Summary
The identification of the mechanism-of-action and therapeutic potential of bioactive small molecules remain considerable challenges in the field of drug discovery and chemical biology. Apart from traditional target identification techniques, new tools have emerged that can significantly aid mechanism elucidation efforts. The development of pattern matching algorithms that compare transcription profile data to analogous data on compounds with known cellular targets allows for mechanistic insights without the need to synthesize chemically modified probes. In addition, such methods can be used to connect small molecules to particular disease states, thus aiding the rational identification of candidate therapeutics. Another method with considerable potential is whole-genome RNAi screening, a technique that can identify critical upstream proteins involved in a small molecule’s mechanism-of-action. Several proof-of-concept studies using compounds with known cellular targets suggest this tool will enable mechanistic characterization of bioactive small molecules with unknown mechanisms. This review highlights recent successes in using these pattern matching and chemical genetic tools, with the goal of uncovering small molecule mechanisms and identifying therapeutic candidates for disease treatment.
Keywords: mechanism of action, target identification, transcript profiling, Connectivity Map database, RNAi, shRNA, whole-genome, disease, cancer
Introduction
The elucidation of the precise biological mode-of-action of small molecules remains a considerable challenge in chemical biology and drug discovery. Information about the mechanism of novel small molecules greatly facilitates the use of such compounds as tools for the investigation of fundamental biological processes. As our understanding of targets and biochemical pathways grows at an ever increasing rate, small molecule “tool” compounds may serve as a starting point for drug development efforts.1–3 Furthermore, at the dawn of the age of personalized medicine, knowledge of the target and availability of companion diagnostics will be critical in the development of novel therapies.4, 5
Traditional approaches using affinity “pull down” reagents have been successful in identifying the biological targets of multiple small molecules.6, 7 While quantitative proteomic mass-spectrometry techniques have significantly aided the distinguishing of low-abundance protein targets from high abundance non-specific small molecule-protein interactions,8 there remain fundamental drawbacks associated with affinity reagent based target identification strategies. Such projects are typically labor-intensive, with no guarantee of success. For success in affinity pull down experiments, the compound must have considerable affinity for its target (and ideally bind to the target covalently), and modified versions of the compound (e.g, with biotin attached) that retain biological activity need to be synthesized. In addition, identification of certain types of targets (i.e. membrane-bound proteins) remains a challenge.
Another set of tools that has recently emerged to facilitate rapid mechanism-of-action studies are genome-wide transcript profiling and RNAi screens. These methods can be successful regardless of compound affinity and are relatively easy to implement as they involve no synthetic overhead (no need for the construction of a probe reagent). Furthermore, even if the information returned does not immediately reveal the molecular target, the data set received is still highly informative. The focus of this review is to highlight how these approaches have been used to 1) aid compound mechanism-of-action studies, and 2) enable the identification of compounds that may be relevant to particular diseases/physiological processes.
Global Transcript Profiling to Assist Target Identification
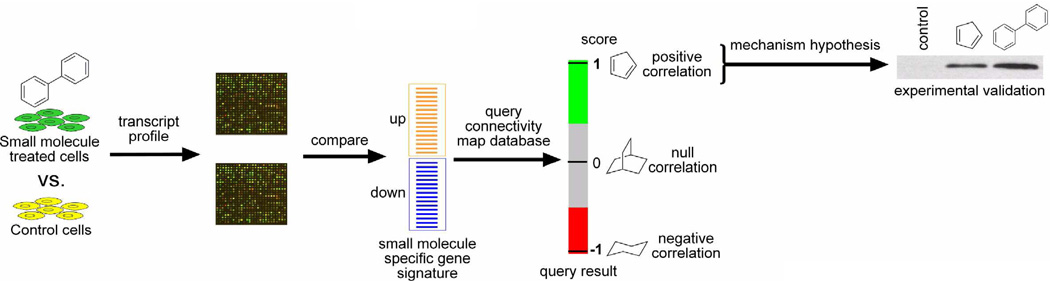
Whole-genome transcript profiling has emerged as a powerful tool to investigate the effects of small molecules on cells. While the list of altered transcripts may shed light on compound mechanism by analysis of the pathways affected, our understanding of pathways, how they interact with one another, and their consequences in various cell types is far from complete. Thus, it is typically not possible to identify the molecular target of a small molecule from a simple analysis of its transcript profile. However, pathway-independent approaches have recently emerged, in which the transcript profile data of a compound of interest is compared to analogous data that has been collected on hundreds of compounds with known molecular targets. The Connectivity Map database, developed at the Broad Institute, is a publicly accessible database comprising gene expression data of cells treated with small molecules.9 In most cases, the MCF-7 (breast cancer) cell line is used, but there are many instances from HL-60 (human leukemia), PC-3 (prostate cancer), and SK-MEL-5 (melanoma) cell lines.9, 10 The expression profiles were obtained by treating cells for six hours with the small molecules (to maximize primary effects) at various doses. The first version (build 01) of the Connectivity Map database contained 164 molecules with 453 signatures, while the second version (build 02) contains 1309 small molecules with 6100 signatures. As the database contains data for compounds that have considerable mechanistic characterization (FDA approved drugs and non-drug bioactive “tool” compounds), matches to the query signature would suggest possible mechanisms-of-action and molecular targets.9, 10
It should be emphasized that this is a pattern matching approach to target identification, with the pattern of up/down regulated transcripts of paramount importance, where (for Connectivity Map purposes) the identity of the transcripts is not taken into consideration. This process is outlined in Figure 1. Users obtain transcript profile data with their compound of interest in cells with a six hour treatment time. A “query signature” of up- and down-regulated genes is generated by the user, input into the Connectivity Map database, and scored based on how well compounds in the database match the input query. Scores of 1 represent a perfect match, 0 represent a null match and −1 indicates perfect anti-correlation to the input signature. The number of genes and the methods for selecting the genes (e.g. based on p value or fold change) for constructing the query signature are at the user’s discretion. Although (as mentioned) several cell lines are represented in the Connectivity Map database, for the purposes of mechanism identification, the use of MCF-7 may be advantageous (provided the compound elicits the desired phenotype) as the profiles in the Connectivity Map are predominantly obtained in this cell line.
Figure 1.
Gene expression signatures of small molecule with unknown mechanisms may be used to query the Connectivity Map database, to identify compounds that elicit similar transcriptional effects. Parts of the figure were adapted from Lamb et al.9
Other pattern matching techniques have been utilized in an attempt to group small molecules according to their mode-of-action.11, 12 Perhaps the most well-known method is the COMPARE analysis, in which the pattern of toxicity of compounds to the NCI 60 cell line panel is compared.11, 13 Compounds that have similar patterns of toxicity are highly correlated in the COMPARE algorithm; such a result suggests the compounds have a similar mechanism-of-action, and indeed COMPARE has been successfully used in target identification studies.14, 15 In practice, however, the nature of the cytotoxicity screens is that there is a significant amount of variability, even amongst replicate evaluations of the same compound. Thus, the correlations returned from COMPARE are usually not high and may therefore limit the utility of this method.
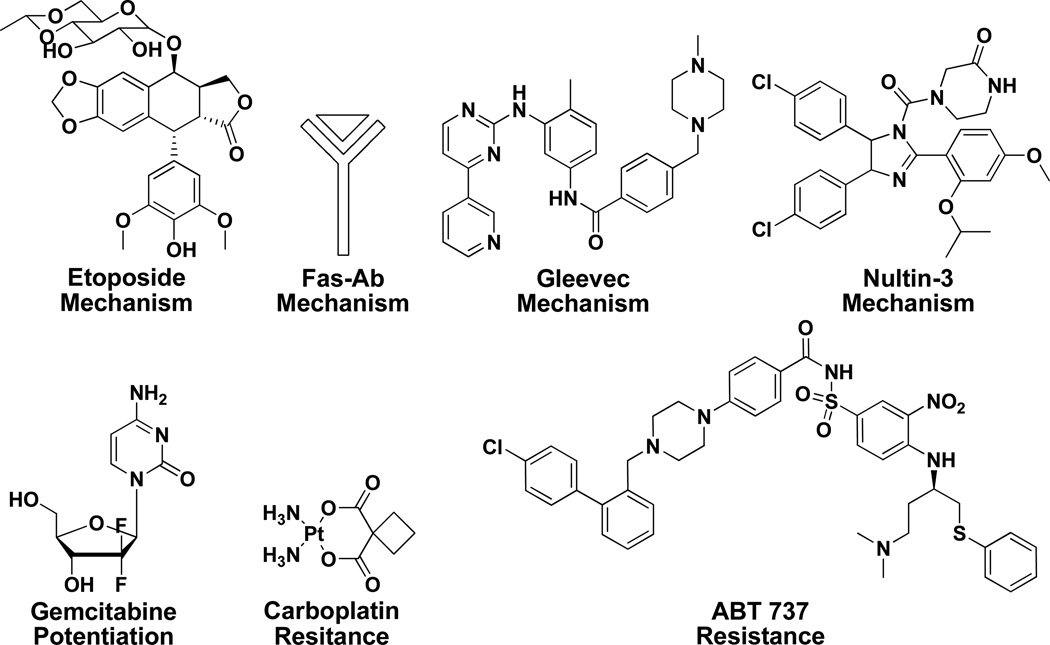
The Connectivity Map database has been utilized in mode-of-action studies of several small molecules. Connectivity Map experiments with six compounds (shown in Figure 2) are described below. These examples have been selected as they correspond to situations that many chemical biologists find themselves in, namely, through high-throughput screening a compound has been identified that elicits an interesting cellular phenotype, but the precise mode-of-action and cellular target is unknown. If the transcriptional profile induced by the compound of interest matches compounds in the Connectivity Map, then downstream biological experiments to confirm the mechanism are typically straightforward.
Figure 2.
Small molecules (listed with their biological mode-of-action) whose mechanisms were identified with the aid of the Connectivity Map database.
Epoxy anthraquinone
The gene expression signature of an epoxy anthraquinone (EAD, Figure 2) in neuroblastoma SK-N-As cells had topoisomerase I/II inhibitors and DNA intercalators as 7 of the top 10 Connectivity Map correlations.16 Follow up experiments confirmed EAD induces DNA alkylation and inhibits topoisomerase I and II in vitro.16
Droxinostat
Droxinostat (Figure 2) was identified from a screen for sensitizers of resistant cells to CH-11 Fas-activating antibody.17 Through use of the Connectivity Map it was revealed that the transcript profile of droxinostat in the resistant cell line matched the HDAC inhibitors vorinostat and trichostatin A. Through a series of cell-based and in vitro assays, it was determined that droxinostat, which contains a hydroxamic acid moiety like other pan-HDAC inhibitors, is a novel HDAC inhibitor that is selective for HDAC3, HDAC6, and HDAC8.17 While other pan-HDAC inhibitors are also able to sensitize resistant cells to Fas induced apoptosis,18 HDAC knockout mice studies suggest isoform selective inhibitors may alleviate undesired toxicity.19–21
15-Deoxy-9Δ-Prostaglandin J2 (PGJ2)
The Connectivity Map has also enabled further mechanistic characterization of molecules in the database itself. 15-Deoxy-Δ-prostaglandin J2 (PGJ2, Figure 2), an endogenous anti-inflammatory cytokine present in the Connectivity Map (Build 02), was found to match the gene expression signature of 3 out of 4 novel small molecules that downregulate HIF2α.22 PGJ2 was found to inhibit translation of HIF2α protein (rather than transcription) through an mTOR-independent manner not affecting global translation. Using luciferase reporter constructs it was discovered that the PGJ2-induced decrease of HIF2α expression is dependent on a 50 bp segment of the 5’-UTR that contains an iron regulatory element (IRE). Downregulation of the IRE binding iron regulatory protein IRP1, but not IRP2, mitigated PGJ2-induced downregulation of HIF2α suggesting an IRP1-dependent mechanism. This study illustrates the first example of an endogenous small molecule regulator of HIF2α translation and assigns a novel mechanism as to how PGJ2 elicits an anti-inflammatory effect.
Gedunin and Celastrol
A high-throughput cell-based screen identified gedunin and celastrol (Figure 2) as inhibitors of androgen receptor (AR)-activated signaling in prostate cancer.23 Transcript profiling of these agents and query of the Connectivity Map database yielded strong matches to the heat shock protein 90 (HSP90) inhibitors, geldanamycin, 17-dimethylamino-geldanamycin and 17-allylaminogeldanamycin.23 As the androgen receptor is a client of HSP90 and inhibition of HSP90 induces AR degradation, gedunin and celastrol may inhibit AR signaling by inhibiting the HSP90 pathway. Consistent with this hypothesis, treatment of LNCaP prostate cancer cells with celastrol and gedunin induced the downregulation of AR and several other HSP90 client proteins (FLT3, EGFR, and BCR-ABL1). Further experiments showed that while celastrol and gedunin inhibit HSP90 ATP binding activity in cells, they do not compete with the ATP-binding site and are therefore mechanistically distinct from the geldanamycin-based HSP90 inhibitors.
Akt Inhibitor Off-target Effects
One of the challenges of drug development is the identification of off-target effects of biologically active small molecules. Inhibition of the Akt pathway is an attractive anticancer strategy for which various compounds have been developed, including SH-5 (Figure 2) and SH-6 (not shown in Figure 2), which are competitive inhibitors of phosphatidyl inositol (3,4,5)-trisphosphate (PIP3). PIP3 is an endogenous product of PI3 kinases that activates AKT signaling. The off-target effect of these inhibitors was investigated by Krech and coworkers using human colorectal cancer cell lines in growth media containing 10% FBS.24 Under these conditions SH-5 and SH-6 are unable to inhibit the AKT pathway but induced the formation of binuclear cells in SW480 cells by inhibiting abscission, the last step of cytokinesis (cell line specific effect). Subjection of the gene transcript profile to Connectivity Map analysis revealed matches to compounds that interfere with PIP2 (Resveratrol), Ca2+ (Thirodiazine) and Protein Kinase C (PKC) signaling (Rottlerin). As PIP2 hydolysis and Ca2+ levels affect the production of diacylglycerol (DAG), a metabolite required for PKC signaling, it was hypothesized that binucleation is related to PKC inhibition. Similar to SH-5 and SH-6, treatment of SW480 cells with resveratrol and rotterlin resulted in the production of the binuclear cells thus supporting interference of PKC signaling as a cause of the AKT-independent binucleation phenotype induced by SH-5 and SH-6.
Using the Connectivity Map to Identify Compounds that Modulate Physiological or Disease Phenotypes
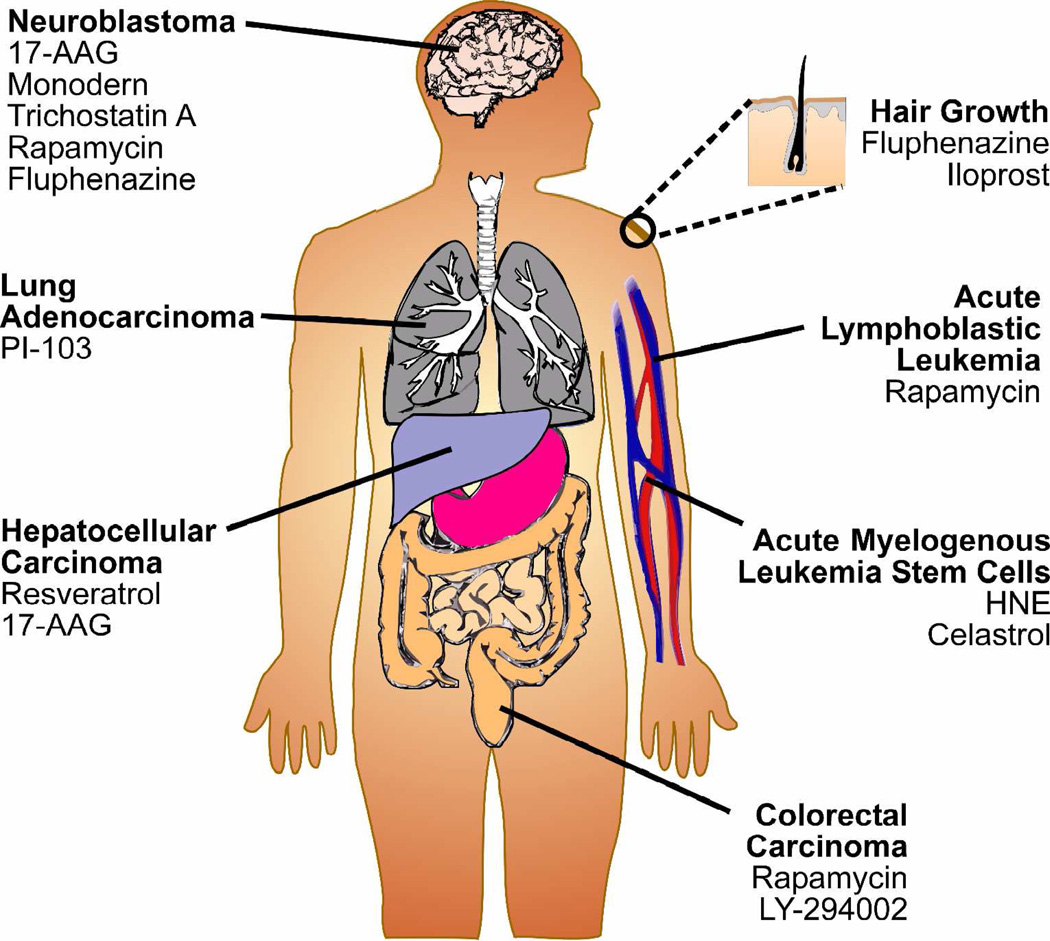
Another major challenge in drug discovery is determining which small molecules may be best suited to treat a disease or modulate a physiological phenotype. Using disease- or physiological process-specific signatures as the query, small molecules in the Connectivity Map database that have positive or negative correlations can be readily identified (Figure 3).9, 10 Small molecules whose signatures negatively (i.e. anti-) correlate with disease-specific signatures may prove useful in reversing disease-specific phenotypes and aid the rational selection of candidate therapeutics for further investigation. On the other hand, compounds whose signatures positively correlate with phenotype-specific signatures may be able to induce the phenotype (Figure 3). Highlighted below are notable examples where this strategy has aided the identification of therapeutic candidates for the treatment of various cancers, and compounds that can modulate other physiological processes, such as hair growth (Figure 4).
Figure 3.
Phenotype-specific genes may be used to query the Connectivity Map database to identify small molecules that may be able to induce or reverse the phenotype. Parts of the figure were adapted from Lamb et al.9
Figure 4.
The use of the Connectivity Map database has led to the identification of several small molecule therapeutic candidates for the treatment of cancers, and compounds that can modulate physiological processes such as hair growth.
Hepatocellular carcinoma vascular invasion
The identification of compounds that modulate hepatocellular carcinoma (HCC) vascular invasion was performed using an invasion-specific 73-gene signature obtained from comparing 38 HCC tumors displaying the vascular invasion phenotype to 43 tumors lacking in this phenotype.25 Input of the 47 up- and 26 down-regulated genes into the Connectivity Map identified two small molecules (resveratrol, and 17-AAG) with significant anti-correlation to the input signature (p value <0.05). These agents were able to inhibit the invasive nature of two HCC cell lines HepG2 and PLC/PRF-5 at non-cytotoxic concentrations (10 µM). Follow up experiments confirmed these compounds revert the expression levels of several genes involved in invasion, adhesion, and angiogenesis.
Neuroblastoma
In order to identify candidate therapeutics for neuroblastoma, an integrative meta-analysis of gene copy number was performed using expression profiles of 146 primary neuroblastoma tumors and normal fetal neuroblasts to construct a list of 132 tumor-specific genes.26 Five of the top six compounds proposed by the Connectivity Map were able to significantly reduce cell viability in five neuroblastoma cell lines; compounds included HSP90 inhibitors (17-AAG and monodern), an HDAC inhibitor (trichostatin A), an mTOR inhibitor (rapamycin) and a phenotriazene (fluphenazine). While the toxicity of these compounds may not be restricted to neuroblastoma, this approach provides a rational basis for the selection of therapeutic candidates for neuroblastoma clinical trials.
Acute myelogenous leukemia stem cells (AML-SC)
The role of cancer stem cells (CSC) in blood cancers (such as acute myelogenous leukemia, AML) has been well characterized.27 Selective toxicity against AML stem cells (AML-SCs) has been observed in vitro using parthenolide (PTL).28, 29 The ability of this agent to kill AML-SCs has been attributed to its ability to simultaneously inhibit NF-κB signaling and induction of oxidative stress.28 PTL’s selective toxicity against AML-SC versus normal hematopoietic stem and progenitor cells served as a tool to identify novel compounds capable of inducing AML-SC death.30 By obtaining gene expression signatures of CD34+ AML cells (isolated from 12 patients) treated with PTL and compared to non-treated cells, a 150-gene signature was used to query the GEO and Connectivity Map databases for agents inducing similar signatures. In addition to agents known for their anti-AML-SC activity (MG-132 and prostaglandin), celastrol was identified in both database searches and three other compounds, hemin, HNE, and gedunin were identified from the GEO database search. Similar to PTL, treatment of CD34+/CD38− AML cells with celastrol and HNE gave selective toxicity at the bulk, progenitor, and stem cell levels versus CD34+/CD38− normal bone marrow cell, normal myeloid, and erythroid cell populations. These agents were shown to simultaneously inhibit NF-κB and induce Nrf2- oxidative stress response. This study further enforced the link between simultaneous NF-κB and oxidative stress induction for efficacy against AML-SCs and identified two novel agents (celastrol and HNE) that exhibit toxicity towards AML-SCs.
Colorectal cancer with microsatellite instability
Microsatellite instability may be a suitable marker to categorize colorectal cancers, as approximately 15% of sporadic colorectal cancers have high frequency instability31, 32 and exhibit differential sensitivity to chemotherapy.33, 34 By analyzing the signature of 38 microsatelite stability (MSS) and 13 high frequency microsatellite instability (MSI-H) tumors, a list of 71 probes were found to be differentially expressed between the two groups.35 This signature was used to query the Connectivity Map, and inhibitors of HDAC, HSP90 and PI3K-mTOR populated the best hit compounds that exhibited anti-correlation to the input signature.35 Subsequent toxicity experiments identified the mTOR inhibitors, rapamycin and LY-294002, to exhibit selective toxicity (4-fold and 1.5-fold respectively) towards MSI-H cell lines versus MSS cell lines in culture while the HDAC and HSP90 inhibitors did not demonstrate selective toxicity. This approach identified MSI-H colorectal cancers as more sensitive to the effects of mTOR inhibition than MSS colorectal cancers, warranting further investigation of the link between the microsatellite instability phenotype and the PI3K pathway.
Aggressive lung adenocarcinoma
Many adenocarcinomas are treated identically even though they exhibit a high degree of morphological and clinical diversity.36 In a study by Ebi and coworkers, gene set enrichment analysis (GSEA) was performed on transcript profile data from two different studies that categorized lung carcinoma based on patient post-surgical survival rates.37 Statistically significant gene sets were identified that correlated with fatal outcome which included a gene set downregulated by rapamycin (an mTOR inhibitor), and those associated with glucose, leucine and glutamine metabolism, processes that are also regulated by the mTOR/PI3K pathway. The Connectivity Map database was queried and revealed several compounds that target the mTOR and PI3K pathway as those that would be able to negatively regulate genes specific to poor prognosis. While several PI3K inhibitors and the mTOR inhibitor rapamycin were active against adenocarcinoma in toxicity assays, a particular PI3K inhibitor, PI-103, was found to exhibit toxicity that correlated with cumulative deregulation scores across the various cell lines investigated in culture. This further supported a functionally relevant connection between adenocarcinoma and the PI3K/mTOR pathway. Transcript profiling of treated cells confirmed PI3K inhibitors were able to revert the GSEA derived signature associated with poor prognosis.
Glucocorticoid resistance
The efficacy of dexamethasone, a glucocorticoid used for the treatment of acute lymphoblastic leukemia (ALL) in children, is compromised by resistance.38 A gene expression signature associated with dexamethasone resistance was obtained by comparing bone-marrow leukemia cells from patients that exhibit resistance to those derived from dexamethasone sensitive patients.39 Using this signature to query the Connectivity Map led to the identification of the mTOR inhibitor rapamycin as an agent capable of reverting resistance in cell culture assays using the dexamethasone-resistant CEM-cl lymphoid cell line.39 This sensitization was dependent on mTOR inhibition and is thought to be mediated by downregulation of the antiapoptotic protein, MCL1. The Connectivity Map thus helped to identify rapamycin as a therapeutic candidate for clinical evaluation in dexamethasone-resistant ALL patients.
Hair growth modulators
Although the Connectivity Map database has been primarily used in the field of cancer, it has also been useful in the identification of compounds that can influence normal physiological processes unrelated to cancer. The discovery of novel hair-growth inducers by Ishimatsu-Tsuji and coworkers utilized the day 2 and day 4 gene expression signature of mouse dorsal skin cells treated topically with cyclosporine A, a known inducer of hair growth.40 Using the 2 day signature to query the Connectivity Map, fluphenazine was identified as agent that induced hair-growth upon topical application. Iloprost, a compound that matched the 4 day signature, was discovered to enhance fluphenazine-induced hair growth. This study demonstrated how investigating different phases of a physiological process may be used to identify phase-specific small molecule modulators.
The use of RNAi screens to aid the identification of mechanism of action
One of the fruits of the genomic age has been the development of powerful genetic techniques that enable interrogation of protein function. Genome-wide, pathway-focused (targeting apoptosis, kinases and phosphatases), and custom made-to-order RNAi libraries are commercially available in siRNA and shRNA formats. Whereas siRNA elicits short-term knockdown (2–5 days) and delivery is limited to cell types that may be easily transfected, shRNA libraries may be transduced (using viral particles) into cell types that are difficult to transfect, enabling stable knockdown and facilitating screens of longer duration.
Commercial RNAi libraries are available in pooled or well formats. The experimental setups for pool- and well-based experiments are illustrated in Figure 5; these experiments can implicate a protein in mediating the effect of a compound. For example, the cells transfeced/transduced with the interfering RNA can be treated with an anticancer compound at a concentration know to give quantitative cell death. Any surviving cells, therefore, must have an mRNA knocked down that was important in the cell death induced by the compound.
Figure 5.
An overview of the (A) pooled- and (B) well-based format RNAi screen experiments.
Well-based experiments may require high-throughput screening intensive methods (depending on the library size), are typically more costly, but allow more freedom in the phenotype being investigated and are suited to both positive and negative selection screens. A variety of read-outs may be employed, including high-content imaging, flow cytometry, ELISA, and cell viability assays (Figure 5B). As an alternative to plate based assays, well-format libraries may be printed onto microarrays upon which adherent cells may be grown, and results observed using high content imaging techniques. In comparison to well-format RNAi, pooled-libraries are more cost-effective and screens can often be performed by a single researcher; however, the phenotypes that may be investigated are limited. In addition, the experimental protocol employed in pooled-library toxicity screens may bias them towards positive selection, although negative selection screens are possible if conditions are optimized.41
Pooled shRNA screens require transduction of cells such that the shRNA multiplicity of infection (MOI) is less than 1. The cells are split into two pools and one is treated with vehicle while the other is treated with the small molecule (Figure 5A). For screens involving toxic molecules, selection is typically performed over 2–3 weeks. The total genomic RNA is isolated, the shRNA constructs are amplified by PCR, and the half-hairpin sequences are then hybridized to complementary DNA microarrays. By comparing the levels of shRNA constructs in treated versus non-treated cells, it is possible to identify shRNAs that are enriched (positive selection) or depleted (negative selection) as a result of small molecule treatment (Figure 5A).
While genome-wide RNAi libraries may allow a systematic and unbiased approach to interrogating cell biology, there are pitfalls associated with RNAi such as lack of target-specificity and knockdown efficiency, which ultimately lead to false positives and false negatives, respectively. Both pooled and well-based libraries typically contain multiple constructs targeting the same gene, increasing the likelihood that a protein will be effectively targeted; this redundancy also helps discern likely true-positives from false-positives arising from off-target effects. Furthermore, as shRNA/siRNA constructs continue to be validated for knockdown efficacy, revised construct design will continue to improve the quality of RNAi libraries. Despite this, it is not always clear if mRNA knockdown is sufficient to modulate the cellular level of a protein such that its function is affected. Protein longevity, expression levels, and contribution to functional pathways in a particular cell line are factors that affect the success of RNAi-based experiments.
Despite these limitations, the use of RNAi libraries may significantly aid investigations of the mechanism of small molecules. As described below, several proof-of-principle studies have investigated compounds with known modes of action using RNAi screens. A particular appeal of this technique is the ability to identify key proteins that are the compound’s direct target, or regulators of the target. Evidence suggests that upstream proteins are more likely to be identified, as proteins considerably downstream of the direct target may be unable confer long-term survival, especially given that selections are performed over 2–3 weeks. Of course, if a protein’s down-regulation itself induces toxicity, this may preclude its identification as a small molecule target. A list of such proteins that may elude identification may be inferred from the study by Luo and coworkers, which identified 268 essential genes common to 12 cell lines.41 These genes were enriched for pathways involving ribosomal proteins, mRNA processing and splicing, translation factors and proteasome degradation.41
Etoposide
The ability of whole genome pooled shRNA barcode screens to reveal the molecular target of the small molecule etoposide (Figure 6) was demonstrated by Luo and coworkers using a 45,000 shRNA library that targeted 9,500 genes.41 H82 small-cell lung cancer cells were transduced with the shRNA library and treated with 1.7 µM etoposide for three weeks (10 independent transductions and treatments). This dose of etoposide was sufficient to kill >99% of treated cells within 7 days. PCR amplification of the shRNA constructs of surviving cells versus control untreated cells revealed 3 of the 5 shRNA constructs targeting topoisomerase II, the known target of etoposide, with >40 fold enrichment.
Figure 6.
Small molecules and proteins that have been investigated in RNAi screens.
Gleevec
A positive selection screen (similar to etoposide) for gleevec-treated K562 cells transduced with a shRNA library was performed over 21 days by Luo and coworkers.41 A dose of 125 nM gleevec (Figure 6) was used, which was sufficient to kill >90% of the treated cells within the first 7 days of treatment. Bcr-Abl, the known target of gleevec, is essential for the survival of K562 cells and thus precluded its identification through this positive selective screen. PTPN1 was one of the genes identified to confer resistance. Encouragingly, this gene was also identified in a shRNA screen to identify genes that confer survival to Bcr-Abl RNAi in K562 cells.41 PTPN1 is a negative regulator of Bcr-Abl and shRNAs targeted against PTPN1 were able to increase phosphorylation of Bcr-Abl thus conferring resistance to gleevec. The shRNA screen thus identified a Bcr-Abl negative regulator that is involved in gleevec induced cell death.
Fas-Activating Antibody (Fas-Ab)
The identification of proteins involved in Fas-Ab induced Jurkat T cell death using a pooled whole genome shRNA positive selection screen was also performed by Luo and coworkers.41 The knockdown of FAS, FADD, and CASP8 genes were found to confer resistance to Fas-Ab. Fas, Fadd and caspase 8 form the death-inducing signaling complex (DISC) that is critical to initiating the extrinsic apoptotic pathway upon binding of Fas-Ab to the Fas receptor. Two novel genes, ARID1A and CBX1, were also identified in this screen whose knockdown prevented caspase-8 activation thus identifying their role upstream of caspase-8 activation in Fas-Ab induced apoptosis. In a separate study, Tsujji and coworkers identified CASP8, BID and FAS as genes whose knockdown prevented Fas-Ab mediated cell death in the D98/AH2 (derived from HeLa) cell line.42 The inability to identify FADD in this study may be due to technical differences in the experiment/RNAi library or cell line-specific effects.
Nultin-3
In order to identify genes that play a role in nutlin-3 mediated cell death, a barcode pooled shRNA screen was conducted in MCF-7 (wild-type p53) breast cancer cells.43 Nutlin-3 (Figure 6) is a small molecule inhibitor of the MDM2-p53 protein-protein interaction. MDM2 binding to p53 inhibits p53-dependent apoptosis by suppressing transcriptional activation of p53 in response to DNA damage, exporting p53 out of the nuclease and targeting p53 for proteasomal degradation due to E3 ligase activity of MDM2. The shRNA screen was performed by treating cells with Nutlin-3 for 14 days at a concentration of 4 µM, was sufficient to induce cell cycle arrest without inducing apoptosis. Despite the use of non-toxic concentrations of nutlin-3, shRNAs that enabled cell proliferation would be amplified under these conditions. Several proteins involved in the p53-pathway were identified which included p53 itself, a p53 binding protein (53BP1), and a MDM2 target known to be a transcriptional activator of p53 (hnRNPK). Further shRNA experiments determined p53BP1 enables p53 transcriptional activity but does not affect induction of p53 protein. As 53BP1 is a component of the ATM-CHK-53BP1 pathway that induces p53 upon activation by DNA double strand breaks, it may explain why cancer cells are more susceptible to nutlin-3 as normal fibroblast cells exhibited considerably less 53BP1-containing nuclear foci than MCF-7 breast cancer cells. This data suggests that combination of nutlin-3 with DNA damaging anticancer agents should be avoided as this may result in undesired toxicity towards normal cells.
Gemcitabine combination therapy
In addition to mechanism elucidation, RNAi libraries may aid the identification of effective combination therapies. Gemcitabine (Figure 6), a nucleoside analog that replaces cytidine during DNA replication and prevents the attachment of other nucleosides, is commonly used for the treatment of pancreatic cancer. Using the well-format Qiagen kinase siRNA library (2 siRNA constructs per kinase targeting 572 kinases), a screen was performed to identify kinases whose knockdown would potentiate gemcitabine toxicity in the MIA PaCa-2 pancreatic cancer cell line.44 Although several kinases were identified that modestly increased toxicity, two siRNA constructs targeting CHK1 were identified that were able to sensitize cells to gemcitabine by >10 fold. Using the small molecule inhibitors of CHK-1, SB 218078 and PD 407824, a 2.6 and 3.5 fold potentiation of gemcitabine was observed, respectively. CHK1, a protein kinase that was known to be activated upon DNA damage by gemcitabine, serves to induce cell cycle arrest and allow DNA repair and inhibition of CHK1 induces apoptosis by preventing DNA repair. The fact that CHK2 did not potentiate gemcitabine suggests that CHK2, unlike CHK1, is not involved in gemcitabine-induced DNA damage response and sheds further light on the mechanism of gemcitabine and functional differences between CHK kinases.
Carboplatin resistance
Apart from investigating the mechanism and potential therapeutic combination strategies, RNAi screens have aided the identification of mechanisms of resistance to the effects of small molecules. The resistance of ovarian cancer cell lines to carboplatin (Figure 6) was investigated in a siRNA screen targeting 90 genes associated with resistance to carboplatin/paclitaxel combination therapy.45 The candidate genes comprised of 39 genes enriched (>2 fold) in post-chemotherapy tumors versus primary tumors and 51 genes enriched (>2 fold) post-chemotherapy versus primary chemoresistant tumors. The screen identified the ENPP2 gene which encodes for autotoxin, a protein with lysophoslipase D activity, as a contributor to carboplatin resistance. Autotaxin produces the pro-survival factors, lysophosphatidic acid (LPA) and sphingosine 1-phosphate (S1P). Experiments using siRNA and a chemical inhibitor confirmed autotoxin inhibition accelerates carboplatin-induced apoptosis in ovarian cancer cells thus supporting the role of autotaxin in conferring resistance to carboplatin.
ABT-737 Resistance
ABT-737 (Figure 6), a potent inhibitor of the antiapoptotic proteins Bcl-2, Bcl-XL and Bcl-w has demonstrated toxicity against small-cell lung carcinomas (SCLC) in culture and preclinical models.46 Some SCLC cell lines and cell lines derived from other cancers are, however, resistant to the effects of ABT-737.46 To identify the mechanism of ABT-737 resistance Lin and coworkers conducted a well-format siRNA screen against 4000 druggable genes using the NCI-H196 SCLC derived cell line.47 RNAi against FGFR2, TNFRSF13B, and PRDM13 were initially identified to confer sensitivity however testing of multiple constructs against these genes revealed the effects to be off-target as no correlation was observed between sensitivity to ABT-737 and the level of target gene knockdown.47 One of the major contributions to off-target effects arises from the complementation of nucleotides 2–8 of the antisense siRNA strand, otherwise known as the ‘seed’ region, and the 3’ UTR of unintended targets.48, 49 A BLAST analysis of the ‘seed’ regions of effective FGFR2, TNFRSF13B, and PRDM13 siRNA constructs suggested an overwhelmingly large number (343) of possible off-targets. However from this list, a particular Bcl-2 antiapoptotic protein, Mcl-1 was identified which had implicated in conferring resistance to ABT-737 in other studies. Mcl-1 is Bcl-2 family member protein that is not susceptible to inhibition by ABT-737. Further experiments confirmed that the effective siRNAs of FGFR2, TNFRSF13B, and PRDM13 induced off-target silencing of Mcl-1 and siRNAs against Mcl-1 were able to confer sensitivity to ABT-737. This study exemplifies one of the pitfalls associated with RNAi-based screening, and emphasizes the necessity of thorough experimental validation of candidate ‘hit’ genes.
Outlook
Pattern matching of small molecule transcript profiles to databases such as the Connectivity Map, which contain profiles of biologically characterized small molecules, is a powerful tool in mode-of-action studies. In addition to its utility in target identification, the Connectivity Map serves as a convenient platform to connect diverse disease or physiological processes to small molecules. The examples highlighted in this review suggest that a single small molecule may be capable of affecting diverse pathways and thus find applications in multiple biological contexts. It is important to note that although the Connectivity Map database contains data from a handful of cancer cell lines, it has proven useful in connecting small molecules to physiological processes and cancers whose cell types are not present in the database. As future versions will incorporate more compounds and transcript profiles obtained from RNAi experiments, the database will strengthen our ability to connect small molecules, diseases and genes. The ease of this method, and the vast amount of data generated from each experiment, are considerable strengths of this technique.
While the Connectivity Map may aid mechanism identification when there are compounds of similar mechanism in the database, it may not be able to address biologically active molecules with unprecedented mechanisms. Several proof-of-principle studies using compounds with known mechanisms have demonstrated that genome-wide shRNA screens are capable of identifying direct targets or upstream proteins involved in the mechanism. Thus, the use of this technology holds considerable promise in aiding characterization of novel small molecules with unknown mechanisms. In addition, RNAi screens may aid the identification of combination therapeutics and resistance mechanisms. The use of the genome expression and genome-wide shRNA screens compliment traditional target identification techniques, and are likely to significantly expedite mechanism elucidation and the applications of small molecules in medicinal chemistry and chemical biology.
Key Sentence.
The emergence of the Connectivity Map database and RNA interference screens as chemical biology tools have aided efforts to elucidate mechanisms of action and identify the therapeutic potential of bioactive small molecules.
Acknowledgment
We are grateful to the National Institutes of Health (R01-CA120439) for supporting some of our work in the area of transcript profiling.
References
- 1.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 2.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 3.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 4.de Bono JS, Ashworth A. Translating cancer research into targeted therapeutics. Nature. 2010;467:543–549. doi: 10.1038/nature09339. [DOI] [PubMed] [Google Scholar]
- 5.Olopade OI, Grushko TA, Nanda R, Huo D. Advances in breast cancer: pathways to personalized medicine. Clin Cancer Res. 2008;14:7988–7999. doi: 10.1158/1078-0432.CCR-08-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leslie BJ, Hergenrother PJ. Identification of the cellular targets of bioactive small organic molecules using affinity reagents. Chem Soc Rev. 2008;37:1347–1360. doi: 10.1039/b702942j. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Hu Y. Identifying the cellular targets of bioactive small molecules with activity-based probes. Curr Med Chem. 2010;17:3030–3044. doi: 10.2174/092986710791959747. [DOI] [PubMed] [Google Scholar]
- 8.Ong S-E, Schenone M, Margolin AA, Li X, Do K, Doud MK, Mani DR, Kuai L, Wang X, Wood JL, Tolliday NJ, Koehler AN, Marcaurelle LA, Golub TR, Gould RJ, Schreiber SL, Carr SA. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc Natl Acad Sci U S A. 2009;106:4617–4622. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 10.Lamb J. The Connectivity Map: a new tool for biomedical research. Nat Rev Cancer. 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- 11.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 12.Muroi M, Kazami S, Noda K, Kondo H, Takayama H, Kawatani M, Usui T, Osada H. Application of proteomic profiling based on 2D-DIGE for classification of compounds according to the mechanism of action. Chem Biol. 2010;17:460–470. doi: 10.1016/j.chembiol.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 13.Holbeck SL, Collins JM, Doroshow JH. Analysis of Food and Drug Administration-approved anticancer agents in the NCI60 panel of human tumor cell lines. Mol Cancer Ther. 2010;9:1451–1460. doi: 10.1158/1535-7163.MCT-10-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stefely JA, Palchaudhuri R, Miller PA, Peterson RJ, Moraski GC, Hergenrother PJ, Miller MJ. N-((1-benzyl-1H-1,2,3-triazol-4-yl)methyl)arylamide as a new scaffold that provides rapid access to antimicrotubule agents: synthesis and evaluation of antiproliferative activity against select cancer cell lines. J Med Chem. 53:3389–3395. doi: 10.1021/jm1000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mezencev R, Kutschy P, Salayova A, Curillova Z, Mojzis J, Pilatova M, McDonald J. Anticancer properties of 2-piperidyl analogues of the natural indole phytoalexin 1-methoxyspirobrassinol. Chemotherapy. 2008;54:372–378. doi: 10.1159/000152027. [DOI] [PubMed] [Google Scholar]
- 16.Gheeya J, Johansson P, Chen QR, Dexheimer T, Metaferia B, Song YK, Wei JS, He J, Pommier Y, Khan J. Expression profiling identifies epoxy anthraquinone derivative as a DNA topoisomerase inhibitor. Cancer Lett. 2010;293:124–131. doi: 10.1016/j.canlet.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood TE, Dalili S, Simpson CD, Sukhai MA, Hurren R, Anyiwe K, Mao X, Suarez Saiz F, Gronda M, Eberhard Y, MacLean N, Ketela T, Reed JC, Moffat J, Minden MD, Batey RA, Schimmer AD. Selective inhibition of histone deacetylases sensitizes malignant cells to death receptor ligands. Mol Cancer Ther. 2010;9:246–256. doi: 10.1158/1535-7163.MCT-09-0495. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe K, Okamoto K, Yonehara S. Sensitization of osteosarcoma cells to death receptor-mediated apoptosis by HDAC inhibitors through downregulation of cellular FLIP. Cell Death Differ. 2005;12:10–18. doi: 10.1038/sj.cdd.4401507. [DOI] [PubMed] [Google Scholar]
- 19.Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, Olson EN. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang S, Young BD, Li S, Qi X, Richardson JA, Olson EN. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell. 2006;126:321–334. doi: 10.1016/j.cell.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 22.Zimmer M, Lamb J, Ebert BL, Lynch M, Neil C, Schmidt E, Golub TR, Iliopoulos O. The connectivity map links iron regulatory protein-1-mediated inhibition of hypoxia-inducible factor-2a translation to the anti-inflammatory 15-deoxy-delta12,14-prostaglandin J2. Cancer Res. 2010;70:3071–3079. doi: 10.1158/0008-5472.CAN-09-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hieronymus H, Lamb J, Ross KN, Peng XP, Clement C, Rodina A, Nieto M, Du J, Stegmaier K, Raj SM, Maloney KN, Clardy J, Hahn WC, Chiosis G, Golub TR. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Krech T, Thiede M, Hilgenberg E, Schafer R, Jurchott K. Characterization of AKT independent effects of the synthetic AKT inhibitors SH-5 and SH-6 using an integrated approach combining transcriptomic profiling and signaling pathway perturbations. BMC Cancer. 2010;10:287. doi: 10.1186/1471-2407-10-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braconi C, Meng F, Swenson E, Khrapenko L, Huang N, Patel T. Candidate therapeutic agents for hepatocellular cancer can be identified from phenotype-associated gene expression signatures. Cancer. 2009;115:3738–3748. doi: 10.1002/cncr.24417. [DOI] [PubMed] [Google Scholar]
- 26.De Preter K, De Brouwer S, Van Maerken T, Pattyn F, Schramm A, Eggert A, Vandesompele J, Speleman F. Meta-mining of neuroblastoma and neuroblast gene expression profiles reveals candidate therapeutic compounds. Clin Cancer Res. 2009;15:3690–3696. doi: 10.1158/1078-0432.CCR-08-2699. [DOI] [PubMed] [Google Scholar]
- 27.Ishikawa F, Yoshida S, Saito Y, Hijikata A, Kitamura H, Tanaka S, Nakamura R, Tanaka T, Tomiyama H, Saito N, Fukata M, Miyamoto T, Lyons B, Ohshima K, Uchida N, Taniguchi S, Ohara O, Akashi K, Harada M, Shultz LD. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol. 2007;25:1315–1321. doi: 10.1038/nbt1350. [DOI] [PubMed] [Google Scholar]
- 28.Guzman ML, Swiderski CF, Howard DS, Grimes BA, Rossi RM, Szilvassy SJ, Jordan CT. Preferential induction of apoptosis for primary human leukemic stem cells. Proc Natl Acad Sci U S A. 2002;99:16220–16225. doi: 10.1073/pnas.252462599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guzman ML, Rossi RM, Karnischky L, Li X, Peterson DR, Howard DS, Jordan CT. The sesquiterpene lactone parthenolide induces apoptosis of human acute myelogenous leukemia stem and progenitor cells. Blood. 2005;105:4163–4169. doi: 10.1182/blood-2004-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassane DC, Guzman ML, Corbett C, Li X, Abboud R, Young F, Liesveld JL, Carroll M, Jordan CT. Discovery of agents that eradicate leukemia stem cells using an in silico screen of public gene expression data. Blood. 2008;111:5654–5662. doi: 10.1182/blood-2007-11-126003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomaki P, Chadwick RB, Kaariainen H, Eskelinen M, Jarvinen H, Mecklin JP, de la Chapelle A. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338:1481–1487. doi: 10.1056/NEJM199805213382101. [DOI] [PubMed] [Google Scholar]
- 32.Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Nakagawa H, Sotamaa K, Prior TW, Westman J, Panescu J, Fix D, Lockman J, Comeras I, de la Chapelle A. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 33.Carethers JM, Chauhan DP, Fink D, Nebel S, Bresalier RS, Howell SB, Boland CR. Mismatch repair proficiency and in vitro response to 5-fluorouracil. Gastroenterology. 1999;117:123–131. doi: 10.1016/s0016-5085(99)70558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fallik D, Borrini F, Boige V, Viguier J, Jacob S, Miquel C, Sabourin JC, Ducreux M, Praz F. Microsatellite instability is a predictive factor of the tumor response to irinotecan in patients with advanced colorectal cancer. Cancer Res. 2003;63:5738–5744. [PubMed] [Google Scholar]
- 35.Vilar E, Mukherjee B, Kuick R, Raskin L, Misek DE, Taylor JM, Giordano TJ, Hanash SM, Fearon ER, Rennert G, Gruber SB. Gene expression patterns in mismatch repair-deficient colorectal cancers highlight the potential therapeutic role of inhibitors of the phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin pathway. Clin Cancer Res. 2009;15:2829–2839. doi: 10.1158/1078-0432.CCR-08-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeuchi T, Tomida S, Yatabe Y, Kosaka T, Osada H, Yanagisawa K, Mitsudomi T, Takahashi T. Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J Clin Oncol. 2006;24:1679–1688. doi: 10.1200/JCO.2005.03.8224. [DOI] [PubMed] [Google Scholar]
- 37.Ebi H, Tomida S, Takeuchi T, Arima C, Sato T, Mitsudomi T, Yatabe Y, Osada H, Takahashi T. Relationship of deregulated signaling converging onto mTOR with prognosis and classification of lung adenocarcinoma shown by two independent in silico analyses. Cancer Res. 2009;69:4027–4035. doi: 10.1158/0008-5472.CAN-08-3403. [DOI] [PubMed] [Google Scholar]
- 38.Tissing WJ, Meijerink JP, den Boer ML, Pieters R. Molecular determinants of glucocorticoid sensitivity and resistance in acute lymphoblastic leukemia. Leukemia. 2003;17:17–25. doi: 10.1038/sj.leu.2402733. [DOI] [PubMed] [Google Scholar]
- 39.Wei G, Twomey D, Lamb J, Schlis K, Agarwal J, Stam RW, Opferman JT, Sallan SE, den Boer ML, Pieters R, Golub TR, Armstrong SA. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell. 2006;10:331–342. doi: 10.1016/j.ccr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 40.Ishimatsu-Tsuji Y, Soma T, Kishimoto J. Identification of novel hair-growth inducers by means of connectivity mapping. Faseb J. 2010;24:1489–1496. doi: 10.1096/fj.09-145292. [DOI] [PubMed] [Google Scholar]
- 41.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, Mermel C, Barbie DA, Awad T, Zhou X, Nguyen T, Piqani B, Li C, Golub TR, Meyerson M, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsujii H, Eguchi Y, Chenchik A, Mizutani T, Yamada K, Tsujimoto Y. Screening of cell death genes with a mammalian genome-wide RNAi library. J Biochem. 2010;148:157–170. doi: 10.1093/jb/mvq042. [DOI] [PubMed] [Google Scholar]
- 43.Brummelkamp TR, Fabius AW, Mullenders J, Madiredjo M, Velds A, Kerkhoven RM, Bernards R, Beijersbergen RL. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat Chem Biol. 2006;2:202–206. doi: 10.1038/nchembio774. [DOI] [PubMed] [Google Scholar]
- 44.Azorsa DO, Gonzales IM, Basu GD, Choudhary A, Arora S, Bisanz KM, Kiefer JA, Henderson MC, Trent JM, Von Hoff DD, Mousses S. Synthetic lethal RNAi screening identifies sensitizing targets for gemcitabine therapy in pancreatic cancer. J Transl Med. 2009;7:43. doi: 10.1186/1479-5876-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vidot S, Witham J, Agarwal R, Greenhough S, Bamrah HS, Tigyi GJ, Kaye SB, Richardson A. Autotaxin delays apoptosis induced by carboplatin in ovarian cancer cells. Cell Signal. 2010;22:926–935. doi: 10.1016/j.cellsig.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 46.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, Joseph MK, Kitada S, Korsmeyer SJ, Kunzer AR, Letai A, Li C, Mitten MJ, Nettesheim DG, Ng S, Nimmer PM, O'Connor JM, Oleksijew A, Petros AM, Reed JC, Shen W, Tahir SK, Thompson CB, Tomaselli KJ, Wang B, Wendt MD, Zhang H, Fesik SW, Rosenberg SH. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 47.Lin X, Morgan-Lappe S, Huang X, Li L, Zakula DM, Vernetti LA, Fesik SW, Shen Y. 'Seed' analysis of off-target siRNAs reveals an essential role of Mcl-1 in resistance to the small-molecule Bcl-2/Bcl-XL inhibitor ABT-737. Oncogene. 2007;26:3972–3979. doi: 10.1038/sj.onc.1210166. [DOI] [PubMed] [Google Scholar]
- 48.Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, Marshall WS, Khvorova A. 3' UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 49.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]