The temperature was maintained at 5 °C. NMR data were recorded on a Bruker Avance 800-MHz spectrometer equipped with a cryoprobe. Spectra were processed using Topspin version 1.3 (Bruker) and analyzed with Sparky version 3.0 (T. D. Goddard and D. G. Koeller).
The linear and intein II-III loop protein fragments were diluted to ∼20 μm for CD measurements, and the pH values and solution conditions were adjusted to pH 6.5 and 10 mm PO42−, respectively. Spectra were recorded on an Applied Photophysics Chirascan spectrometer at 20 °C. A cell with a 0.1-cm path length was used for spectra recorded between 190 and 250 nm with a spectral bandwidth of 1 nm, a 0.5-nm step size at 0.5 s/point. Each spectrum was obtained by averaging several scans, and the protein CD spectra were corrected for buffer contributions. The temperature was controlled by a Melcor peltier temperature controller. Fluoresecence binding experiments and analysis of loop binding to the recombinant SPRY2 protein were carried out as described previously (11).
SR Vesicle Preparation and Single Channel Recording and Analysis
Back and leg muscle from New Zealand White rabbits was prepared as described (19–21). Artificial planar bilayers separating two baths (cis and trans) were formed, and SR vesicles were incorporated as described previously (20–22). In general, SR vesicles incorporated in such a manner that the cytoplasmic surface of the SR and RyRs faced the cis solution. The orientation was confirmed by characteristic changes in channel activity with changes in cis (cytoplasmic) [Ca2+]. For channel incorporation, the solutions were as follows: cis, 230 mm CsMS, 20 mm CsCl, 1 mm CaCl2, and 10 mm TES (pH 7.4); trans, 30 mm CsMS, 20 mm CsCl, 1 mm CaCl2, and 10 mm TES (pH 7.4). After channel incorporation, the trans [Cs+] was raised to 250 mm with the addition of 200 mm CsMS. The cis solution was altered by the addition of ∼4.5 mm BAPTA (1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid) (free [Ca2+] = 100 nm; confirmed using a Ca2+ electrode). Trans [Ca2+] was maintained at 1 mm under all conditions. Channels were identified as RyRs by their characteristic conductance and block by 20 μm ruthenium red at the end of the experiment. Voltage is expressed as cytoplasm relative to lumen. Channel currents were filtered at 1 kHz and sampled at 5 kHz (22). Voltage was changed between +40 and −40 mV every 30 s. Measurements were carried out at 23 ± 2 °C.
Single channel parameters, open probability (Po), mean open time (To), and mean closed times (Tc) were obtained using the Channel 2 program (developed by P. W. Gage and M. Smith, John Curtin School of Medical Research) (20–22). Open and closed times were collected into logged bins, and the square root of the relative frequency of events (P½) was plotted against the logarithm of the open (open circles) or closed times (filled circles) in milliseconds (23). Channel activity was assessed from 60 s of recording, 30 s at −40 mV and 30–90 s at +40 mV under each condition. Because all effects were independent of voltage (Table 1), data from the two potentials were pooled and included in calculations of average channel activity. The effect of the linear II-III loop, the cyclized II-III loop, and the linker proteins on native RyR1 channel activity was examined.
TABLE 1.
There is no voltage dependence in the effects of the linear or cyclic loop on RyR1 channel Po
Average relative Po at +40 and −40 mV (relative to activity prior to construct addition), in the presence of 10, 50, 100 and 200 nm concentrations of each construct (linear, cyclic, and linker) and after washout by perfusion of the cis chamber.
| Construct | Construct concentration |
Washout | |||
|---|---|---|---|---|---|
| 10 nm | 50 nm | 100 nm | 200 nm | ||
| Linear | |||||
| +40 mV | 1.75 ± 0.03 | 3.77 ± 0.24 | 0.34 ± 0.09 | 0.33 ± 0.07 | 1.01 ± 0.07 |
| −40 mV | 1.95 ± 0.30 | 3.65 ± 0.50 | 0.62 ± 0.04 | 0.56 ± 0.05 | 0.88 ± 0.04 |
| Cyclic | |||||
| +40 mV | 2.98 ± 0.56 | 5.73 ± 0.83 | 1.49 ± 0.26 | 1.07 ± 0.35 | 2.24 ± 0.16 |
| −40 mV | 2.94 ± 0.64 | 5.63 ± 1.56 | 1.65 ± 1.06 | 1.18 ± 0.67 | 2.10 ± 0.25 |
| Linker | |||||
| +40 mV | 0.80 ± 0.15 | 0.77 ± 0.13 | 0.63 ± 0.27 | 1.27 ± 0.49 | 0.90 ± 0.19 |
| −40 mV | 0.92 ± 0.11 | 0.94 ± 0.06 | 0.90 ± 0.14 | 0.84 ± 0.15 | 0.82 ± 0.09 |
Statistics
Average data are presented as mean ± S.E. The significance of differences was tested using Student's t test for paired or unpaired data or the non-parametric sign test, as appropriate. A p value of <0.05 was considered significant.
RESULTS
Gel Analysis of II-III Loop Proteins
SDS-polyacrylamide gels of the purified linear and cyclic skeletal DHPR II-III loop proteins are shown in Fig. 1B. The theoretical molecular mass for the linear skDHPR II-III loop is 14.1 kDa; however, it has a relatively slow electrophoretic mobility and appears to run at a high molecular mass as noted previously (9, 24). The inclusion of the His6 tag and the 9-residue linker in the cyclic protein (Fig. 1A) increases its theoretical molecular mass to ∼16 kDa. However, the change in conformation of the cyclized protein increases its electrophoretic mobility, as seen with other cyclized proteins (25–27), so that it appears to be smaller than the linear protein on the gel (Fig. 1B). The change in mobility is consistent with cyclization of the protein.
The cyclical nature of the intein loop was confirmed by exposure to Cp-Y. Because cyclic proteins lack both N and C termini, they should be insensitive to exopeptidase-catalyzed proteolysis (25), whereas linear proteins are susceptible to digestion. Fig. 1C shows a gradual reduction in size and intensity of the linear loop during a 4-h incubation, indicating that, as expected, the linear protein was susceptible to Cp-Y proteolysis. In marked contrast, the cyclic II-III loop maintained a similar intensity and position during the 4-h incubation period, indicating that it was not susceptible to digestion and implying that the C-terminal end was not accessible. The undigested linear protein did not show any change in density during the 4-h incubation (not shown).
Cyclizing the Skeletal DHPR II-III Loop Promotes Structural and Dynamic Change
The linear DHPR II-III loop is an intrinsically unstructured protein that consists of several nascent α-helical secondary structures and turns (11). The unstructured nature of the linear II-III loop is reflected in its CD profile, where the spectrum displays a distinct minimum at 198 nm and a slight dip at 222 nm (Fig. 2A). Upon cyclization of the II-III loop, however, a marked structural difference can be observed in the CD profile (Fig. 2A). The CD spectrum for the cyclic II-III loop is significantly broader than its linear counterpart and shows two shallow minima at 208 and 222 nm that are indicative of a protein containing mainly α-helical and some β-sheet elements (28). The marked difference in structure suggests that cyclization may induce substantial secondary structure and potentially tertiary structural changes throughout the II-III loop protein.
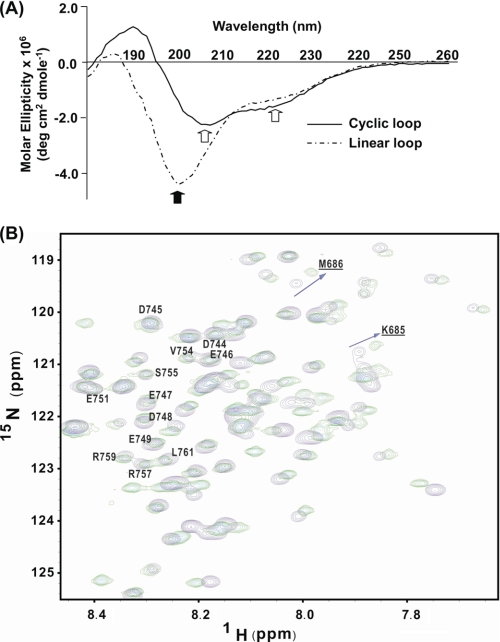
FIGURE 2.
Spectroscopic comparison of the linear and cyclic II-III loops. A, CD spectra for the cyclized and linear skDHPR II-III loop. The linear loop has a negative peak (filled arrow) at 198 nm, and the cyclized loop has two negative peaks (open arrows) at 208 and 222 nm. B, 15N HSQC NMR spectra for the cyclized loop (green contours) overlaid on the spectrum for the linear II-III loop (purple contours) spectra showing the distribution of amino acid residues. Residues not affected by cyclization in the C region are labeled, whereas some residues adjacent to the linker (Lys685 (K685) and Met686 (M686)) are also labeled (underscored).
In order to more closely investigate the structural implications of cyclization of the II-III loop protein, we carried out a series of multinuclear NMR experiments on the 15N/13C uniformly labeled cyclized loop. A set of experiments that included 15N edited total correlation spectroscopy and NOESY-HSQC, HCCONH, and HNCA protocols were used to assign the NMR resonances of cyclized loop. The complete sequential assignment for the non-proline residues of the original II-III loop residues, including the inserted intein linker, was performed, and the backbone assignment is shown in supplemental Fig. 1. The numbering system relating to the II-III loop residues has been maintained (i.e. residues 666–790), whereas residues comprising the intein linker and the six histidine residues in the sequence have been assigned an alternative numbering system as specified in the legends to Figs. 2B and supplemental Fig. 1.
Structural Comparison of the Linear and Cyclized DHPR II-III Loop
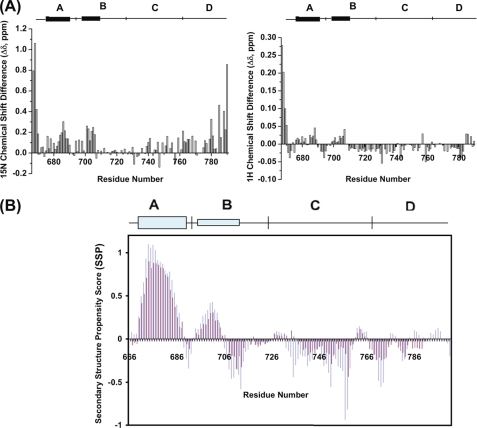
Chemical shift mapping is a valuable and sensitive indicator of changes in structure, and based on the sequential backbone assignment, 1H/15N HSQC NMR chemical shift analysis was initially used to directly compare the chemical shift resonances of the two II-III loop constructs. The core region of the 1H/15N HSQC spectrum of the linear loop is shown in Fig. 2B with the cyclized DHPR II-III loop superimposed. Apart from the presence of additional linker and histidine residues, there are clear resonance shifts (Δδ) between the linear and cyclized spectra. However, it is also clear that the chemical shifts of numerous residues remained unchanged. All clearly identified changes are summarized in Fig. 3A. For orientation and historical purposes, the II-III loop has been assigned four arbitrary regions (29) (i.e. the A (residues Glu666–Leu690), B (residues Pro691–Glu724), C (residues Phe725–Gln765), and D regions (residues Leu766-Leu791)). Unsurprisingly, the N- and C-terminal residues underwent the largest chemical shift changes (Δδ >0.1 for 1H or 0.5 for 15N), whereas the remaining residues exhibited only modest (Δδ <0.1 for 1H or 0.5 for 15N) or no chemical shift changes. Aside from the terminal residues, the major 15N chemical shift differences were clustered in the A region, the adjacent B region, and the D region. Of these, the A and B regions had been previously identified as forming nascent α-helical structures in the linear II-III loop (11, 30). In order to expand upon this chemical shift analysis, we used the backbone α-carbon and side chain β-carbon chemical shift values, which are regarded as a more accurate predictor of secondary structure for intrinsically unstructured proteins (31). This analysis was performed by comparing the difference in secondary structure propensity (SSP) of the two constructs using a program that combines different chemical shifts into a single residue-specific secondary structure propensity score (31). Due to the severe overlap for Hα chemical shift values, only the Cα and Cβ chemical shift values were incorporated into this program, and the SSP for the two constructs was assessed. Fig. 3B shows that as previously determined in our studies, the A and B regions have a tendency to adopt α-helical structures, with the A region in particular showing almost a fully formed α-helix. More interestingly, however, is that cyclization resulted in an increase in the SSP for the A and B regions of the intein compared with the linear loop (72% versus 59% for residues 670–706). Another region of interest is the C region (labeled residues in Fig. 2B), which is the “critical region” for skeletal EC coupling (32). For residues 733–760 of the C region, we observe a significant increase in β-strand or extended structure from 11% (linear) to 29.6% (intein). To explore these changes in secondary structure further, 15N and 13C NOESY-based two- and three-dimensional experiments were carried out for the intein II-III loop protein. This set of experiments revealed little change in the NOE profile for the intein compared with the linear II-III loop protein (data not shown) and detected no additional medium or long range NOEs. Furthermore, careful examination of the A and B helical regions or the C region showed little difference in the NOE profile, suggesting that despite increases in secondary structure propensity, the residual dynamic nature of these regions is not reflected in NOE effects. These sets of NMR experiments indicate that conventional NOE measurements alone are not sufficient to detect slight changes in structure.
FIGURE 3.
Spectral shifts in assigned residues occurring as a result of cyclization of the II-III loop. The A–D regions are shown at the top of A, with line thickness reflecting α-helical strength. A, the top graph shows the 1HN chemical shift perturbations, and the lower graph shows 15NH chemical shift perturbations for the II-III loop residues indicated on the lower axis. The divisions of the loop into A, B, C, and D regions are indicated along the central bar. The magnitude of change was calculated by comparing the 1H and 15N HSQC spectra of the cyclized II-III loop and the linear II-III loop. It is notable that the residues most strongly affected by the cyclization are clustered in the A, B, and D regions of the II-III loop. B, SSP scores for the two II-III loop constructs. The purple bars denote the linear whereas the green bars represent the intein II-III loop. Positive values denote α-helical structure propensity, and negative values represent β-strand or extended structure propensity. A SSP score of 1 or −1 corresponds to a fully formed α- or β-structure, respectively.
Dynamic Properties of the Cyclized II-III Loop
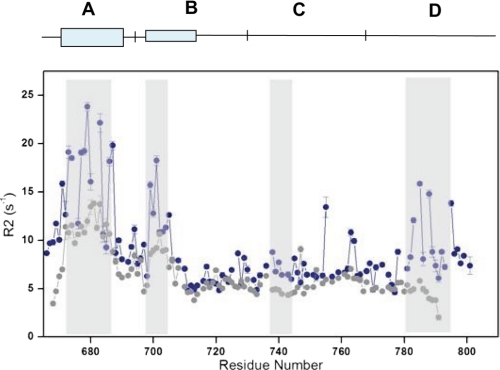
Spin-lattice relaxation rates (R2) are an effective tool in evaluating the dynamic properties of proteins and are capable of detecting motional fluctuations within a protein occurring within a microsecond to picosecond time scale. The dynamic consequences of cyclizing the II-III loop were examined by comparing the R2 values for the two II-III loop constructs and are shown in Fig. 4. It is evident from this comparison that, overall, the cyclized loop displays larger R2 values than its linear counterparts, indicating a global reduction in motional freedom. This result is particularly prominent at the N- and C-terminal ends of the loop, where R2 differences of ∼5 s−1 were observed. This result is not unexpected, however; additional segments throughout the cyclized protein also displayed elevated R2 values of ∼4–10 s−1 compared with the linear loop. The locations of these residues coincide with α-helical regions (i.e. regions A and B) previously found in the linear loop and maintained in the cyclized construct. Notably, these helical regions are the key interaction elements responsible for binding to the II-III loop to the SPRY2 domain of RyR1. Several of the peaks belonging to the A region of the cyclized loop, such as Lys675, Lys677, Glu682, Lys685, and Met686, were broadened in the HSQC spectrum and display larger R2 values compared with their linear loop counterparts. In addition to these regions, one segment located in the center of the C region of the II-III loop (residues 737–745) showed an increase in the R2 of ∼40% (all regions with increased R2 values are highlighted in gray in Fig. 4). Therefore, the dynamic changes upon cyclization are not confined to the N and C termini or the A and B regions and, importantly, also occur in the critical region for EC coupling.
FIGURE 4.
Transverse relaxation rates (R2) of the circular and the linear loop as a function of residue number. A–D regions are shown at the top, with line thickness reflecting α-helical strength. The black points denote the linear R2 values, whereas the blue points represent the intein II-III loop. The shaded regions cover consecutive residues that undergo significant increase for the cyclized loop.
The II-III loop binds to the SPRY2 domain of RyR1 and activates RyR1 in in vitro studies through its A and B regions, and the efficacy of both interactions depends on the helical nature of these regions (11, 30). Therefore, we predicted that these in vitro functions of the loop would be enhanced as a consequence of the enhanced structure in the A and B regions. These predictions are examined below.
The Cyclic skDHPR II-III Loop Binds More Tightly to the SPRY2 Domain of RyR1 in Vitro than the Linear Loop
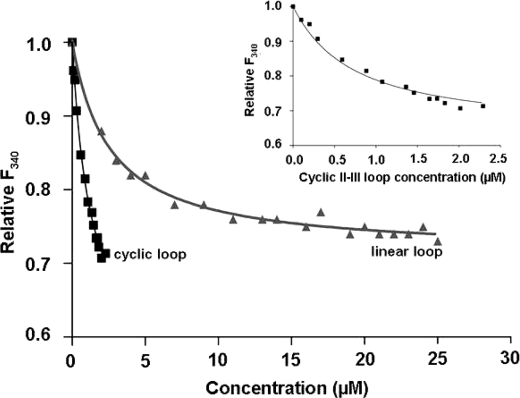
The SPRY domain belongs to a class of structural domains that was first identified in the Dictyostelium discoideum tyrosine kinase spore lysis A (SplA) and all three isoforms of the mammalian RyR (33). Three SPRY domains are conserved in the RyR across species (34), and we have previously shown that the DHPR II-III loop binds with low micromolar affinity to the second of these domains (SPRY2; Ser1085–Val1208) (11). Indeed, the helical nature of the N-terminal residues of the II-III loop is essential for the interaction between the loop and the SPRY2 domain. Thus, we predicted that, with a modified helical composition in its N-terminal part, the cyclic loop may bind with different affinity than the linear loop to the recombinant SPRY2 domain. Binding of the linear and cyclic loops to the SPRY2 domain was compared using intrinsic tryptophan fluorimetry (Fig. 5). A KD value of 0.6 ± 0.05 μm was measured for the cyclic II-III loop binding to SPRY2, indicating an ∼4-fold higher affinity than the linear construct (2.3 ± 0.1 μm).
FIGURE 5.
The relative fluorescence values of SPRY2 (F340) plotted as a function of the concentration (μm) of the linear loop (triangles) and cyclic (squares) skDHPR II-III loop. The fluorescence values were corrected for dilution effects and the fluorescence contribution arising from the quenchers. The data were analyzed as described previously (11), using a non-linear regression fit to the relative fluorescence intensity at 340 nm (F340) as a function of quencher concentration (μm). The curve fitting was performed using the GraphPad Prism software (GraphPad Software), from which the Kd measurements were derived. The inset at the top shows the cyclic loop data and curve using an expanded y axis.
The Cyclic skDHPR II-III Loop Is a More Effective Activator of RyR1 than the Linear Loop
The activity of channels was recorded for ≥10 min after the addition of recombinant protein to the cytoplasmic (cis) solution. The usual effect of the linear loop (9, 10) was seen, with channel activity increasing with 10 and 50 nm, decreasing with 100 and 200 nm, and returning toward the initial control levels after perfusion to remove the protein (Fig. 6A). Similar changes in activity were seen with the cyclic loop (Fig. 6B), although activation was greater and inhibition less than with the linear loop. The linker peptide did not alter activity at concentrations of 10–100 nm (Fig. 6C). A small increase in activity with 200 nm linker peptide in Fig. 6C was not reflected in the average data (Fig. 7).
FIGURE 6.
Effects of the linear (A) and cyclic (B) II-III loops and the linker peptide (C) on single RyR1 channel activity. In each panel, the recordings show 3-s segments of representative channel activity recorded at −40 mV under control conditions before the addition of the construct and then after the addition of 10, 50, 100, and 200 nm concentrations of each construct, as indicated on the left. The final record in each panel was obtained after perfusion of the cis chamber to remove the protein from the solution.
FIGURE 7.
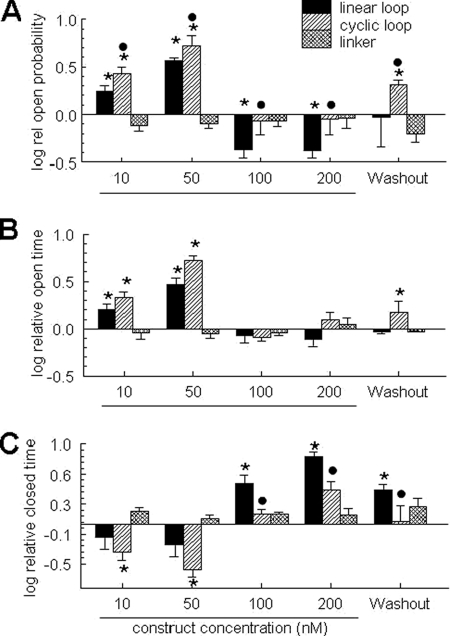
Average relative single channel parameters measured with each concentration of the linear loop (filled bins), cyclic loop (hatched bins), and the linker peptide (cross-hatched bins). Data are shown for relative open probability (A), mean open time (B), and mean closed time (C). Each bar indicates the average ± S.E. (error bars) of 4–8 measurements. The asterisk indicates a significant difference from the control data, the filled symbol indicates a significant difference between the linear and cyclic loops at the particular concentration. Data are presented as the log10 of the relative values in order to better display values that were less than control.
The average values in Fig. 7 include data at +40 and −40 mV (because the effects were not voltage-dependent; Table 1) and are presented as relative changes in activity (calculated for each channel) to reduce effects of variability between individual RyR1 channels (35, 36), which is illustrated in Table 2. The cyclized II-III loop was ∼2-fold more effective in increasing the open probability than the linear loop, indicating a stronger high affinity activating effect after cyclization. The decrease in activity when loop concentration was increased from 50 to 100 nm was ∼4-fold with each construct, suggesting that cyclization did not alter the loop's ability to inhibit the channel. Channel activity returned to control levels after removal of the linear loop, whereas removal of the cyclic loop was followed by an increase in activity to approximately double that of the initial control value. This higher activity may have reflected stronger binding and slower dissociation of the cyclic loop.
TABLE 2.
Control parameter values for RyR1 channels prior to exposure to each of the constructs
The variability between average values for each set of experiments illustrates normal variability between RyR1 channels.
| Po | To | Tc | |
|---|---|---|---|
| ms | ms | ||
| Linear loop | 0.11 ± 0.07 | 10.1 ± 6.4 | 122 ± 38 |
| Cyclized loop | 0.09 ± 0.03 | 3.1 ± 0.6 | 80 ± 21 |
| Linker | 0.22 ± 0.06 | 2.8 ± 0.4 | 23 ± 6 |
Effect of the Cyclic skDHPR II-III Loop on RyR1 Gating Parameters
The changes in To and Tc underlying the changes in Po are shown in Fig. 7, B and C. In general, the significantly greater increase in Po with the cyclic loop (Fig. 7A) was due to trends toward a greater increases in To and reductions in Tc. The decrease in the relative Po at higher loop concentrations (100 and 200 nm) was due to a significant increase in Tc, which was significantly greater in the linear loop. These effects on channel gating suggest that the cyclic loop interacted most strongly with the channel opening mechanism.
Effect of the Linear and Cyclic Loops on Open and Closed Time Distributions
We examined the effects of the loop constructs on the distribution of open and closed times to further explore the effects of the linear and cyclic loops on RyR1 gating. The exponential fits to data from individual channels exposed to the linear or cyclic loop constructs at three different concentrations are shown in Fig. 8, and the average time constants for each construct over the range of concentrations are shown in Fig. 9.
FIGURE 8.
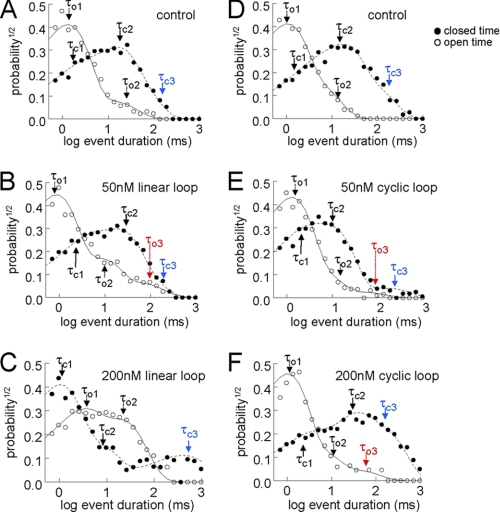
Effects of the linear and cyclic loop constructs on the distribution of open and closed times. Exponential open and closed time constants were determined. Open and closed times were collected into logged bins, and the square root of the relative frequency of events (P½) was plotted against the logarithm of the open (open circles) or closed times (filled circles) in ms (23). Examples are shown for data from individual channels under control conditions (A and D) and after exposure to 50 nm linear and cyclic loops (B and E) and then 200 nm linear and cyclic loops (C and F). The numbers of events generally varied between 500 and 1000. The broken lines in each graph show the fit of multiple exponential functions to the data. The arrows indicate individual open time constants (τo1, τo2, and τo3) and individual closed time constants (τc1, τc2, and τc3). The variables τo3 and τc3 (see description under “Results”) are shown in red and blue, respectively. A–C, exposure to linear loop. D–F, exposure to cyclic loop.
FIGURE 9.
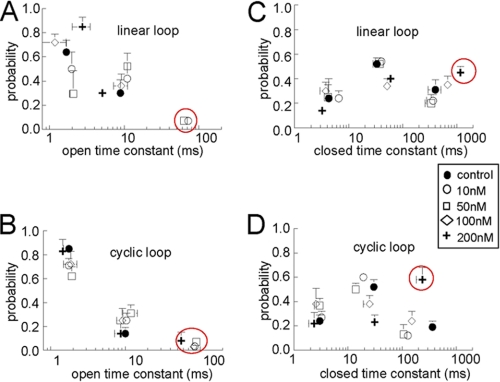
Effects of the linear and cyclic loops on the average open (A and B) and closed (C and D) time constants. The probability of events falling into each time constant is plotted against the time constant in ms (open time or closed time). Time constants are shown for control data (filled circles), 10 nm loop (open circles), 50 nm loop (open squares), 100 nm loop (open diamonds), and 200 nm loop (crosses). The red circles indicate regions of major difference between the effects of the linear and the cyclic loops, which are primarily in τo3 and τc3. Data from four bilayers were analyzed for the linear loop and seven bilayers for the cyclic loop. Each data point is the average of 8–16 measurements (at +40 and −40 mV) for the linear and cyclic loops, respectively. Error bars indicate S.E.
The data were fitted with either two or three exponential components, and the appropriate fit was determined using least squares analysis. In all cases where three exponentials were used rather than two, the sum of the squares for triple fit was lower than that for the double fit, and the differences were on average significant (Table 3).
TABLE 3.
Average values for the sum of least squares differences between data and curves fitted to the data assuming either two (τo2, τc2) or three (τo3, τc3) exponential components for open and closed time distributions
The asterisks and boldface type indicate instances in which the use of three components resulted in a significantly better fit to the data.
| Open time distribution |
Closed time distribution |
|||
|---|---|---|---|---|
| τo2 | τo3 | τc2 | τc3 | |
| Linear loop | ||||
| Control | 0.0263 ± 0.0056 | 0.0253 ± 0.0055 | 0.0497 ± 0.0150 | 0.0413 ± 0.0145* |
| 10 nm | 0.0205 ± 0.0055 | 0.0147 ± 0.0035 | 0.0462 ± 0.0094 | 0.0274 ± 0.0059* |
| 50 nm | 0.0229 ± 0.0052 | 0.0184 ± 0.0043* | 0.0387 ± 0.0095 | 0.0274 ± 0.0073* |
| 100 nm | 0.0292 ± 0.0084 | 0.0323 ± 0.0101 | 0.2850 ± 0.1235 | 0.2272 ± 0.0976* |
| 200 nm | 0.0449 ± 0.0008 | 0.0439 ± 0.0001 | 0.6674 ± 0.3633 | 0.6254 ± 0.3582* |
| Cyclic loop | ||||
| Control | 0.0146 ± 0.0019 | 0.0151 ± 0.0019 | 0.0368 ± 0.0076 | 0.0211 ± 0.0037* |
| 10 nm | 0.0139 ± 0.0034 | 0.0099 ± 0.0002* | 0.0241 ± 0.0065 | 0.0114 ± 0.0026* |
| 50 nm | 0.0185 ± 0.0032 | 0.0107 ± 0.0023* | 0.0181 ± 0.0051 | 0.0131 ± 0.0053* |
| 100 nm | 0.0239 ± 0.0062 | 0.0217 ± 0.0063* | 0.0244 ± 0.0085 | 0.0186 ± 0.0068* |
| 200 nm | 0.0229 ± 0.0056 | 0.0188 ± 0.0055* | 0.0532 ± 0.0244 | 0.0264 ± 0.0117* |
Open times for RyR1 under control conditions fell into two exponential components, and a third longer component was introduced after the addition of 10 and 50 nm concentrations of the loop constructs and was maintained with the cyclic loop, but not the linear loop, at higher concentrations of 100 and 200 nm. The shortest open time constant, τo1, was 1–3 ms, and the second time constant, τo2, was 5–12 ms. The longest time constant, τo3, seen only in channels in the presence of the loop constructs, was 50–100 ms. The increase in mean open time in the presence of the loop (Fig. 7) was largely due to the appearance of the τo3 component. The mean open time tended to remain higher with higher loop concentrations (≥100 nm) of the cyclic loop than the linear loop (Fig. 7). This can be attributed to the continued presence of τo3 with higher cyclic loop concentrations.
Closed times for RyR1 channels fell into three exponential components under all conditions. The shortest closed time constant, tc1, was 2–4 ms, and the second time constant, tc2, was 10–40 ms. The longest time constant, tc3, was 300–400 ms under control conditions and increased to ∼1000 ms with a 200 nm concentration of the linear loop. In marked contrast, tc3 decreased to 100–200 ms in the presence 10–100 nm cyclic loop and then increased toward control values with 200 nm cyclic loop, where it contained the majority (60%) of events. The very long tc3 with 200 nm linear loop is largely responsible for the long closed times with this construct. The shorter tc3 values contribute to the briefer closed times with 200 nm cyclic loop than with the linear loop. These results again indicate that the cyclized loop may interact more strongly with the channel opening mechanism than the linear loop.
DISCUSSION
Until now, in vitro structural and functional investigations of the II-III loop fragment of the DHPR α1S subunit have been limited to a linear construct of this loop with untethered ends (8–11, 24). However, the unrestrained ends of the loop are not a true representation of the physiological state, where the termini are anchored to membrane-spanning segments. We have used intein-mediated technology to effectively fuse the N and C termini of the loop with the aid of a linker peptide. We show for the first time that cyclization results in significant structural changes in several parts of the loop, including the critical region for EC coupling and in other regions essential for the in vitro function of the loop. Changes in the in vitro function of the protein were observed and were consistent with the structural changes.
Functional Changes in the Cyclic II-III Loop Are Correlated with Structural Changes
In vitro, the linear skeletal DHPR II-III loop is an activator of RyR1 (8, 9, 24). The ability of the linear II-III loop to activate native skeletal RyR channels was clearly enhanced with the cyclic loop. We have previously demonstrated that the helical content of a peptide with a sequence corresponding to the A region of the II-III loop is inextricably linked to the peptide's ability to activate RyR1 channels (37, 38) and show here that the cyclic II-III loop is 2–4-fold more potent in increasing the open probability of the RyR1 channel than its linear counterpart. This increased ability to activate the RyR1 channel is consistent with the stabilization of the helical components of the A and B regions.
High Affinity Binding of the Cyclized Loop to the SPRY2 Domain of RyR1
SPRY domains contribute to protein-protein interactions (39, 40), and several studies suggest that the RyR1 SPRY2 domain (Ser1085–Val1208) plays a role in the in vitro interactions between the II-III loop and RyR1. The skeletal II-III loop can pull down a 37-amino acid residue fragment of the RyR1, containing Arg1076– Asp1112 (41), and the A region of the skeletal DHPR II-III loop interacts with residues Val1021–Glu1631 of the RyR1 (42). These two sequences overlap with the N-terminal end of the RyR1 SPRY2 domain, and the linear II-III loop binds to the recombinant SPRY2 domain through the A and B region (11, 30, 34) with a Kd of 2.3 ± 0.1 μm, which is critically dependent on the presence of five basic residues in the A region α-helix (Arg681–Lys685) and two adjacent lysine residues (Lys703 and Lys704) in the B region α-helix (30).
The cyclized wild type II-III loop, with a Kd of 0.6 ± 0.05 μm, has a higher affinity for the RyR1 SPRY2 than its linear counterpart. Because the A and B regions are major contributors to the increased α-helical content of the cyclized II-III loop, it is likely that this structural stabilization in the cyclized II-III loop is correlated with the 4-fold higher affinity for the SPRY2 domain. It is also likely that the tighter binding of the cyclized loop to the SPRY2 domain is linked to the enhanced activation of RyR1 channels. Thermodynamically, the increase in RyR1 binding and activity by the cyclic construct can be explained by a smaller entropic loss upon binding brought about by the stabilization of the helical interaction sites (A & B regions).
Structural and Dynamic Changes with Cyclization of the Loop
The marked shift in the CD profile of the cyclized protein provides compelling evidence of a change in the structure of the II-III loop following cyclization. We have previously reported that the linear II-III loop is an intrinsically unstructured protein with at least two nascent helical elements located in the A and B regions (11). The NMR 15N and 1H chemical shift data presented in Fig. 3a show that the most significant chemical shift changes occur not only in the N- and C-terminal residues adjacent to the linker residues but also in the regions adjacent to the directly modified region, such as the A (Glu666–Leu690) and B (Pro691–Glu724) regions of the loop. A more comprehensive chemical shift comparison using Cα and Cβ chemical shifts (Fig. 3b) showed not only an increase in helical structural propensity for this region but a marked increase in extended β-strand structural propensity for the C region.
A series of three-dimensional NOESY-based NMR experiments revealed stable structural changes promoted by cyclization of the II-III loop. Notably, apart from new NOEs associated with the linker region, no discernable overall difference in the NOE profile between the linear and intein loop was evident. Close inspection of signature NOEs related to the α-helical secondary structure elements (A and B regions) and extended β-strand structure (C region) also showed a similar profile for the linear and intein loops. Therefore, cyclization does not promote alternative, stable structures within the II-III loop but rather results in subtle shifts in the equilibrium that exits between random coil and secondary structure elements. These subtle changes are detectable by spin-spin relaxation rate (R2) measurements, and the increased R2 values for the A/B helical and C regions are evidence of stabilization of the secondary structure elements within the intein protein. In summary, the CD and NMR data suggest that cyclization of the intrinsically unstructured DHPR II-III loop does not involve major structural modifications but instead induces subtle structural changes to the secondary structure elements of specific regions in the molecule and can dramatically influence the function of the protein.
Functional and Physiological Implications of Cyclization
It is a generally held view that upon depolarization, movement of charged residues in the membrane-spanning segments of α1s DHPR is responsible for a conformational change in the II-III loop, which in turn is transmitted to the RyR1. We convincingly demonstrate by altering the geometry of the N- and C-terminal ends of the II-III loop that the structural and dynamic consequences are sensed throughout the entire loop. This observation has significant implications for EC coupling, where movement of charged residues in the S4 voltage sensor, by analogy with other voltage-gated cation channels, is likely to induce a major structural change in the membrane-spanning domains linked to the S4 segment (43). This structural reorientation of the S1 to S4 helices could dramatically alter the structural relationship between the N- and C-terminal ends of the II-III loop and alter the conformation of the II-III loop during EC coupling to either increase or decrease its structural content.
Although the importance of the II-III loop in EC coupling and the critical region for EC coupling identified in the central C region (residues 720–765) has been long been established (44), knowledge of the interactive partners of the critical region within the loop in the coupling process is less clear. Concerted attempts by several groups to define the RyR1 binding partner for the critical C region have not been successful, suggesting either that the RyR1 binding partner is complex and remains elusive or that interactions of the C region may proceed through some intermediate protein/factor. In contrast, the in vitro binding between the A and B regions of the loop and the SPRY2 domain of RyR1 has been well characterized (11, 30, 34). Indeed, an unpublished cryoelectron microscopy study of the location of the SPRY2 domain on the RyR1 has mapped this region on domain 6 of the RYR1.6 Although this location is within contact distance with DHPR II-III loop, in vivo chimeric studies show that when the A region has been removed or disrupted, EC coupling is not significantly affected and thus is not critical (5, 45). It remains possible that, although not critical in a dysgenic myotube background, the II-III loop A region/SPRY2 interaction may play a supportive role in EC coupling, which may be revealed in transgenic animals in which a full range of muscle function can be explored. Such supportive roles have been found to be life-supporting in maintaining normal muscle function under stress in both skeletal muscle and the heart, in studies of other non-essential proteins, such as calsequestrin, triadin, and junctin (46–48).
The major finding of this study is that both the helical A/B region and the C region of the II-III loop are structurally altered by cyclization and that this influences the biophysical properties of the loop. The A/B region of the loop interacts in vitro with the SPRY2 domain, and we demonstrated that a modest increase (∼13%) in helical content in this region of the loop was associated with a 3–4-fold increase in RyR1 channel activity and binding affinity. The most likely explanation for this increase is thermodynamic. The stabilized α-helical elements in the intein loop would be more favorably predisposed to interact with SPRY2 RyR1 binding sites. This finding is consistent with our previous findings that substituting strategic residues in the isolated A peptide (residues 671–690) led to the enhancement of its helical structure and subsequently its increased ability to activate RyR1 (37, 49). Because the interactive binding partner of the central C region of the II-III loop has yet to be established, it is not possible to perform a comparable structural and functional analysis for this region. It is possible that the changes in channel gating induced by cyclization are imposed by an interaction between the more structured C region and a binding partner in RyR1. However, the substantial increase in β-strand or extended structure found in the C region may allow the identification of new RyR1 binding partners using this improved construct.
Conclusion
We find that cyclization of the skeletal DHPR II-III loop affects the secondary structure and the dynamic properties of the helical A/B region as well as the critical C region. These structural effects are correlated with a change in the in vitro activation profile of the RyR1 and with an interaction with DHPR II-III loop α-helical recognition sites in the SPRY2 domain of RyR1. The results provide a clear example of how subtle structural and dynamic changes in an intrinsically unstructured protein may affect molecular recognition. Also, the critical region of the loop that supports skeletal in vivo EC coupling demonstrated an increased β-strand or extended structure upon cyclization, indicating that this portion of the loop is susceptible to structural and dynamic changes, with functional consequences, induced by t-tubule membrane depolarization.
Supplementary Material
Acknowledgments
We are grateful to Prof. Nick Dixon and Dr. Pavel Prosselkov for providing the pNW1120 intein vector. We also acknowledge Dr. Derek Laver for providing the logged bin software as well as Suzy Pace and Joan Stivala for preparation of SR vesicles for bilayer studies.
This work was supported by National Health and Medical Research Council Grant 471418.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
M. Samso, personal communication.
- EC
- excitation contraction
- skDHPR
- skeletal dihydropyridine receptor
- Cp-Y
- carboxypeptidase-Y
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- SR
- sarcoplasmic reticulum
- TES
- 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid
- RyR
- ryanodine receptor
- SSP
- secondary structure propensity.
REFERENCES
- 1. Dulhunty A. F., Haarmann C. S., Green D., Laver D. R., Board P. G., Casarotto M. G. (2002) Prog. Biophys. Mol. Biol. 79, 45–75 [DOI] [PubMed] [Google Scholar]
- 2. Carbonneau L., Bhattacharya D., Sheridan D. C., Coronado R. (2005) Biophys. J. 89, 243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kugler G., Weiss R. G., Flucher B. E., Grabner M. (2004) J. Biol. Chem. 279, 4721–4728 [DOI] [PubMed] [Google Scholar]
- 4. Tanabe T., Beam K. G., Adams B. A., Niidome T., Numa S. (1990) Nature 346, 567–569 [DOI] [PubMed] [Google Scholar]
- 5. Wilkens C. M., Kasielke N., Flucher B. E., Beam K. G., Grabner M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5892–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kugler G., Grabner M., Platzer J., Striessnig J., Flucher B. E. (2004) Arch. Biochem. Biophys. 427, 91–100 [DOI] [PubMed] [Google Scholar]
- 7. Meissner G. (1994) Annu. Rev. Physiol. 56, 485–508 [DOI] [PubMed] [Google Scholar]
- 8. Stange M., Tripathy A., Meissner G. (2001) Biophys. J. 81, 1419–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dulhunty A. F., Karunasekara Y., Curtis S. M., Harvey P. J., Board P. G., Casarotto M. G. (2005) Biochem. J. 385, 803–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dulhunty A. F., Karunasekara Y., Curtis S. M., Harvey P. J., Board P. G., Casarotto M. G. (2005) Front. Biosci. 10, 1368–1381 [DOI] [PubMed] [Google Scholar]
- 11. Cui Y., Tae H. S., Norris N. C., Karunasekara Y., Pouliquin P., Board P. G., Dulhunty A. F., Casarotto M. G. (2009) Int. J. Biochem. Cell Biol. 41, 677–686 [DOI] [PubMed] [Google Scholar]
- 12. Derbyshire V., Wood D. W., Wu W., Dansereau J. T., Dalgaard J. Z., Belfort M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 11466–11471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paulus H. (2000) Annu. Rev. Biochem. 69, 447–496 [DOI] [PubMed] [Google Scholar]
- 14. Perler F. B. (2005) IUBMB Life 57, 469–476 [DOI] [PubMed] [Google Scholar]
- 15. Williams N. K., Liepinsh E., Watt S. J., Prosselkov P., Matthews J. M., Attard P., Beck J. L., Dixon N. E., Otting G. (2005) J. Mol. Biol. 346, 1095–1108 [DOI] [PubMed] [Google Scholar]
- 16. Wu H., Xu M. Q., Liu X. Q. (1998) Biochim. Biophys. Acta 1387, 422–432 [DOI] [PubMed] [Google Scholar]
- 17. Williams N. K., Prosselkov P., Liepinsh E., Line I., Sharipo A., Littler D. R., Curmi P. M., Otting G., Dixon N. E. (2002) J. Biol. Chem. 277, 7790–7798 [DOI] [PubMed] [Google Scholar]
- 18. Catanzariti A. M., Soboleva T. A., Jans D. A., Board P. G., Baker R. T. (2004) Protein Sci. 13, 1331–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saito A., Seiler S., Chu A., Fleischer S. (1984) J. Cell Biol. 99, 875–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beard N. A., Sakowska M. M., Dulhunty A. F., Laver D. R. (2002) Biophys. J. 82, 310–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wei L., Varsányi M., Dulhunty A. F., Beard N. A. (2006) Biophys. J. 91, 1288–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beard N. A., Wei L., Cheung S. N., Kimura T., Varsányi M., Dulhunty A. F. (2008) Cell Calcium 44, 363–373 [DOI] [PubMed] [Google Scholar]
- 23. Sigworth F. J., Sine S. M. (1987) Biophys. J. 52, 1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu X., Xu L., Meissner G. (1994) J. Biol. Chem. 269, 6511–6516 [PubMed] [Google Scholar]
- 25. Iwai H., Plückthun A. (1999) FEBS Lett. 459, 166–172 [DOI] [PubMed] [Google Scholar]
- 26. Scott C. P., Abel-Santos E., Wall M., Wahnon D. C., Benkovic S. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13638–13643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iwai H., Lingel A., Pluckthun A. (2001) J. Biol. Chem. 276, 16548–16554 [DOI] [PubMed] [Google Scholar]
- 28. Whitmore L., Wallace B. A. (2008) Biopolymers 89, 392–400 [DOI] [PubMed] [Google Scholar]
- 29. el-Hayek R., Antoniu B., Wang J., Hamilton S. L., Ikemoto N. (1995) J. Biol. Chem. 270, 22116–22118 [DOI] [PubMed] [Google Scholar]
- 30. Tae H. S., Norris N. C., Cui Y., Karunasekara Y., Board P. G., Dulhunty A. F., Casarotto M. G. (2009) Clin. Exp. Pharmacol. Physiol. 36, 346–349 [DOI] [PubMed] [Google Scholar]
- 31. Marsh J. A., Singh V. K., Jia Z., Forman-Kay J. D. (2006) Protein Sci. 15, 2795–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grabner M., Dirksen R. T., Suda N., Beam K. G. (1999) J. Biol. Chem. 274, 21913–21919 [DOI] [PubMed] [Google Scholar]
- 33. Ponting C., Schultz J., Bork P. (1997) Trends Biochem. Sci. 22, 193–194 [DOI] [PubMed] [Google Scholar]
- 34. Tae H., Casarotto M. G., Dulhunty A. F. (2009) Eur. Biophys. J. 39, 51–59 [DOI] [PubMed] [Google Scholar]
- 35. Copello J. A., Barg S., Onoue H., Fleischer S. (1997) Biophys. J. 73, 141–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wei L., Gallant E. M., Dulhunty A. F., Beard N. A. (2009) Int. J. Biochem. Cell Biol. 41, 2214–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Casarotto M. G., Gibson F., Pace S. M., Curtis S. M., Mulcair M., Dulhunty A. F. (2000) J. Biol. Chem. 275, 11631–11637 [DOI] [PubMed] [Google Scholar]
- 38. Casarotto M. G., Green D., Pace S. M., Curtis S. M., Dulhunty A. F. (2001) Biophys. J. 80, 2715–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rhodes D. A., de Bono B., Trowsdale J. (2005) Immunology 116, 411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Woo J. S., Imm J. H., Min C. K., Kim K. J., Cha S. S., Oh B. H. (2006) EMBO J. 25, 1353–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Leong P., MacLennan D. H. (1998) J. Biol. Chem. 273, 7791–7794 [DOI] [PubMed] [Google Scholar]
- 42. Altafaj X., Cheng W., Estève E., Urbani J., Grunwald D., Sabatier J. M., Coronado R., De Waard M., Ronjat M. (2005) J. Biol. Chem. 280, 4013–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shrivastava I. H., Durell S. R., Guy H. R. (2004) Biophys. J. 87, 2255–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nakai J., Tanabe T., Konno T., Adams B., Beam K. G. (1998) J. Biol. Chem. 273, 24983–24986 [DOI] [PubMed] [Google Scholar]
- 45. Ahern C. A., Bhattacharya D., Mortenson L., Coronado R. (2001) Biophys. J. 81, 3294–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dainese M., Quarta M., Lyfenko A. D., Paolini C., Canato M., Reggiani C., Dirksen R. T., Protasi F. (2009) FASEB J. 23, 1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Oddoux S., Brocard J., Schweitzer A., Szentesi P., Giannesini B., Brocard J., Fauré J., Pernet-Gallay K., Bendahan D., Lunardi J., Csernoch L., Marty I. (2009) J. Biol. Chem. 284, 34918–34929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shen X., Franzini-Armstrong C., Lopez J. R., Jones L. R., Kobayashi Y. M., Wang Y., Kerrick W. G., Caswell A. H., Potter J. D., Miller T., Allen P. D., Perez C. F. (2007) J. Biol. Chem. 282, 37864–37874 [DOI] [PubMed] [Google Scholar]
- 49. Green D., Pace S., Curtis S. M., Sakowska M., Lamb G. D., Dulhunty A. F., Casarotto M. G. (2003) Biochem. J. 370, 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.