Abstract
Objective
MicroRNA (miRNA) play important roles in vascular biology, but the regulation of endothelial specific miRNA is not well characterized. MiR-126 is highly expressed in endothelial cells, and it regulates angiogenesis and vascular inflammation. Here we show that the transcription factors Ets-1 and Ets-2 regulate miR-126 expression.
Methods and Results
A genomic region between −71 and −100 bp upstream of the miR-126 transcriptional start site is critical for transactivation of the gene containing miR-126. This genomic region contains a potential Ets binding site. Mutations within the Ets binding site block transactivation, and Ets-1 and Ets-2 interact with this critical genomic region. Knockdown of endogenous Ets-1 and Ets-2 decreases miR-126 expression. Finally, knockdown of miR-126 alters regulation of an Ets-1 target gene.
Conclusions
Taken together, these data show that the transcription factors Ets-1 and Ets-2 play a key role in controlling the expression of miR-126, and suggest that miR-126 may mediate some of their vascular effects.
MiRNA in endothelial cell biology
MicroRNA (miRNA) are short, non-coding RNA molecules that post-transcriptionally regulate the expression of target genes, and play a role in diverse cellular, physiological, and pathophysiological processes.1–8 Endogenously expressed miRNA molecules are transcribed in the nucleus by RNA polymerase II as long primary transcripts (pri-miRNA), and then further processed by the nuclear enzyme Drosha into shorter precursor species (pre-miRNA). The pre-miRNA can then be exported from the nucleus by the ran-GTP dependent dsRNA binding protein, exportin-5.9–11 Once in the cytoplasm, the ribonuclease Dicer processes the pre-miRNA into the mature 20–24 nt miRNA species. The mature miRNA forms a complex with the RNA induced silencing complex (RISC) and represses the expression of target mRNA transcripts via binding to the 3′ UTR.
MiRNA may play an important role in angiogenesis.12–15 Endothelial cells express a set of miRNA at high levels.16–19 Specific endothelial miRNA may regulate angiogenesis.20, 21 For example, miR-221 inhibits endothelial migration in vitro by repressing c-kit,18 miR-130 induces endothelial migration in vitro through inhibition of the homeobox gene HoxA5,22 and let-7f may decrease expression of the angiogenesis inhibitor thrombospondin-1.17 Additional in vivo evidence supports the theory that miRNA regulate angiogenesis. For example, global knockdown of the miRNA processing enzyme Dicer causes embryonic lethality associated with impaired embryonic angiogenesis, and endothelial specific deletion of Dicer decreases post-natal angiogenesis in vivo.17, 23, 24 Furthermore, miR-296 increases tumor angiogenesis by indirectly boosting endothelial levels of VEGFR2.25 Thus specific miRNA play an important role in angiogenesis.
MiR-126 in endothelial cell biology
MiR-126 is one of the miRNA most abundantly expressed in endothelial cells, and it is most prominent in the heart, lung, and other highly vascularized murine tissue.19, 26, 27 Knockdown of miR-126 in zebrafish leads to hemorrhage during embryogenesis.28 Furthermore, targeted deletion of miR-126 in mice leads to partial embryonic lethality with hemorrhagic defects during development.27 The mice that do survive show increased mortality after myocardial infarction and inadequate wound healing due to impaired neovascularization.27 MiR-126 promotes angiogenesis in part through repression of Spred-1 and PIK3R2.27, 28 We have also shown that miR-126 plays a role in vascular inflammation through the regulation of the adhesion molecule VCAM-1.19 Although miR-126 plays a prominent role in vascular biology, the transcription factors that regulate miR-126 in endothelial cells have not been defined.
Ets-1 and Ets-2 in endothelial cell biology
The ETS (E26 Transformation-specific Sequence) factors are a family of transcription factors that share a highly conserved DNA binding domain and regulate cell development, senescence, death, and tumorigenesis.29–31 The conserved Ets domain is a winged helix-turn-helix DNA binding domain that interacts with a core GGAA/T consensus sequence found within genomic regions of target genes. Several members of the ETS family are expressed in endothelial cells and have been shown to play a role in vasculogenesis, angiogenesis, inflammation, and remodeling.29, 31, 32 For example, Ets-1−/− mice have impaired inflammatory responses to angiotensin II,33 Tel1−/− mice show vascular defects in the developing yolk sack,34 Fli−/− have defects in megakaryopoiesis, homeostasis, and vascular integrity,35 Net −/− mice show a defect in vascular integrity and lymphatic development, and removal of both Ets-1 and Ets-2 in the developing chicken leads to impaired cardiac development.36 Finally, a recent set of studies with mice lacking both Ets-1 and Ets-2 reveal overlapping functions of both factors in regulating embryonic angiogenesis.37, 38 Here we show that the Ets family members Ets-1, and Ets-2 induce the expression of miR-126 in endothelial cells.
Methods
Reagents
Human umbilical vein endothelial cells (HUVEC) and endothelial cell basal media (EBM-2) and growth factors were purchased from Cambrex (East Rutherford, NJ). RNA oligonucleotides for pre-miRNA were purchased from Integrative DNA technologies (IDT, Coralville, IA). SiRNA oligonucleotides were purchased from Applied Biosystems (Foster City, CA) and Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies to Ets-1 and Ets-2 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Luciferase Reporter plasmid pGL3Basic was purchased from Promega. Plasmids expressing Ets family members were obtained from the lab of Dr. Peter Oettgen (Beth Israel Deaconess Medical Center, Boston, MA).
Northern Analysis
Total RNA was harvested from HUVEC using Trizol reagent (Invitrogen, Carlsbad, California) according to the manufacturer’s protocol and ran on a 15% TBE-Urea gel (Invitrogen, Carlsbad, California) and transferred to a Nytran nylon transfer membrane (Schleicher & Schuell, Keene, N.H.). A [32P]-probe was synthesized from miR-126 anti-sense oligonucleotides (IDT, Coralville, IA) and hybridized using UltraHyb reagents (Ambion, Austin, TX) according to the manufacturer’s protocol.
Quantitative RT-PCR (qRT-PCR)
Total RNA was harvested from endothelial cells, HeLa, and HEK293 cells as described above. RNA was diluted and reverse transcribed using microRNA RT Kit for miR-126, miR-126* and RNU66 (Applied Biosystem, Foster City, CA). microRNA RT products were then amplified using the TaqMan microRNA kit (Applied Biosystem, Foster City, CA). For EGFL7 qRT-PCR, RNA was isolated as above. RNA was diluted and reverse transcribed using High Capacity cDNA kit (Applied Biosystems, Foster City, CA). Following RT, EGFL7 and GAPDH transcripts were amplified using the TaqMan assay (Applied Biosystems, Foster City, CA).
Generation of −1670 promoter construct
Primers were generated to clone the region upstream of the EGFl7 transcriptional start site. The 5′ primers included from −1670: GCCTGCTGCCAACTTGTTCT; from −1081: AGGGAAATGGGGGTGTCCCA; from −659: AGCTCTTTTAGGGGAGAGAG; from −483: TGCTGTGTCACACACATCTG; from −383: CAAAGATGCAGCAGCTCCCTT; from −150: CTCAGCCTCCTGTTTGTCCGA; and from −71: ATCCCAATCCCGATTACCCA. The 3′ primer at +100 was: ATGGACCCTAGCCCTTGCTG. The PCR products were then inserted into the multiple cloning site of the promoter expression vector pGL3Basic upstream of the cDNA for Photinus luciferase (Promega, Madison, WI). Each vector, along with various siRNA (Santa Cruz biotechnology, Santa Cruz, CA), and the RL-TK (Promega, Madison, WI) Renilla luciferase vector was transfected into various primary cells and cell lines using the Lipofectin reagent (Invitrogen, Carlsbad, California) according to the manufacturer’s protocol. Cells were cultured for 2–3 d and assayed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI). We used a computer to search for potential transcription factor binding sites to the EGFL7 5′UTR. (http://www.cbrc.jp/research/db/TFSEARCH.html, Parallel Application TRC Laboratory, RWCP Japan).
Cell Culture and Transfection
Human umbilical vein endothelial cells (HUVEC) were obtained from Cambrex (East Rutherford, NJ) and grown in EBM-2 media supplemented with essential growth factors. HUVEC were transfected with siPort NeoFX reagent (Ambion, Austin, TX) and siRNA or with Lipofectin reagent (Invitrogen) and plasmids.
Western Blotting
Western blotting was performed as described previously 39. In brief, HUVEC were lysed with Laemmli Sample Buffer (Bio-Rad, Hercules, CA), boiled, fractionated on a 7.5% Tri-HCl gel (Bio-Rad) and transferred to a nitrocellulose membrane, which was hybridized with antibodies to Ets-1, Ets-2, Erk, and GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA).
Chromatin Immunoprecipitation
Chromatin proteins were cross linked to chromatin with formaldehyde and sheared into 400 – 1000 bp fragments. Nucleoprotein complexes were immunoprecipitated using antibody to Ets-1, Ets-2, or control IgG antibody. The precipitated DNA fractions were analyzed by Quantitative-PCR for the presence of the miR-126 proximal regulatory region encompassing the EBS1 and EBS2 (region −150 to + 100 bp). The regulatory region from −1670 to −1070 was used as a negative control. Input DNA was used as a positive control.
Statistical analyses
Data are expressed as the mean ± S.D. Statistical comparisons were made between two groups with the t-test and between multiple groups by analysis of variance. A P value < 0.05 was considered significant.
Results
Endothelial cells express miR-126/miR-126* and the host gene EGFL7
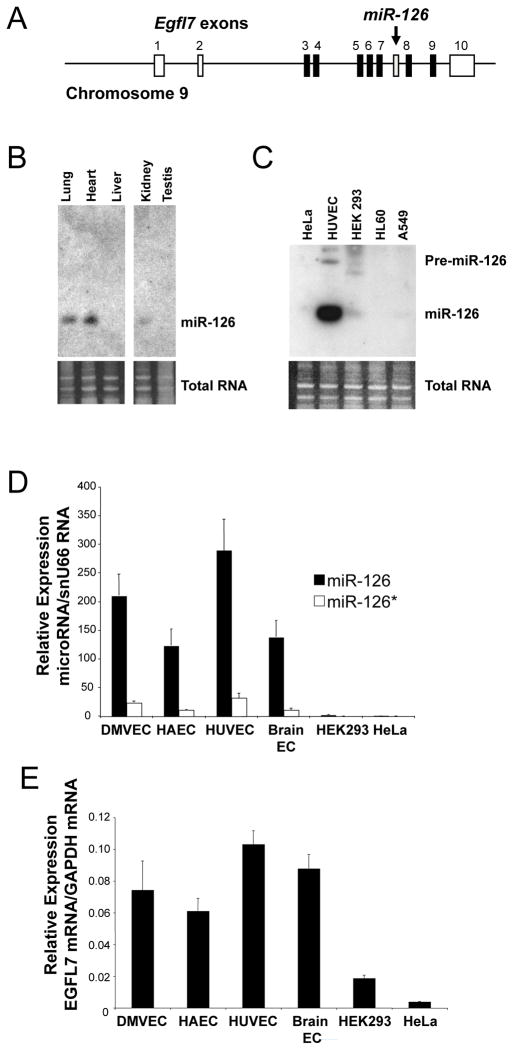
In order to explore the regulation of miR-126 and its host gene EGF-like domain 7 (Egfl7) (Fig. 1A), we first confirmed its expression in endothelial cells. We harvested RNA from a variety of murine tissues, endothelial cells, and control cell lines and analyzed it by Northern blot for miR-126 and by qRT-PCR for miR-126, miR-126* and EGFL7 expression. MiR-126, miR-126* and their host gene EGFL7 share a similar expression profile: highly vascular tissues such as lung and heart express miR-126 (Fig. 1B). Endothelial cells from umbilical vein, aorta, skin, and brain all express miR-126 and miR-126* (Fig. 1C–D). In contrast, non-endothelial cell lines such as HeLa, HL60, and A549 cells do not express detectable levels of these transcripts. Furthermore, endothelial cells express the parent gene Egfl7 but HeLa cells do not (Fig. 1E). Taken together, these data suggest that endothelial cells express higher levels of miR-126 and EGFL7 than non-endothelial cells.
Figure 1.
Endothelial cells express miR-126 and its host gene Egfl7.
(A) Schematic of Egfl7/miR-126 locus. The host gene encoding EGFL7 also encodes the microRNA miR-126. Arrow points to intronic location of hsa-miR-126. White boxes: untranslated Egfl7 exons. Black boxes: translated Egfl7 exons. Gray box: miR-126 locus. (B) Tissue expression of miR-126 by Northern blotting. Total RNA was harvested from various mouse tissues and analyzed by Northern blotting for miR-126 RNA (top). Ethidium bromide staining of total RNA (below). (C) Cell expression of miR-126 by Northern blotting Total RNA was harvested from various cell types and analyzed by Northern blotting for miR-126 RNA (top). Ethidium bromide staining of total RNA (below). (D) Cell expression of miR-126 by Q-RT-PCR. Total RNA was harvested from various cell types and analyzed by quantitative-RT-PCR for miR-126 and miR-126* expressed relative to snu66 RNA (n = 2 ± S.D.). (E) Cell expression of EGFL7 mRNA by Q-RT-PCR. Total RNA was harvested from various cell types and analyzed by Q-RT-PCR for EGFL7 and GAPDH mRNA.
A 1.7 kb luciferase promoter construct recapitulates the miR-126 expression profile
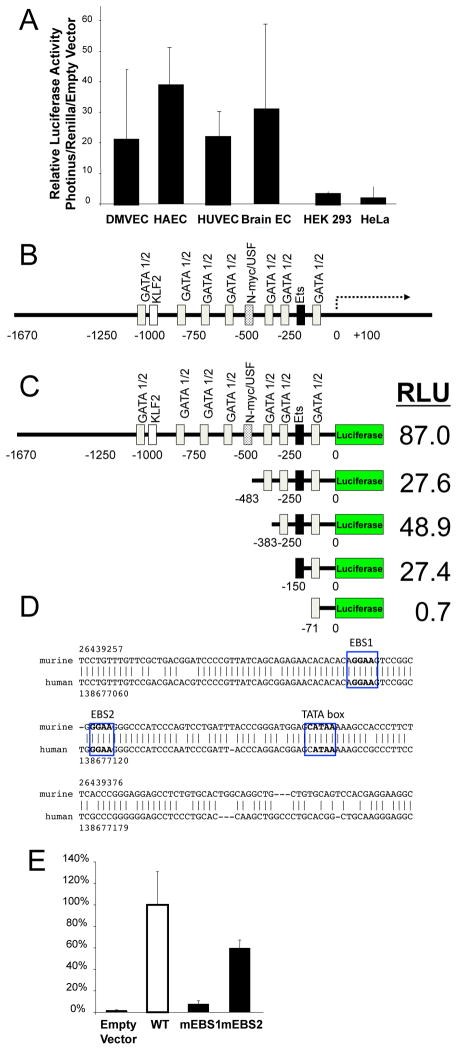
We next defined a region upstream of the Egfl7/miR-126 locus that regulates expression of miR-126. Using PCR, we amplified 1.7 Kb upstream of the Egfl7/miR-126 transcriptional start site and cloned it into the luciferase expression plasmid pGL3-Basic. This genomic fragment is designated as the Egfl7/miR-126 5′ flanking region, and it is directly upstream of exon 1b, the first exon encoding the major splice variant EGFL7b.40 This construct was then transfected into a variety of cells, including HUVEC, HAEC, dermal, and brain endothelial cells, as well as HEK 293 and HeLa cells. The Egfl7/miR-126 5′ flanking region was transactivated at high levels in HUVEC, HAEC, dermal and brain endothelial cells, and transactivated at lower levels in HEK293 cells and HeLa cells (Fig. 2A). These data suggest that the Egfl7/miR-126 5′ flanking region is transactivated in endothelial cells but not in some other cell types.
Figure 2.
Ets binding site is a critical domain within the miR-l26 regulatory region.
(A) A genomic fragment region upstream of the Egfl7/miR-126 locus is transactivated in endothelial cells, but not in other cell types. The genomic region extending 1670 bp upstream of the Egfl7 variant b transcriptional start site was cloned upstream of Photinus luciferase in the pGL3-Basic reporter vector. The reporter vector was transfected into various cell lines along with a control vector constitutively expressing Renilla luciferase, and the cells were analyzed for luciferase expression. (B) Computer analysis of 1670 bp upstream of the Egfl7/miR-126 transcriptional start site reveals potential binding sites for GATA-1 and GATA-2 (gray), KLF2 (white), N-myc (spots), and Ets (black). (C) The genomic fragment extending 150 bp upstream from the Egfl7 transcriptional start site is a critical domain in the regulatory region. Truncated domains of the Egfl7 regulatory region were cloned into the luciferase reporter vector, and luciferase activity in HUVEC was analyzed as above (n=3 ± S.D. *P < 0.05 for 150 vs. 71). (D) Ets binding sites are evolutionarily conserved between human (above) and mouse (below). EBS1 extends from −93 to −97, and EBS2 extends from −80 to −84, in the genomic regions upstream of miR-126/Egfl7. (E) EBS1 is required for transactivation of the miR-126 regulatory region. Mutations were made to either of the two Ets binding sites, EBS1 or EBS2, in a reporter vector containing the region of the EGFL7 5′ untranslated region extending to −383 bp upstream of the luciferase gene, and transfected into HUVEC with RL-TK control vector.
Deletion analysis highlights critical −150 base pair region in Egfl7/miR-126 5′ flanking region
In order to identify transcription factors that could potentially bind the EGFL7 5′UTR and regulate the expression of miR-126, we used a bioinformatic approach (Methods). In silico analysis revealed several potential binding sites for transcription factors known to play important roles in endothelial cells biology (Fig. 2B). In particular, there is a potential TATA box at the −40 position and several potential Ets, GATA, and KLF2 binding sites in the Egfl7/miR-126 5′ flanking region.
In order to determine which portion of the Egfl7/miR-126 5′ flanking region might be most critical for transactivation, we created a series of deletion constructs. Truncated regions of the Egfl7/miR-126 5′ flanking region were cloned upstream of a cDNA for Photinus luciferase, transfected into HUVEC, and analyzed for the expression of luciferase (measured in relative light units or RLU). These 5′ flanking region constructs extended 1670, 483, 383, 150, and 71 base pairs upstream from the transcriptional start sites. Maximal expression of luciferase is conferred by the 1670 bp fragment (Fig. 2C). Deletion from the 5′ end decreases expression of luciferase. However, deletion of the region between −483 and −383 leads to a small but statistically significant increase in luciferase expression. Furthermore, the largest percentage change in luciferase expression occurs when the region from −150 to −71 is deleted. In fact, the 71 bp construct was not transactivated in HUVEC (Fig. 2C). These data suggest that there is a critical region in between −150 bp and −71 bp that is necessary for transactivation of the promoter constructs in HUVEC.
Computer analysis showed two potential Ets binding sites (EBS) between the critical −150 and −71 bp region (Fig. 2D). Additionally, the 200 bp upstream of the Egfl7/miR-126 5′ transcriptional start site is evolutionarily conserved between mice and human (Fig. 2D). In order to determine if these Ets binding sites were necessary for transactivation of the promoter in endothelial cells, we created constructs with mutations in the Ets binding sites (EBS), the distal site that we designate as EBS1 or the proximal site EBS2 (Supplemental Fig. S I). Mutations in EBS1 lead to a 90% reduction in the transactivation of a promoter reporter construct (Fig. 2E). Mutations in EBS2 lead to a 30% reduction in the transactivation of the promoter reporter construct (Fig. 2E). Taken together, these data imply that EBS1 is required for the transactivation of the Egfl7/miR-126 5′ region in endothelial cells.
Ets-1 and Ets-2 regulate mir-126 expression
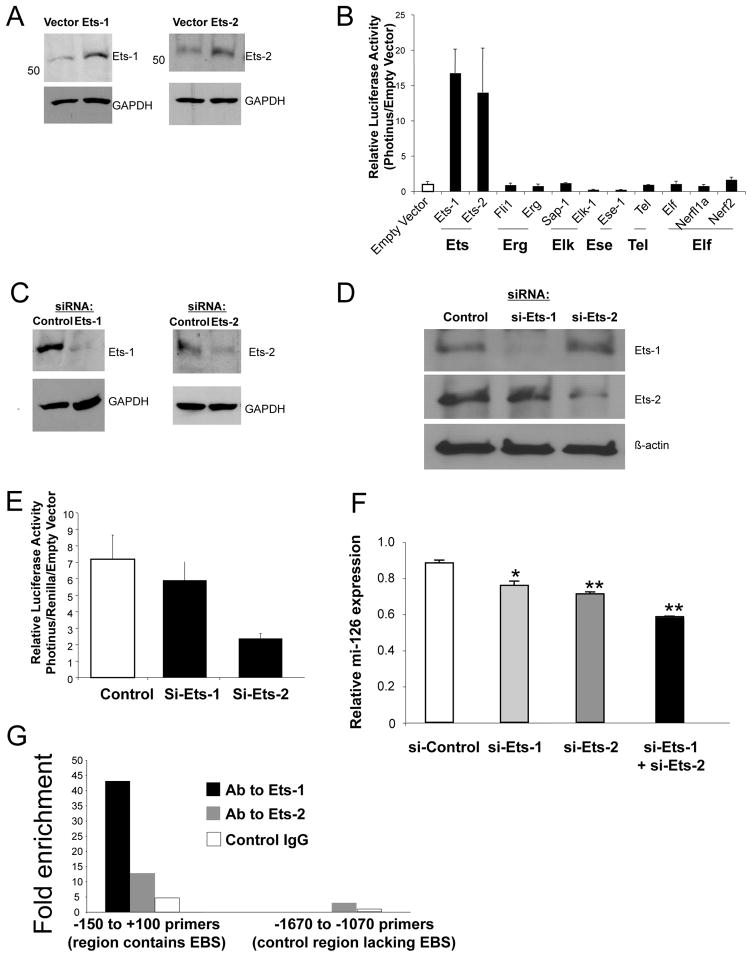
Several members of the Ets-family of proteins are known to be expressed in endothelial and bind to Ets binding sites like the ones present in the Egfl7/miR-126 5′ region.29 In order to identify which member of the Ets family member could regulate the expression of the mir-126 we over-expressed several Ets-family members in endothelial cells. We transfected endothelial cells with expression plasmids for Ets-1 or Ets-2 and measured the levels of each protein by Western blots. Ets-1 and Ets-2 expression plasmids increased the expression of Ets-1 and Ets-2 in endothelial cells (Fig. 3A). We next wanted to test the ability of Ets transcription factors to transactivate the Egfl7/miR-126 5′ region. We co-transfected the Egfl7/miR-126 5′ reporter construct along with Ets1, Ets2, Fli1, Etf, Elk-1, Erp, Sap-1, Nerf1a, Nerf2, Elkf, Tel, Ese1 expression vector, or an empty vector into HUVEC, and analyzed cell lysates for luciferase expression. Over-expression of Ets-1 or Ets-2 leads to a 15 fold increase in the transactivation of the mir-126 promoter construct. (Fig. 3B). Over-expression of other Ets-factors leads to smaller changes in the expression of the Egfl7/miR-126 5′ reporter construct. (Over-expression of Ets-1 or Ets-2 also transactivates the miR-126 promoter construct in HEK293 cells, a cell line which normally does not express miR-126 (Supplemental Fig. S II).) Taken together these data suggest that Ets-1 and Ets-2 can transactivate the Egfl7/miR-126 5′ region in endothelial cells.
Figure 3.
Ets-1 and Ets-2 regulate the expression of miR-126.
(A) Over-expression of Ets-1 and Ets-2. HUVEC were transfected with 200 ng of an expression vector that was empty or expressed Ets-1 or Ets-2 for 2 d. Total protein was harvested and analyzed by immunoblotting for Ets-1 or Ets-2 (representative of 2 experiments). (B) Ets-1 and Ets-2 over-expression increases transactivation of the EGFL7 proximal promoter in HUVEC. The EGFL7 proximal promoter −1670 construct and PCI-Ets factor expression constructs were co-transfected into HUVEC (*P < 0.05 vs. empty vector). (C) Knockdown of Ets-1 and Ets-2. HUVEC were transfected with siRNA to Ets-1, Ets-2, or with siRNA control; and then cultured for 2 days. Total protein was harvested and analyzed by immunoblotting for Ets-1 or Ets-2. (D) Knockdown of Ets-1 transcription factors is specific for Ets isoforms. Knockdown of Ets-1 decreases Ets-1 but not Ets-2 levels by immunoblotting. Knockdown of Ets-2 only decreases Ets-2 levels. (E) Ets-1 and Ets-2 knock-down decreases transactivation of the EGFL7 proximal promoter in HUVEC. The EGFL7 proximal promoter -383 construct were co-transfected along with Ets-1, Ets-2, or control siRNA into HUVEC, and luciferase activity measured as above (n = 3 ± S.D. *P < 0.05 vs. control). (F) Knockdown of Ets-1 and Ets-2 decreases miR-126. HUVEC were transfected with 50nM of Ets-1, Ets-2, control siRNA or both and cultured for 3 d. Total RNA was harvested and analyzed by QRT-PCR for miR-126 expression. (n=3 ± S.D., *< 0.05 vs control, **<0.005 vs control). (G) Ets-1 and Ets-2 occupy the miR-126 promoter. Chromatin immunoprecipitations (ChIP) were performed on resting HUVEC. Chromatin proteins were cross-linked to chromatin with formaldehyde and sheared into 400 – 1000 bp fragments. Nucleoprotein complexes were immunoprecipitated using anti-Ets-1, anti-Ets-2, or control antibody. The precipitated DNA fractions were analyzed by quantitative-PCR for the presence of the miR-126 proximal regulatory region encompassing the EBS1 and EBS2 (region −150 to + 100 bp). The regulatory region from −1670 to −1070 was used as a negative control. Input DNA was used as a positive control.
To explore the effect of endogenous Ets-1 and Ets-2 on mir-126 expression, we altered the levels of endogenous Ets-1 and Ets-2 in endothelial cells. Endogenous Ets-1 and Ets-2 are expressed in endothelial cells and other cell types (Supplemental Fig. S III). In order to decrease endothelial Ets-1 and Ets-2 levels, we transfected HUVEC with siRNA directed to Ets-1, Ets-2, or control. siRNA to Ets-1 or Ets-2 decreased the expression of Ets-1 and Ets-2 respectively in HUVEC (Fig. 3C–D). Using this approach, we found that knockdown of Ets-2 decreased transactivation of the promoter construct in HUVEC (Fig. 3E). Knockdown of Ets-1 also lead to a smaller decrease in the transactivation of the Egfl7/miR-126 5′ reporter construct.
To explore the ability of Ets-1 and Ets-2 to alter the expression of mir-126 in endothelial cells, we transfected HUVEC with siRNA directed against Ets-1, Ets-2, or control, and analyzed cellular RNA for mir-126 expression by qRT-PCR. Knockdown of Ets-1 decreases miR-126 expression in HUVEC by approximately 15% (Fig. 3F). Knockdown of Ets-2 decreases expression of mir-126 by approximately 20%. Knockdown of Ets-1 and Ets-2 together decreases expression of miR-126 by about 35%. Taken together, these data suggest that both Ets-1 and Ets-2 regulate mir-126 expression, with Ets-2 playing a more significant role. These data also imply that other transcription factors play a role in the endogenous expression of mir-126, or that miR-126 is expressed at very stable levels that are only partially affected by temporary knockdown of Ets-1 and Ets-2.
To confirm that Ets-1 and Ets-2 regulate mir-126 expression, we measured Ets-1 and Ets-2 interactions with the mir-126/EGFL7 5′UTR at the EBS binding sites of genomic DNA inside cells. We used formaldehyde to cross-link transcription factors to chromatin, sheared the chromatin into small fragment, used antibodies to Ets-1 or Ets-2 or control IgG to immunoprecipitate the nucleoprotein complexes, and analyzed the DNA by qPCR for the region containing the EBS1 and EBS2 sites between −150 and −71 bp upstream from the transcriptional start site. Immunoprecipitation with the Ets-1 antibody leads to approximately a 40-fold enrichment of the region containing EBS1 and EBS2 (Fig. 3G). Immunoprecipitation with Ets-2 leads to a 10-fold enrichment of the same region. Taken together, these data show that both Ets-1 and Ets-2 interact with the mir-126/EGFL7 5′UTR.
The Ets-miR-126 pathway suppresses target genes
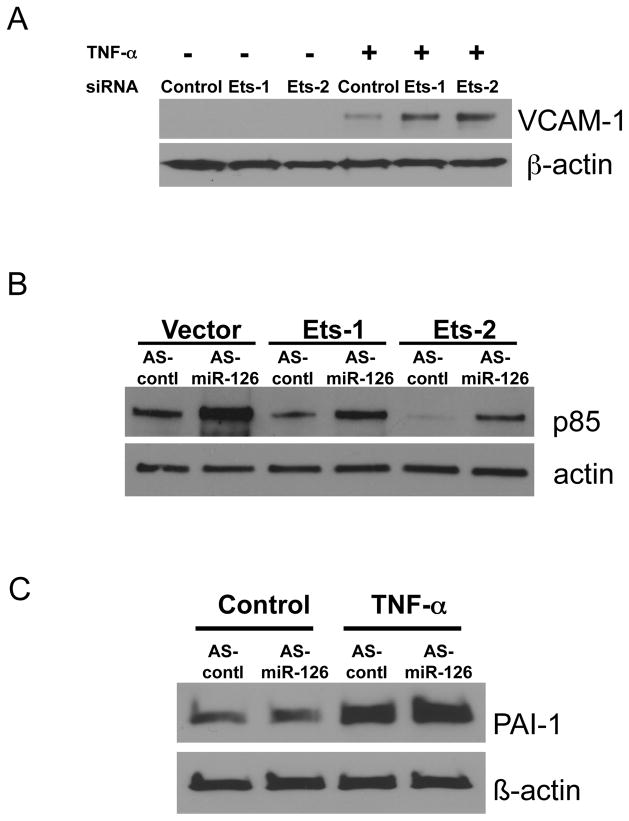
In order to test the idea that Ets isoforms regulate miR-126, we explored Ets signal transduction pathways inside endothelial cells. First we tested the idea that Ets boosts miR-126 suppression of a miR-126 target gene. We had previously shown that miR-126 suppresses VCAM-1. Silencing Ets would decrease miR-126 and decrease miR-126 suppression of VCAM-1. To test this idea we induced VCAM-1 in endothelial cells with TNF-a, and then silenced Ets isoforms. Silencing Ets-1 or Ets-2 permits an increase in VCAM-1 (Fig. 4A).
Figure 4.
MiR-126 mediates the suppressive effects of Ets-1
(A) Ets-1 and Ets-2 suppress VCAM-1. Endothelial cells were transected with siRNA directed against Ets isoforms, and then stimulated with TNF-a. Knockdown of Ets isoforms leads to an increase in VCAM-1 expression, suggesting that the Ets pathway includes a suppressor of VCAM-1 such as miR-126. (B) The Ets-miR-126 pathway suppresses PI3K-p85 expression. Ets-1 or Ets-2 were over-expressed in HUVEC, and then miR-126 was knocked down. Ets isoforms decrease p85 expression, but silencing miR-126 partially relieves this inhibition. These data suggest that miR-126 partially mediates Ets suppression of p85. (C) The miR-126 pathway does not suppress PAI-1. Endothelial cells were stimulated with TNF-a and miR-126 was silenced. MiR-126 does not affect TNF-a activation of PAI-1.
Next we tested the idea that miR-126 mediates Ets suppression of PI3K-p85. We over-expressed Ets isoforms and then silenced miR-126 in endothelial cells. Ets isoforms decrease p85 levels, but silencing miR-126 limits the effect of Ets upon p85 (Fig. 4B). These data support the hypothesis that Ets regulates miR-126.
Finally we explored the effect of miR-126 upon PAI-1, a target of Ets that Ets increases rather than decreases. TNF-a increased PAI-1 levels, and silencing miR-126 had no effect upon PAI-1 (Fig. 4C). These data support the idea that miR-126 does not play a role in regulation of PAI-1.
Discussion
Summary
The major finding of our study is that Ets-1 and Ets-2 regulate miR-126 expression. Ets-1 and Ets-2 interact with an Ets binding element in genomic regions upstream of the mir-126/Egfl7 gene. Mutation of the Ets binding element decreases promoter transactivation and decreases miR-126 expression. Silencing miR-126 limits the ability of Ets to suppress a target gene.
Ets-1 and Ets-2 regulate miR-126
Several lines of evidence suggest that both Ets-1 and Ets-2 regulate miR-126 expression in endothelial cells. Over-expression of Ets-1 or Ets-2 in endothelial cells transactivated a reporter construct consisting of 1.7 kb of genomic DNA upstream of the miR-126/Egfl7 gene (Fig. 3A–B). Conversely, knockdown of Ets-1 or Ets-2 suppresses the reporter construct (Fig. 3C–d). Finally, knockdown of endogenous Ets-1 or Ets-2 decreases miR-126 levels, and knockdown of both isoforms together decreases miR-126 levels more than either individually (Fig. 3E). The Ets-1 and Ets-2 isoforms are important regulators of immune responses and of angiogenesis: Ets-1 knockout mice show partial perinatal lethality and surviving mice have immune defects,41 Ets-2 knockout mice die in utero,42 and inhibition of both isoforms disrupts coronary artery formation in chick embryos.36. It is possible that a lack of miR-126 or EGFL7 are responsible for some of the vascular defects associated with lack of Ets-1 and Ets-2.
Our data raise several issues about transcriptional regulation of miR-126. First, deletion of the genomic region extending 383 to 483 bp upstream of miR-126/Egfl7 increases reporter construct transactivation (Fig. 2C), suggesting that this region may contain a binding element that mediates repression of miR-126 expression. Second, knockdown of both Ets isoforms does not completely eliminate miR-126 expression, suggesting that other ETS family members or unidentified transcription factors may also activate miR-126 expression. For example, we found that the upstream genomic region contains putative binding elements for the KLF2 transcription factor, and deletion of this region decreases miR-126 reporter levels by 3 fold (Fig. 2). A recent study showed that KLF2 mediates the genetic effects of flow upon endothelial cells in part by controlling miR-126 expression.43 This other study extends our findings by identifying other regulatory elements in the miR-126 regulatory region. Furthermore, even though HEK293 cells express Ets-2 (Supplemental Data), levels of miR-126 are low in HEK293 cells (Fig. 2A), emphasizing that other transcription factors in addition to Ets isoforms are necessary to induce miR-126 expression.
Ets binding sites regulating miR-126/Egfl7
Our work extends a prior study that explored the effect of Ets upon miR-126 expression. The pre-mRNA for miR-126/Egfl7 is alternatively spliced to produce the minor transcript variant EGFL7a and the major transcript EGFL7b.40 Olson and colleagues first showed that a genomic fragment extending 5.4 kb upstream of the Egfl7b transcript that includes the first exon of Egfl7a could direct endothelial specific expression of a reporter gene in mouse embryos.27 Olson and colleagues then studied Ets transactivation of the genomic region upstream of the Egfl7a transcript, using COS cells. This regulatory region contained an EBS, and ectopic expression of Ets-1 transactivated this regulatory region.27 In contrast, we studied Ets transactivation of the genomic region upstream of the Egfl7b transcript, and used endothelial cells. Our data emphasize the functional importance of an ETS binding site (EBS) extending between −93 to −97 bp upstream from the transcriptional start site of miR-126/Egfl7b. Both studies emphasize the importance of Ets-1, while our data extend the work of Olson and colleagues by showing that endogenous Ets isoforms Ets-1 and Ets-2 regulate miR-126 in endothelial cells, and by characterizing a distinct miR-126/Egfl7 transcript. Furthermore, our studies emphasize the importance of the Ets binding sites: deletion of the region containing the EBS or mutation of the EBS1 site decreases miR-126 reporter transactivation by over 20-fold (Fig. 2).
MiR-126 and vascular biology
Our studies suggest a connection between the phenotypes of Ets knockout mice and miR-126 knockout mice. Mutations in both Ets-1 and Ets-2 lead to abnormalities in vascular formation.36, 41, 42 Mutations in miR-126 also lead to vascular abnormalities, partial embryonic lethality, and defective angiogenesis.27, 28, 44 It is possible that loss of expression of Ets-1 and Ets-2 transcribed genes such as miR-126 might play a role in abnormal vasculogenesis or angiogenesis. Targets of miR-126 such as Spred-1 and PI3KR may mediate the effects of Ets-1 and Ets-2 upon vascular development.27, 28
Our data also suggest a negative feedback loop through which Ets and miR-126 influence vascular inflammation. Pro-inflammatory agonists such as TNF-α or angiotensin II induce Ets-1 expression, which in turn activates transcription of pro-inflammatory mediators such as MCP-1 and VCAM-1.33 However, Ets-1 also induces miR-126 which inhibits VCAM-1 translation.19 Thus the net effect of Ets-1 upon vascular inflammation might depend in part upon the balance between Ets-1 induced pro-inflammatory factors and Ets-1 induced anti-inflammatory factors such as miR-126.
Supplementary Material
Acknowledgments
We acknowledge the assistance of Jackie Hewitt for help preparing the manuscript.
Sources of Funding
Supported by grants from the NIH (R01 HL63706-04, R01 HL074061, P01 HL65608, P01 HL56091), AHA (EIG 0140210N), the Paul Yu Professorship to CJL; by a training grant from The Cellular and Molecular Medicine Program to TAH; and by grants from the NIH (R01 HL-082717) to PO.
Abbreviations
- miR
MicroRNA
- EGFL7
Epidermal growth factor-like domain 7
Footnotes
Disclosures
All authors declare no competing interests.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 3.Mello CC, Conte D., Jr Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 4.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 5.Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- 6.Latronico MV, Catalucci D, Condorelli G. Emerging role of microRNAs in cardiovascular biology. Circ Res. 2007;101:1225–1236. doi: 10.1161/CIRCRESAHA.107.163147. [DOI] [PubMed] [Google Scholar]
- 7.van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res. 2008;103:919–928. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim VN. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol. 2004;14:156–159. doi: 10.1016/j.tcb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. Rna. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonauer A, Boon RA, Dimmeler S. Vascular microRNAs. Curr Drug Targets. doi: 10.2174/138945010791591313. [DOI] [PubMed] [Google Scholar]
- 13.Suarez Y, Sessa WC. MicroRNAs as novel regulators of angiogenesis. Circ Res. 2009;104:442–454. doi: 10.1161/CIRCRESAHA.108.191270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fish JE, Srivastava D. MicroRNAs: opening a new vein in angiogenesis research. Sci Signal. 2009;2:pe1. doi: 10.1126/scisignal.252pe1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S, Olson EN. AngiomiRs--key regulators of angiogenesis. Curr Opin Genet Dev. 2009;19:205–211. doi: 10.1016/j.gde.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 17.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 18.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 19.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci U S A. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 21.Fish JE, Srivastava D. MicroRNAs: opening a new vein in angiogenesis research. Sci Signal. 2008;1:pe1. doi: 10.1126/scisignal.252pe1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280:9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 24.Suarez Y, Fernandez-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS, Iruela-Arispe ML, Merkenschlager M, Sessa WC. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci U S A. 2008;105:14082–14087. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wurdinger T, Tannous BA, Saydam O, Skog J, Grau S, Soutschek J, Weissleder R, Breakefield XO, Krichevsky AM. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;14:382–393. doi: 10.1016/j.ccr.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Tuschl T. A Mammalian microRNA Expression Atlas Based on Small RNA Library Sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oettgen P. Regulation of vascular inflammation and remodeling by ETS factors. Circ Res. 2006;99:1159–1166. doi: 10.1161/01.RES.0000251056.85990.db. [DOI] [PubMed] [Google Scholar]
- 30.Turner DP, Findlay VJ, Moussa O, Watson DK. Defining ETS transcription regulatory networks and their contribution to breast cancer progression. J Cell Biochem. 2007;102:549–559. doi: 10.1002/jcb.21494. [DOI] [PubMed] [Google Scholar]
- 31.Dejana E, Taddei A, Randi AM. Foxs and Ets in the transcriptional regulation of endothelial cell differentiation and angiogenesis. Biochim Biophys Acta. 2007;1775:298–312. doi: 10.1016/j.bbcan.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 32.Sato Y. Role of ETS family transcription factors in vascular development and angiogenesis. Cell Struct Funct. 2001;26:19–24. doi: 10.1247/csf.26.19. [DOI] [PubMed] [Google Scholar]
- 33.Zhan Y, Brown C, Maynard E, Anshelevich A, Ni W, Ho IC, Oettgen P. Ets-1 is a critical regulator of Ang II-mediated vascular inflammation and remodeling. J Clin Invest. 2005;115:2508–2516. doi: 10.1172/JCI24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang LC, Kuo F, Fujiwara Y, Gilliland DG, Golub TR, Orkin SH. Yolk sac angiogenic defect and intra-embryonic apoptosis in mice lacking the Ets-related factor TEL. Embo J. 1997;16:4374–4383. doi: 10.1093/emboj/16.14.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spyropoulos DD, Pharr PN, Lavenburg KR, Jackers P, Papas TS, Ogawa M, Watson DK. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol Cell Biol. 2000;20:5643–5652. doi: 10.1128/mcb.20.15.5643-5652.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lie-Venema H, Gittenberger-de Groot AC, van Empel LJ, Boot MJ, Kerkdijk H, de Kant E, DeRuiter MC. Ets-1 and Ets-2 transcription factors are essential for normal coronary and myocardial development in chicken embryos. Circ Res. 2003;92:749–756. doi: 10.1161/01.RES.0000066662.70010.DB. [DOI] [PubMed] [Google Scholar]
- 37.Wei G, Srinivasan R, Cantemir-Stone CZ, Sharma SM, Santhanam R, Weinstein M, Muthusamy N, Man AK, Oshima RG, Leone G, Ostrowski MC. Ets1 and Ets2 are required for endothelial cell survival during embryonic angiogenesis. Blood. 2009;114:1123–1130. doi: 10.1182/blood-2009-03-211391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oettgen P. Functional redundancy of Ets1 and Ets2. Blood. 2009;114:934–935. doi: 10.1182/blood-2009-05-221812. [DOI] [PubMed] [Google Scholar]
- 39.Matsushita K, Morrell CN, Cambien B, Yang SX, Yamakuchi M, Bao C, Hara MR, Quick RA, Cao W, O’Rourke B, Lowenstein JM, Pevsner J, Wagner DD, Lowenstein CJ. Nitric oxide regulates exocytosis by S-nitrosylation of N-ethylmaleimide-sensitive factor. Cell. 2003;115:139–150. doi: 10.1016/s0092-8674(03)00803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soncin F, Mattot V, Lionneton F, Spruyt N, Lepretre F, Begue A, Stehelin D. VE-statin, an endothelial repressor of smooth muscle cell migration. Embo J. 2003;22:5700–5711. doi: 10.1093/emboj/cdg549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, Leiden JM. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–563. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto H, Flannery ML, Kupriyanov S, Pearce J, McKercher SR, Henkel GW, Maki RA, Werb Z, Oshima RG. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 1998;12:1315–1326. doi: 10.1101/gad.12.9.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE, Lawson ND. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 464:1196–1200. doi: 10.1038/nature08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuhnert F, Mancuso MR, Hampton J, Stankunas K, Asano T, Chen CZ, Kuo CJ. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135:3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.