Abstract
Nucleation of promoter melting in bacteria is coupled with RNA polymerase (RNAP) binding to a conserved −10 promoter element located at the upstream edge of the transcription bubble. The mechanism of downstream propagation of the transcription bubble to include the transcription start site is unclear. Here we introduce new model downstream fork junction promoter fragments that specifically bind RNAP and mimic the downstream segment of promoter complexes. We demonstrate that RNAP binding to downstream fork junctions is coupled with DNA melting around the transcription start point. Consequently, certain downstream fork junction probes can serve as transcription templates. Using a protein beacon fluorescent method, we identify structural determinants of affinity and transcription activity of RNAP-downstream fork junction complexes. Measurements of RNAP interaction with double-stranded promoter fragments reveal that the strength of RNAP interactions with downstream DNA plays a critical role in promoter opening and that the length of the downstream duplex must exceed a critical length for efficient formation of transcription competent open promoter complex.
Keywords: Fluorescence, Promoters, RNA Polymerase, Transcription, Transcription Initiation Factors
Introduction
Formation of the transcription-competent open promoter complex (RPo) by DNA-dependent RNA polymerase (RNAP)3 is a critical checkpoint on the pathway of gene expression. In bacteria, the transition from initial recognition of the promoter by RNAP to RPo proceeds through a series of short-lived intermediate complexes (1–3). In RPo, the DNA duplex is disrupted over a stretch of 12–15 base pairs, which leads to the formation of the transcription bubble and makes the transcription start point (position +1) accessible to the RNAP catalytic center (4, 5). An important source of energy driving this strand separation process is the interaction of the RNAP σ70 subunit region 2 with the non-template strand of the −10 promoter element at the upstream edge of the transcription bubble (6–9). The interaction with the −10 element adenine at position −11 of the non-template strand is of special importance for nucleation of promoter melting (6, 10). RNAP interaction with the template segment of the transcription bubble was estimated to be considerably weaker than that with the non-template segment (8). Although formation of intermediates on the pathway to RPo has been characterized kinetically in many studies (1–3, 11, 12), it is still unclear which energetically favorable RNAP-promoter interactions are coupled with downstream propagation of the transcription bubble and determine its final boundary position at +2/+3.
The crystal structures of prokaryotic RNAP indicate that establishment of RNAP interactions with the downstream duplex upon RPo formation must introduce a sharp kink in DNA, which may facilitate melting and stabilize the open complex (13, 14). Indeed, RNAP mutations that change RNAP contacts with downstream DNA reduce the longevity of open complexes formed at certain promoters (15, 16). On highly supercoiled DNA, the downstream interactions may not be obligatory, as an RNAP subassembly containing an N-terminal fragment of the Escherichia coli RNAP β′ subunit (lacking β′ amino acids involved in contacts with downstream DNA) and a fragment of σ70 subunit was reported to recognize and melt an extended −10 consensus promoter (17). However, melting observed in these experiments might not necessarily mirror steps in RPo formation by RNAP (3, 18). Downstream RNAP-promoter contacts are targeted by low molecular weight inhibitors and protein repressors that affect transcription initiation (16, 19–21).
Unraveling the role of downstream RNAP-promoter interactions in open complex formation and regulation of transcription initiation is hindered by the lack of experimental tools to directly measure their strength and specificity. Upon formation of the open promoter complex, two DNA fork junction structures are created around positions −12 and +2. Studies of RNAP interactions with model DNA fragments mimicking the upstream fork junction provided important structural (22) and functional insights into the processes of promoter recognition and melting (8, 9, 23–26). Here, we introduce downstream fork junction fragments as a tool to study the structure and function of the downstream segment of the RPo. The downstream fork junction promoter fragments used consist of stretches of short single-stranded non-template DNA containing the −10 promoter element followed by double-stranded downstream segments of various lengths. We find that specific binding of RNAP to downstream fork junctions can be quantitatively measured using a protein beacon assay method (9). We demonstrate that RNAP complexes with downstream forks mimic RNAP complexes with native promoters in several discriminative biochemical tests. Consequently, certain downstream fork junctions can be used as transcription templates. Our experiments reveal that RNAP interaction with the downstream promoter duplex is strong and provides ∼5.5 kcal/mol binding energy in the context of downstream fork junction probes. We identify structural determinants of affinity and transcription activity of RNAP-downstream fork junction complexes. Our data show that RNAP interactions with downstream promoter DNA play a critical role in open promoter complex formation and may be subject to regulation.
EXPERIMENTAL PROCEDURES
Proteins
RNA polymerase holoenzyme containing the σ70 derivative labeled at position 211 with fluorescent label 5-tetramethylrhodamine ((211Cys-TMR) σ70) was prepared as in Mekler et al. (9). T7 phage gp2 and T4 phage AsiA proteins were prepared as described previously (27, 28). E. coli core RNAP containing a deletion (1149–1190) in the β′ subunit was a generous gift of Dr. Wigneshweraraj (Imperial College, London).
DNA Probes
DNA oligonucleotides and modified oligonucleotides containing the SP18 spacer were synthesized by Integrated DNA Technologies. Fork junction and double-stranded DNA probes were prepared as in Mekler et al. (9).
Fluorometric Assays
Fluorescence measurements were performed using a Quanta-Master QM4 spectrofluorometer (PTI) in transcription buffer (40 mm Tris-HCl, pH 8.0, 100 mm NaCl, 5% glycerol, 1 mm DTT, and 10 mm MgCl2) containing 0.02% Tween 20 at 25 °C. Final assay mixtures (800 μl) contained 1 nm labeled RNAP holoenzyme and DNA probes at various concentrations. The TMR fluorescence intensities were recorded with an excitation wavelength of 550 nm and an emission wavelength of 578 nm. Time-dependent fluorescence changes were monitored after manual-mixing of RNAP beacon (800 μl) and a DNA probe (<20 μl) in a cuvette; the mixing dead-time was 15 s.
To obtain equilibrium dissociation constants (Kd) of the RNAP beacon with downstream fork junctions bearing a relatively short double-stranded segment (hairpin downstream fork junctions and (−11/+14) (+2/+14) probe) and with an upstream fork junction probe, the experimental dependence of the fluorescent signal amplitude on DNA probe concentration was fitted to a chemical equilibrium equation (i.e. titration assay) as described in Mekler et al. (9). A dissociation constant of ATATTAGATTCA oligo, corresponding to −11 to +1 non-template T5N25 promoter sequence but containing A for T substitution at −8 (−11/+1, −8T), was obtained from equilibrium competition binding experiments using the consensus TATAATAGATTCA oligo (−12/+2) as a reference, as in Mekler et al. (9). Relative occupancy of the RNAP beacon by −8T substituted downstream fork junctions, and double-stranded probes were determined by a competition binding assay using −12/+2 oligo as a reference, as described in the supplemental material. Beacon assays of transcription activity were carried out as in Mekler et al. (9); the final concentration of each NTP was 0.5 mm.
In Vitro Abortive Initiation Assay
Abortive transcription reactions were performed in a final volume of 10 μl and contained 200 nm E. coli σ70-holoenzyme (Epicenter) and 0.5 μm concentrations of various DNA templates prepared as described above in a transcription buffer (30 mm Tris-HCl (pH 7.9), 40 mm KCl, 10 mm MgCl2, 2 mm β-mercaptoethanol). Reactions were incubated for 30 min at 25 °C followed by the addition of RNA dinucleotide primers (200 μm), [α-32P]UTP (3000 Ci/mmol), and cold NTPs (100 μm). GpA and UTP were used as substrates in the reaction with a partially non-complementary fork junction shown in Fig. 6A, and ATP+UTP and ApU+ATP+UTP and were used in experiments with the [−12/+16][+1/+16] fork junction. In all other reactions, CpA and UTP were used as substrates. The reactions were incubated for a further 10 min at 25 °C before being terminated by the addition of an equal volume of urea-formamide loading buffer. The reaction products were resolved on a 20% (w/v) polyacrylamide denaturing gel and visualized using phosphorimaging.
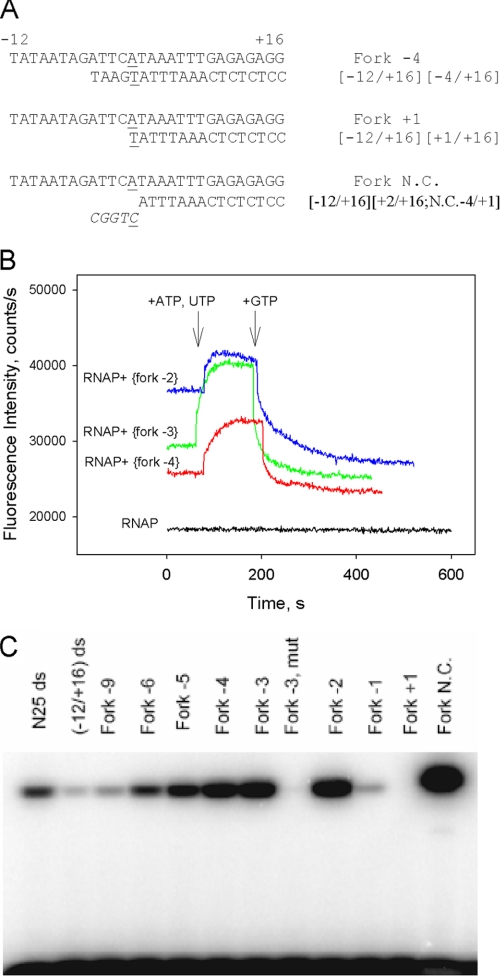
FIGURE 6.
Transcription activity of downstream fork junction probes. A, downstream fork junction probes used. B, shown is a beacon assay of transcription activity of [−12/+16][−2/+16], [−12/+16][−3/+16], and [−12/+16][−4/+16] downstream fork junctions. Time-dependent change of the fluorescence signal was measured upon the addition of ATP and UTP followed by the addition of GTP (final concentration of 0.5 mm each) to complexes of the fork junctions (2 nm) with 1 nm (211Cys-TMR) σ70 holoenzyme. C, shown is an abortive transcript synthesis by RNAP complexes with downstream fork junction probes. The Fork −3 mut template is a derivative of [−12/+16][−3/+16] fork junction with −11A and −7T substituted by non-consensus C; N25 ds and (−12/+16) ds represent double-stranded −60/+30 and −12/+16 fragments of the T5N25 promoter DNA.
RESULTS
RNAP Binding to a Downstream Fork Junction DNA Probe Can Be Detected Readily Using a Protein Beacon Assay
We have recently developed a new fluorometric beacon assay to quantitatively monitor site-specific interactions of promoter DNA and various promoter fragments with bacterial RNAP holoenzyme (9). The assay relies on the detection of an increase in fluorescence intensity upon DNA binding to RNAP σ subunit region 2. The σ subunit contains a site-specifically attached tag whose fluorescence is quenched through interaction with σ region 2 Trp and Tyr residues. Upon interaction with promoter DNA or promoter fragments, these aromatic amino acids lose contact with the fluorescent probe, decreasing the quenching efficiency (9). Here, we used this assay to measure RNAP interaction with downstream fork junction promoter fragments based on the sequence of the N25 promoter of bacteriophage T5 and consisting of a non-template strand segment of the transcription bubble followed by a double-stranded downstream segment. The likely location of the downstream fork junction promoter segment relative to a model of an RNAP-promoter open complex is shown in Fig. 1A.
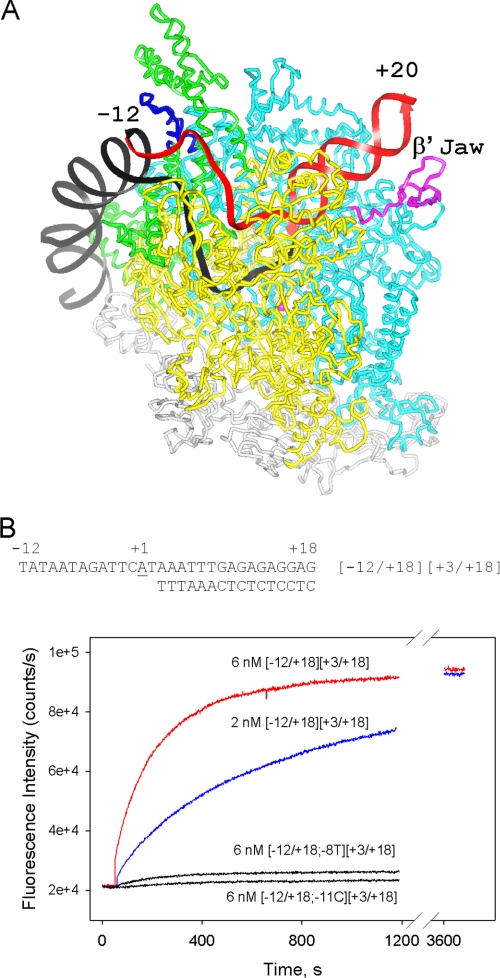
FIGURE 1.
Measuring RNAP-downstream fork junction interactions using the protein beacon assay. A, the likely location of a downstream fork junction promoter segment (in red) relative to a model of Thermus aquaticus RNAP-promoter open complex is shown. The colors of the RNAP subunits contacting the downstream fork junction segment are: yellow, β; cyan, β′; green, σ. The β′ jaw and conserved region 2.3 of σ are shown in magenta and blue, respectively. The model of the open complex structure is from Lawson et al. (51). B, time dependence of the increase in fluorescence upon mixing 1 nm (211Cys-TMR) σ70 holoenzyme beacon with downstream fork junction probes. The structure of the parental fork junction based on the T5N25 sequence is shown in the panel. In probe names, numbers in the left and the right brackets correspond to upper and bottom strands fork junction ends with respect to the transcription start position (+1); −8T and −11 C stand for a T at the −8 position and a C at the −11 position, respectively.
An RNAP beacon based on a σ70 derivative labeled at amino acid position 211 with tetramethylrhodamine-5-maleimide (211Cys-TMR) σ70 (9) was used throughout this work. Binding of a T5 N25-based downstream fork probe to RNAP was readily detected using the beacon assay (Fig. 1B). The RNAP beacon signal generated by the downstream fork was saturated at 2 nm probe concentration, as seen from comparison of saturation signals generated by 2 and 6 nm of the probe (Fig. 1B). Similar kinetics of a signal increase were observed upon RNAP beacon binding to downstream fork junctions based on lacUV5 and T7 A1 promoter sequences (data not shown).
RNAP Binding to a Downstream Fork Junction Mimics RNAP Interactions with the Downstream Part of the Open Promoter Complex
RNAP binding to short DNA oligos containing sequences corresponding to the non-template strand of the −10 promoter element is specific and mimics RNAP-promoter interaction (6, 7). RNAP beacon binding to such oligos generates a fluorescent signal dependent on the −10 element sequence (9). As shown in Fig. 1A, only the single-stranded part of a specifically bound downstream fork junction probe interacts with σ region 2. Thus, RNAP interaction with the single-stranded segment should account for a signal produced by such a specifically bound probe, whereas the double-stranded segment is expected to modulate the probe affinity. We compared binding of RNAP to downstream fork junction shown in Fig. 1B and to the −12/+2 oligo corresponding to the single-stranded part of the fork junction. The amplitude of a fork junction fluorescence signal increase at saturation (∼4-fold) was similar to that generated by the −12/+2 oligo; however, a much higher oligo concentration was required to reach the binding saturation (data not shown; see also Ref. 9). Downstream forks similar to that shown in Fig. 1B but bearing a non-consensus C at position −11 or T at position −8 produce a very low signal upon addition to the RNAP beacon (Fig. 1B). Similar behavior was observed for derivatives of the −12/+2 oligo (Ref. 9 and data not shown). These results indicate that RNAP interacts similarly with the −10 element containing a single-stranded segment in the context of single-stranded oligonucleotides and in the context of downstream fork probes.
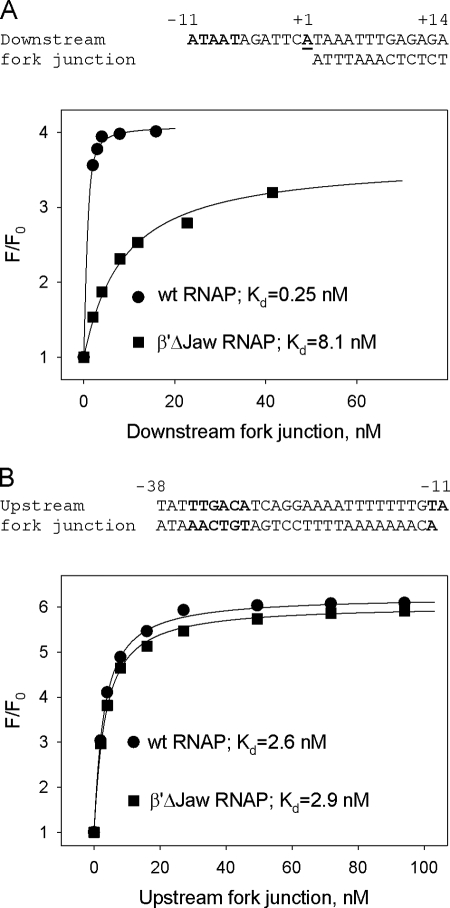
To confirm that the location of the downstream fork duplex segment in RNAP complex is similar to that of downstream DNA in RPo, we compared the affinities of a downstream fork to (211Cys-TMR) σ70 holoenzymes containing wild-type RNAP core enzyme and β′Δ(1149–1190) E. coli core RNAP mutant. The mutant lacks the downstream jaw domain thought to be part of a trough that binds downstream DNA (Fig. 1A). The mutant holoenzyme is known to form comparatively unstable transcription initiation complexes (16, 29). Thus, a significant difference in the affinity of the downstream forks to the mutant and wild-type RNAP beacons is expected if the downstream fork were a faithful mimic of downstream DNA in the RPo. Indeed, we found that the Kd of the mutant RNAP beacon for the downstream fork was ∼40-fold greater than that of the wild-type RNAP beacon, whereas affinities of either enzyme to an upstream fork junction probe were similar (Fig. 2, A and B).
FIGURE 2.
Binding of downstream and upstream fork junction probes to wild-type and to β′Δ(1149–1190) RNAP beacons. Titration of (211Cys-TMR) σ70 holoenzymes containing the wt or mutant β′Δ(1149–1190) subunit with a downstream (A) or upstream (B) fork junction probes is shown. The solid lines correspond to a nonlinear regression fit of the data.
In the RNAP holoenzyme, σ70 subunit region 1.1 (σR1.1) is positioned inside the RNAP active-site channel from where it is displaced by the downstream promoter segment upon RPo formation (13, 30). The displacement of σR1.1 can be monitored by fluorescence resonance energy transfer (FRET) (30); the efficiency of FRET between a fluorescent label attached to region 1.1 and rifampicin, an antibiotic binding near the RNAP active center, is higher in RNAP holoenzyme than in the T5 N25 promoter-RNAP complex (supplemental Fig. S1). The same decrease in FRET efficiency was observed upon RNAP binding to the downstream but not the upstream fork junction probe (supplemental Fig. S1). Thus, the downstream fork probe displaces σ region 1.1 in the same way as does the downstream promoter segment in RPo.
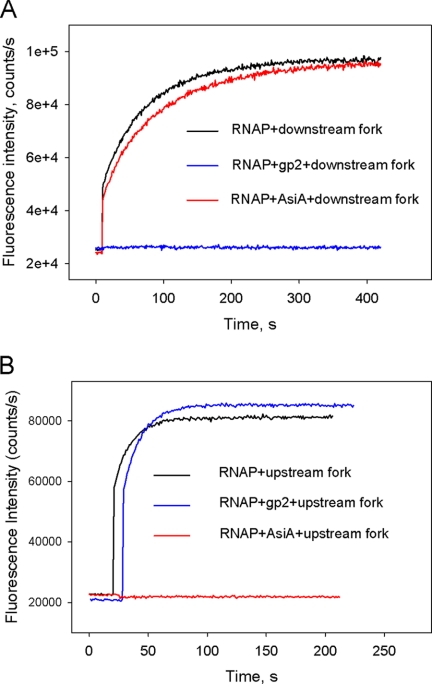
Further verification of a correct location for the downstream fork junction template was obtained by studying the effects of two phage transcription factors on RNAP binding to the downstream fork. T7 bacteriophage gp2 protein inhibits RPo formation by binding to the RNAP downstream jaw domain and interfering with RNAP interactions with downstream promoter DNA (21, 31). Another well studied inhibitor of open complex formation, T4 bacteriophage AsiA, binds to σ70 region 4 and inhibits transcription from −35 element-dependent promoters by preventing region 4 interactions with the −35 promoter element (27, 32). We detected no signal increase upon the addition of a downstream fork junction probe to RNAP beacon preincubated with gp2, whereas AsiA produced a negligible effect in a similar experiment (Fig. 3A). In contrast, AsiA, but not gp2, inhibited RNAP binding to the upstream fork junction probe, which was used as a control (Fig. 3B). Overall, data obtained with phage-encoded transcription factors support a view that the double-stranded segment of downstream fork junction templates occupies an RNAP site that is occupied by downstream DNA in RPo.
FIGURE 3.
The effect of phage T7 gp2 and phage T4 AsiA transcription initiation inhibitors on RNAP binding to downstream and upstream fork junction probes. The 8 nm [−12/+18][+3/+18] downstream fork junction shown in Fig. 1A or the 2 nm upstream fork junction shown in Fig. 2B was combined with RNAP beacon (1 nm) preincubated for 15 min with or without 0.2 μm T7 gp2 (A) or T4 AsiA (B) and increase in fluorescence was monitored.
Quantitative Dissection of the Downstream RNAP-Promoter Interaction Using Downstream Fork Junction Probes
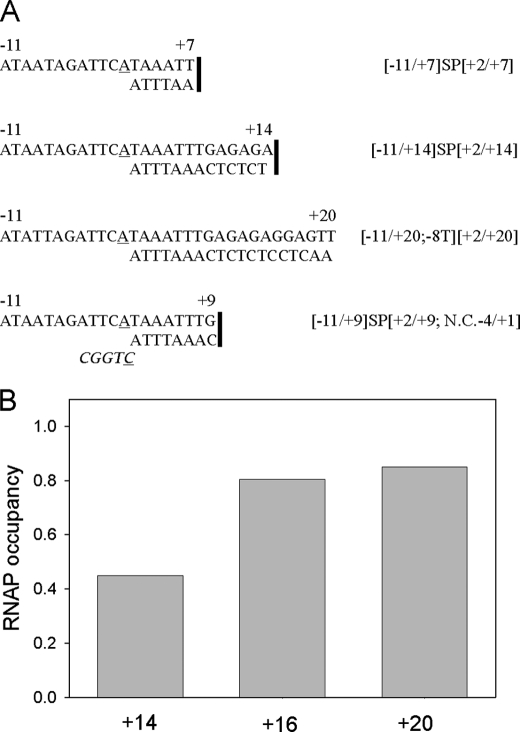
We evaluated the effect of the double-stranded segment length on RNAP-downstream fork junction affinity by using a set of probes whose upstream edges were the same (−11 for non-template and +2 for template strands), whereas the duplex segment length varied from 6 to 19 bp. The expectation was that increasing the length of the double-stranded segment should stimulate binding up to the point where the DNA duplex exits the complex. Representative probes are shown in Fig. 4A. To prevent dissociation of short duplexes, some template and non-template oligos were linked with a flexible ethylene oxide SP18 spacer, which allows the formation of B-form DNA (33). The Kd values for SP18-linked forks were obtained using a titration assay (similar to that shown in Fig. 2) and are presented in Table 1. As can be seen, the Kd of linked downstream fork junction probes gradually decreased as the duplex length increased from 6 to 13 bp. The highest, 6-fold stimulatory effect on RNAP binding was observed when the duplex was extended from base pair +8 to +9.
FIGURE 4.
RNAP affinity to downstream fork junction probes depends on the length of the duplex segment. A, shown are representative structures of downstream fork junctions probes used. The SP18 linker that joins non-template strand oligos with short template strand oligos in some probes is shown as a vertical line. In probe names, SP stands for the SP18 linker; N.C.−4/+1 stands for a non-complementary extension of the template strand from −4 to +1. B, RNAP occupancy was measured in samples containing 1 nm RNAP beacon and 2 nm [−11/+14;−8T][+2/+14], [−11/+16;−8T][+2/+16], and [−11/+20;−8T][+2/+20] probes abbreviated as +14, +16, and +20, respectively.
TABLE 1.
Dissociation constants for the binding of RNAP to downstream fork junction probes with varying lengths of downstream duplex segment
The Kd values presented are averages from 2-3 individual experiments; the error is ±15%.
| Probe | Kd |
|---|---|
| nm | |
| −11/+1 | 290 |
| [−11/+7]SP[+2/+7] | 61 |
| [−11/+8]SP[+2/+8] | 13 |
| [−11/+9]SP[+2/+9] | 1.9 |
| [−11/+10]SP[+2/+10] | 0.53 |
| [−11/+12]SP[+2/+12] | 0.30 |
| [−11/+14]SP[+2/+14] | <0.2 |
| −11/+1;−8T | 3700 |
| [−11/+14;−8T][+2/+14] | 3.0 |
| [−11/+16;−8T][+2/+16] | 0.55 |
| [−11/+20;−8T][+2/+20] | 0.38 |
The Kd values for fork junction probes with duplexes longer than 14 bp were lower than the limit for quantitative determination of the titration assay (Kd ∼ 0.2 nm). Therefore, affinities of probes with 13-, 15-, and 19-bp duplex segments were determined on the background of a non-consensus T in the −8 position, which weakened the interaction. The occupancy of RNAP beacons was measured in the presence of 2 nm concentrations of each of the probes using a competition binding assay as shown in Fig. 4B and supplemental Fig. S2 and described in the supplemental material. The values of Kd calculated from these measurements are shown in Table 1. The data demonstrate that extension of the duplex from base pair +16 to +20 leads to only a small (1.4-fold) increase of affinity, suggesting that essential RNAP contacts with downstream promoter duplex are located upstream of base pair +17. The total contribution of the +2/+20 duplex segment to downstream fork junction probe affinity can be evaluated by comparing the Kd values of the −11/+1;−8T oligo and the [−11/+20][+2/+20;−8T] fork junction; the duplex segment affords a dramatic (∼9000-fold) affinity increase (Table 1), corresponding to 5.5 kcal/mol gain of binding free energy.
We also estimated the strength of RNAP interaction with a segment of template strand adjacent to the transcription start point. To this end, RNAP beacon affinities to the [−11/+9]SP[+2/+9] fork junction and to a derivative bearing a 5-nucleotide-long overhang of the bottom strand (shown in Fig. 4A) were compared. The overhang increased affinity only ∼3-fold, indicating that the gain of free energy of binding afforded by this segment of the transcription bubble is relatively minor, at least in the context of downstream fork junction probes.
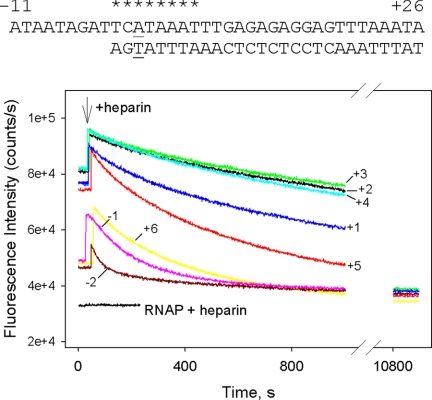
The Effect of the Position of the Double-strand/Single-strand Boundary on the Affinity of Downstream Fork Junction Probes for RNAP
The maximal affinity of upstream fork junctions to RNAP is reached when the double-stranded/single-stranded boundary is located at base pairs −11 or −12, i.e. in the same place as in RPo (8). We tested the effect of junction position on downstream fork junction affinity for an RNAP beacon using binding substrates based on a probe shown in Fig. 5 (asterisks indicate the junction position in different probes). Probes with upstream boundaries between +1 and +4 bound RNAP very strongly (Kd estimated at below 0.2 nm), which prevented comparisons of Kd values. A highly sulfated glycosaminoglycan heparin is known to compete with DNA for binding to RNAP. Heparin resistance is the hallmark of stable intermediates in the formation of the open complex (8, 25, 34). Thus, to access relative affinities of these probes, stabilities of RNAP complexes in the presence of heparin were compared. Heparin also promotes dissociation of free holo-RNAP to free σ70 and core RNAP (35, 36). Indeed, in the presence of heparin the signal of holo RNAP beacon was found to be equal to that of free (211Cys-TMR) σ70. In contrast, heparin had no effect on fluorescence of free (211Cys-TMR) σ70. Because of these effects, in the presence of heparin the fluorescence signal from RNAP beacon complexes with downstream fork junctions ultimately decayed to free (211Cys-TMR) σ70 levels. For reasons unknown, heparin also caused some increase in fluorescence intensity of the downstream fork junction complexes immediately after its addition (Fig. 5). This effect may indicate that RNAP can simultaneously bind heparin and downstream fork junctions, but the resulting complex is short-lived. The data in Fig. 5 demonstrate that rates of dissociation of RNAP complexes with probes having template strand edges at +2, +3, and +4 are very similar and comparatively slow, although fork junction with junction point at +1 dissociates 2.3-fold faster. Moving the edge of the duplex to base pairs −1, −2 or +5, +6 results in considerable acceleration of the decay.
FIGURE 5.
The effect of junction point position on the strength of RNAP interaction with downstream fork junction probes. Time-dependent changes of the fluorescence signal were measured upon the addition of 50 μg/ml heparin to RNAP beacon complexes with fork junctions probes whose junction points were located between positions −2 to +6. The concentrations of RNAP beacon and fork junctions were 1 and 2 nm, respectively, except that concentration of the [−11/+26][−2/+26] probe was 4 nm.
Further movement of the junction position downstream of base pair +6 resulted in probes that exhibited complicated kinetics of signal intensity change upon binding to the beacon, first reaching a maximal amplitude followed by intensity decrease (the behavior of the [−12/+26][+9/+26] probe shown in supplemental Fig. S3 is typical). We speculate that this behavior may be related to DNA scrunching, a process in which single-stranded DNA is generated during abortive initiation, as RNAP moves the downstream boundary of the transcription bubble without releasing upstream promoter contacts (37–40). The decrease in stability upon moving the edge of the duplex to base pairs +5 or +6 is also consistent with the “scrunching” hypothesis (40).
Overall, our data show that RNAP preferentially binds downstream forks with junctions located at base pairs +2 to +4, which corresponds to the location of downstream boundary of the transcription bubble in RPo. Importantly, this result indicates that RNAP binding to downstream forks with junction points located upstream of base pair +2 should be coupled with a shift of the equilibrium between duplexed and melted conformations of the DNA segment surrounding the transcription start point in the direction of melting.
Transcription Activity of Downstream Fork Junction Probes
The ability of various fork junctions to serve as transcription templates was determined using a beacon assay and an abortive transcription initiation assay. Some representative fork junctions used are shown in Fig. 6A. A considerable decrease in fluorescence intensity was observed when an RNAP beacon escapes from the promoter into elongation (9) as σ contacts with the −10 element are disrupted upon promoter escape (41). Transcription activity of downstream fork junction probes also can be monitored using this effect (Fig. 6B, supplemental Fig. S4). The initial transcribed segment of the T5 N25 promoter lacks C and G up to position +9; therefore, in the presence of ATP and UTP only, synthesis of RNA products up to 8 nucleotides in length can occur. As can be seen (Fig. 6B) comparatively low fluorescence generated by forks with junctions at −2, −3, and −4 increased upon the addition of ATP and UTP. We surmise that fluorescence enhancement is due to the increase in complex stability in the presence of a limited set of NTPs, as has been observed with other transcription complexes (42). As expected, the addition of GTP to reactions containing ATP and UTP led to a rapid 2–3-fold drop in fluorescence intensity (Fig. 6B) due to RNAP escape from the promoter. Fluorescence signal generated upon RNAP beacon interactions with fork junctions bearing 3–5 nucleotides non-complementary extensions of template strand upstream from the +2 position decreased rapidly upon the addition of NTPs, suggesting high transcription activity of these probes. Signals from a fork bearing a 5-nucleotide non-complementary extension (shown in Fig. 6A) dropped down particularly rapidly (supplemental Fig S4). As expected, a negligible decrease in signal intensity was detected upon the addition of NTPs to RNAP beacon-fork junction complexes preincubated with 1 μm rifampicin, an RNAP inhibitor that prevents synthesis of transcripts longer than three nucleotides long (supplemental Fig. S4).
The addition of all possible combinations of NTPs (up to 0.5 mm each) did not lead to a fluorescence intensity change in RNAP beacon complexes with a probe whose junction point was at base pair +1 (data not shown). The lack of signal change was not caused by the RNAP inability to melt the +1 base pair in this probe, as there was also no fluorescence change with similar probes harboring mismatches at positions +1, +2, or +1 and +2 (data not shown). Downstream fork junctions with upstream edges of the template strand located at base pairs +1 to −6, the −9 base pair, and a fully double-stranded [−12/+16] probe were tested in abortive transcription initiation assay. A high concentration of templates (0.5 μm) was used in these experiments to ensure RNAP binding to low affinity probes. RNAP catalyzed efficient synthesis of CpApU on fork junction probes with upstream edges of template strand located at base pairs −2, −3, and −4, whereas a fork junction having the junction point at −1 supported weak abortive initiation activity (Fig. 6C). Transcription activity progressively decreased upon shifting the junction point upstream from the −4 position. A derivative of the [−12/+16][−3/+16] probe in which highly conserved −11A and −7T bases were substituted for C showed no activity. In agreement with results obtained using the beacon assay, the fork junction bearing a 5-nucleotide-long non-complementary extension of the template supported a very high transcription level, whereas no abortive initiation product was detected with a fork junction with the bottom strand edge at +1 (Fig. 6C).
Effect of DNA Downstream from the −10 Element on the Binding of Double-stranded Promoter Fragments with RNAP
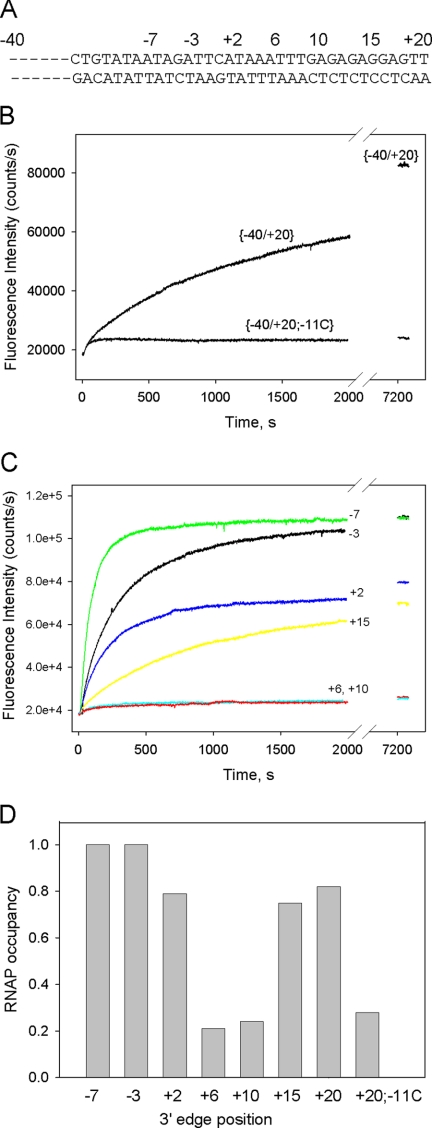
A set of double-stranded probes shown in Fig. 7A was used to assess the influence of promoter segments downstream of the −10 element on the affinity and kinetics of binding to RNAP. The parent probe was a [−40/+20] fragment of the T5 N25 promoter (supplemental Fig. S5). This fragment bound RNAP specifically, as a control bearing a C to A substitution at the critically important −11 position [−40/+20;−11C] generated a much lower signal than the wild-type probe (Fig. 7B). The binding of six probes based on [−40/+20] but truncated at base pairs −7, −3, +2, +6, +10, and +15 was assessed by measuring relative occupancies of 1 nm RNAP beacon by 2 nm concentrations of each probe (Fig. 7C). The occupancies were determined using a competition binding assay described in supplemental material. Extension of the downstream end from the −7 to the −3 base pair slowed down the rate of complex formation (Fig. 7C). The binding assay sensitivity was insufficient to detect the effect of this extension on affinity as the [−40/−7] and [−40/−3] fragments bound very strongly, in fact better than the parental [−40/+20] probe; no free beacon could be detected in the presence of probes [−40/−7] and [−40/−3], whereas 82% of the beacon was bound by probe [−40/+20] (Fig. 7D). The [−40/+2] and [−40/+15] fragments demonstrated comparable affinities (79 and 75% occupancy, respectively). The weakest binding was observed with the [−40/+6] and [−40/+10] probes (21 and 24% occupancy, respectively). In fact, the binding of these probes is similar to negative control (probe [−40/+20;-11C]) binding, which suggests that the measured affinities of [−40/+6] and [−40/+10] may partially reflect their nonspecific interaction with RNAP. The decrease in affinity caused by extension of the downstream end from the −3 to +2 base pairs is in agreement with previously observed inhibition of RNAP binding to double-stranded lacUV5 promoter fragments when the downstream boundary was extended from base pair −6 to the transcription start site (26). The inhibition of RNAP binding becomes more pronounced when the downstream boundary is moved from +2 to +6. This effect can be explained at least in part by the coupling of double-stranded promoter DNA binding with transcription bubble formation. Indeed, because the downstream edge of the transcription bubble is at the end of the [−40/+2] probe but is internally located in the [−40/+6] probe, melting of the former should be easier. Extension of the duplex beyond +10 results in a considerable improvement in binding. We propose that this effect is due to formation of a site necessary for interaction with the RNAP trough that binds downstream DNA. This interaction leads to a change in the DNA trajectory and promotes the formation of the transcription bubble.
FIGURE 7.
Measuring RNAP interaction with double-stranded promoter fragments using protein beacon assay. A, shown is the primary sequence of a double-stranded [−40/+20] probe, based on the sequence of the T5N25 promoter. Other probes are derivatives of [−40/+20] truncated downstream at positions −7, −3, +2, +6, +10, or +15. B, shown is time dependence of the increase in fluorescence upon mixing 1 nm (211Cys-TMR) σ70 holoenzyme with 2 nm [−40/+20] and a derivative bearing a C to A substitution at position −11 [−40/+20;−11C]. C, time dependence of fluorescence upon mixing 1 nm (211Cys-TMR) σ70 holoenzyme with 2 nm [−40/−7] (green), [−40/−3] (black), [−40/+2] (blue), [−40/+6] (cyan), [−40/+10] (red), and [−40/+15] (yellow). D, RNAP occupancies measured in samples containing 1 nm RNAP beacon and 2 nm probes truncated at the indicated positions. The occupancies were determined using a competition binding assay described in the supplemental material.
DISCUSSION
Here we quantitatively investigated downstream RNAP-promoter interactions using a new class of model promoter fragments, which we call a downstream fork junction. We found that these new DNA probes mimicked the corresponding downstream part of promoter DNA in all respects and function as the −10 element-dependent transcription templates. Specific affinity of short downstream fork junction templates to RNAP and real-time binding kinetics can be measured readily with the beacon assay. Thus, the downstream fork junctions are a useful new tool for studying the structure and function of the downstream RPo segment.
It has been previously proposed that RNAP contacts with an ∼20-nt-long promoter DNA duplex located downstream of the transcription start can play a significant role in stabilization of RPo (16, 43, 44). Our direct measurements of RNAP binding to a set of downstream fork junctions reveal that RNAP affinity to the downstream duplex is quite high. The data in Table 1 show that this tight binding apparently results from individually rather modest contributions of many RNAP-DNA contacts. Indeed, every consecutive 1-base pair extension of the duplex is estimated to increase the binding by less than 7-fold. The lack of strongly localized contacts may favor smooth RNAP sliding along the downstream DNA duplex during transcription.
We have investigated the effect of the double-stranded/single-stranded boundary position on downstream fork junction affinity to RNAP and found that the maximal affinity is reached when the boundary is located between bp +2 to +4. Therefore, the stability of RNAP complex with downstream fork junction is one of the factors that determines the position of the downstream boundary of the transcription bubble. Conversely, RNAP binding to downstream forks whose junction point is located upstream of position +2 should destabilize the DNA duplex segment surrounding the transcription start point. Specific transcription activity of downstream forks confirms the existence of such promoter DNA melting activity that is driven by RNAP interactions with the downstream junction.
In principle, strand separation in RPo can be maintained solely through strong RNAP interactions with single-stranded DNA, both template and non-template strands, within the bubble. However, the non-template strand fragment surrounding the transcription start point and the entire template strand part of the transcription bubble interact with RNAP comparatively weakly in the context of model promoter fragments (8, 9). Thus, the energy of these interactions may be insufficient to support the downstream propagation of promoter melting, in particular when the DNA sequence around the transcription start is G/C reach. Coupling melting with interaction of RNAP with the downstream promoter duplex, a highly energetically favorable process, may provide additional driving force for promoter melting.
Applying the beacon assay to a set of double-stranded promoter fragments allowed further evaluation of RNAP interactions with various downstream promoter segments (shown in Fig. 7). The [−40/−7] probe truncated just downstream of the −10 element showed most rapid and very tight binding. Extension of the downstream edge from base pair −7 to +2 inhibited the binding, in agreement with previous data (26), and extension from position +2 to +6 led to further inhibition. However, the binding affinity was recovered upon further extension of the downstream double-stranded segment, indicating that a critical length required for effective RNAP binding had been reached. These results are consistent with a view that RNAP binding to a promoter segment surrounding the transcription start site represents a barrier on the pathway to RPo (13, 14, 45, 46, 52). Favorable interactions established upon filling in the RNAP downstream DNA binding site with DNA of sufficient length overcome this barrier and thus lead to formation of RPo.
Some downstream fork junctions support highly efficient transcription with a start site located within the duplex DNA and matching a start site expected on double-stranded promoters. This is in contrast with results obtained with 3′ tailed templates composed of duplex DNA with a 3′ single-stranded extension. Such templates support transcription initiation by bacterial and eukaryotic RNA polymerases (47, 48), but initiation occurs at the boundary of single-stranded and double-stranded segments of the template (48). Neither in vitro transcription nor beacon assays revealed transcription activity of the [−12/+16][+1/+16] fork junction bearing no template strand bases upstream from +1, whereas a fork junction having the junction point at −1 showed only weak transcription activity. Similarly, E. coli σ70 RNAP holoenzyme transcription from a minimal M13 bacteriophage DNA fragment was found to be dependent on a three-nucleotide stretch upstream of the transcription start point (49). Shortening of this single-stranded extension abolished transcription (49). These data correlate with results on yeast RNAP III transcription from model non-transcribed strand promoter templates (50). The templates resemble downstream fork junctions used in this work but are much longer because they contain a non-transcribed strand comprising the entire upstream promoter segment and bear a long downstream duplex. Such templates bearing bottom strands truncated at the +1 position yielded a low amount of transcription product (50), similarly to the result that we observed with the [−12/+16][+1/+16] fork junction. This effect has been explained by the existence of requirements for anchoring of the transcribed strand of the transcription bubble relative to the enzyme active site (50). Transcription from downstream fork junctions may similarly depend on proper positioning of short bottom strand segment just upstream of the transcription start site in a specific RNAP site. Overall, the data presented here strongly argue that establishment of specific contacts between a segment of double-stranded DNA located 2–4 nucleotides downstream of the transcription initiation start point is a major force that governs the formation and controls stability of transcription-competent promoter complex in bacteria and may thus be a subject of genetic regulation. The use of model substrates mimicking the downstream portion of DNA in the open complex opens a way for functional dissection of these interactions and for direct structural analysis of RNAP interactions with downstream promoter DNA.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM64530. This work was also supported by a Molecular and Cell Biology Program grant from the Russian Academy of Sciences Presidium and Federal Program “Scientific and Scientific-Pedagogical Personnel of Innovative Russia 2009–2013,” State Contract 02.740.11.0771 (to K. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- RNAP
- RNA polymerase
- TMR
- tetramethylrhodamine.
REFERENCES
- 1. Buc H., McClure W. R. (1985) Biochemistry 24, 2712–2723 [DOI] [PubMed] [Google Scholar]
- 2. deHaseth P. L., Zupancic M. L., Record M. T., Jr. (1998) J. Bacteriol. 180, 3019–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sclavi B. (2009) in RNA Polymerases as Molecular Motors (Buc H., Strick T. eds) pp. 38–68, RCS Publishing, Cambridge, UK [Google Scholar]
- 4. Kirkegaard K., Buc H., Spassky A., Wang J. C. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 2544–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sasse-Dwight S., Gralla J. D. (1989) J. Biol. Chem. 264, 8074–8081 [PubMed] [Google Scholar]
- 6. Roberts C. W., Roberts J. W. (1996) Cell 86, 495–501 [DOI] [PubMed] [Google Scholar]
- 7. Marr M. T., Roberts J. W. (1997) Science 276, 1258–1260 [DOI] [PubMed] [Google Scholar]
- 8. Guo Y., Gralla J. D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mekler V., Pavlova O., Severinov K. (2011) J. Biol. Chem. 286, 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lim H. M., Lee H. J., Roy S., Adhya S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14849–14852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saecker R. M., Tsodikov O. V., McQuade K. L., Schlax P. E., Jr., Capp M. W., Record M. T., Jr. (2002) J. Mol. Biol. 319, 649–671 [DOI] [PubMed] [Google Scholar]
- 12. Sclavi B., Zaychikov E., Rogozina A., Walther F., Buckle M., Heumann H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4706–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murakami K. S., Masuda S., Darst S. A. (2002) Science 296, 1280–1284 [DOI] [PubMed] [Google Scholar]
- 14. Vassylyev D. G., Sekine S., Laptenko O., Lee J., Vassylyeva M. N., Borukhov S., Yokoyama S. (2002) Nature 417, 712–719 [DOI] [PubMed] [Google Scholar]
- 15. Bartlett M. S., Gaal T., Ross W., Gourse R. L. (1998) J. Mol. Biol. 279, 331–345 [DOI] [PubMed] [Google Scholar]
- 16. Ederth J., Artsimovitch I., Isaksson L. A., Landick R. (2002) J. Biol. Chem. 277, 37456–37463 [DOI] [PubMed] [Google Scholar]
- 17. Young B. A., Gruber T. M., Gross C. A. (2004) Science 303, 1382–1384 [DOI] [PubMed] [Google Scholar]
- 18. Davis C. A., Bingman C. A., Landick R., Record M. T., Jr., Saecker R. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7833–7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mukhopadhyay J., Das K., Ismail S., Koppstein D., Jang M., Hudson B., Sarafianos S., Tuske S., Patel J., Jansen R., Irschik H., Arnold E., Ebright R. H. (2008) Cell 135, 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rutherford S. T., Villers C. L., Lee J. H., Ross W., Gourse R. L. (2009) Genes Dev. 23, 236–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cámara B., Liu M., Reynolds J., Shadrin A., Liu B., Kwok K., Simpson P., Weinzierl R., Severinov K., Cota E., Matthews S., Wigneshweraraj S. R. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 2247–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murakami K. S., Masuda S., Campbell E. A., Muzzin O., Darst S. A. (2002) Science 296, 1285–1290 [DOI] [PubMed] [Google Scholar]
- 23. Matlock D. L., Heyduk T. (2000) Biochemistry 39, 12274–12283 [DOI] [PubMed] [Google Scholar]
- 24. Guo Y., Lew C. M., Gralla J. D. (2000) Genes Dev. 14, 2242–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsujikawa L., Tsodikov O. V., deHaseth P. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 3493–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fenton M. S., Gralla J. D. (2003) J. Biol. Chem. 278, 39669–39674 [DOI] [PubMed] [Google Scholar]
- 27. Severinova E., Severinov K., Fenyö D., Marr M., Brody E. N., Roberts J. W., Chait B. T., Darst S. A. (1996) J. Mol. Biol. 263, 637–647 [DOI] [PubMed] [Google Scholar]
- 28. Nechaev S., Severinov K. (1999) J. Mol. Biol. 289, 815–826 [DOI] [PubMed] [Google Scholar]
- 29. Ederth J., Isaksson L. A., Abdulkarim F. (2002) Mol. Genet. Genomics 267, 587–592 [DOI] [PubMed] [Google Scholar]
- 30. Mekler V., Kortkhonjia E., Mukhopadhyay J., Knight J., Revyakin A., Kapanidis A. N., Niu W., Ebright Y. W., Levy R., Ebright R. H. (2002) Cell 108, 599–614 [DOI] [PubMed] [Google Scholar]
- 31. Hesselbach B. A., Nakada D. (1975) Nature 258, 354–357 [DOI] [PubMed] [Google Scholar]
- 32. Colland F., Orsini G., Brody E. N., Buc H., Kolb A. (1998) Mol. Microbiol. 27, 819–829 [DOI] [PubMed] [Google Scholar]
- 33. McCullagh M., Zhang L., Karaba A. H., Zhu H., Schatz G. C., Lewis F. D. (2008) J. Phys. Chem. B 112, 11415–11421 [DOI] [PubMed] [Google Scholar]
- 34. Craig M. L., Tsodikov O. V., McQuade K. L., Schlax P. E., Jr., Capp M. W., Saecker R. M., Record M. T., Jr. (1998) J. Mol. Biol. 283, 741–756 [DOI] [PubMed] [Google Scholar]
- 35. Pfeffer S. R., Stahl S. J., Chamberlin M. J. (1977) J. Biol. Chem. 252, 5403–5407 [PubMed] [Google Scholar]
- 36. Brodolin K., Zenkin N., Mustaev A., Mamaeva D., Heumann H. (2004) Nat. Struct. Mol. Biol. 11, 551–557 [DOI] [PubMed] [Google Scholar]
- 37. Carpousis A. J., Gralla J. D. (1985) J. Mol. Biol. 183, 165–177 [DOI] [PubMed] [Google Scholar]
- 38. Hsu L. M. (2002) Biochim. Biophys. Acta 1577, 191–207 [DOI] [PubMed] [Google Scholar]
- 39. Kapanidis A. N., Margeat E., Ho S. O., Kortkhonjia E., Weiss S., Ebright R. H. (2006) Science 314, 1144–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Revyakin A., Liu C., Ebright R. H., Strick T. R. (2006) Science 314, 1139–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mooney R. A., Darst S. A., Landick R. (2005) Mol. Cell 20, 335–345 [DOI] [PubMed] [Google Scholar]
- 42. Gourse R. L. (1988) Nucleic Acids Res. 16, 9789–9809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wigneshweraraj S. R., Burrows P. C., Severinov K., Buck M. (2005) J. Biol. Chem. 280, 36176–36184 [DOI] [PubMed] [Google Scholar]
- 44. Kontur W. S., Saecker R. M., Capp M. W., Record M. T., Jr. (2008) J. Mol. Biol. 376, 1034–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Murakami K. S., Darst S. A. (2003) Curr. Opin. Struct. Biol. 13, 31–39 [DOI] [PubMed] [Google Scholar]
- 46. Kontur W. S., Saecker R. M., Davis C. A., Capp M. W., Record M. T., Jr. (2006) Biochemistry 45, 2161–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dedrick R. L., Chamberlin M. J. (1985) Biochemistry 24, 2245–2253 [DOI] [PubMed] [Google Scholar]
- 48. Helmann J. D., deHaseth P. L. (1999) Biochemistry 38, 5959–5967 [DOI] [PubMed] [Google Scholar]
- 49. Zenkin N., Severinov K. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4396–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schroder O., Geiduschek E. P., Kassavetis G. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 934–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lawson C. L., Swigon D., Murakami K. S., Darst S. A., Berman H. M., Ebright R. H. (2004) Curr. Opin. Struct. Biol. 14, 10–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saecker R. M., Record M. T. J., Dehaseth P. L. (2011) J. Mol. Biol. doi: 10.1016/j.jmb.2011.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.