Abstract
The ability to control the activity of intracellular signaling processes in live cells would be an extraordinarily powerful tool. Ideally, such an intracellular input would be (i) genetically encoded, (ii) able to be turned on and off in defined temporal or spatial patterns, (iii) fast to switch between on and off states, and (iv) orthogonal to other cellular processes. The light-gated interaction between fragments of two plant proteins—termed Phy and PIF—satisfies each of these constraints. In this system, Phy can be switched between two conformations using red and infrared light, while PIF only binds one of these states. This chapter describes known constraints for designing genetic constructs using Phy and PIF and provides protocols for expressing these constructs in mammalian cells, purifying the small molecule chromophore required for the system’s light responsivity, and measuring light-gated binding by microscopy.
1. Introduction
In recent years, tremendous strides have been made in developing quantitative readouts that report on live cell activity at the molecular scale. Time-lapse microscopy has been combined with fluorescent detection of protein concentration (Heim and Tsien, 1996; Michalet et al., 2005) and protein–protein association (Truong and Ikura, 2001), enabling studies of the temporal dynamics (Lahav et al., 2004; Nelson et al., 2004) and spatial organization (Ilani et al., 2009) of complex signaling pathways. These techniques have also been instrumental in characterizing complex emergent properties such as perfect adaptation (Cohen-Saidon et al., 2009; Shimizu et al., 2010). All of these advances rely on the ability to quantitatively measure signaling outputs—the concentration or activity of various pathway components—with precision in living cells.
The ability to quantitatively vary intracellular signaling inputs in time and space, in a user controlled way, would be equally revolutionary, allowing researchers to better manipulate and probe the cellular processes they study. However, comparatively few technologies are available to achieve the goal of manipulation as compared to measurement in living cells. Microfluidic devices are limited to controlling the spatial and temporal pattern of extracellular inputs (Paliwal et al., 2007; Tay et al., 2010). For intracellular inputs, the rapamycin-inducible FKB/FRBP protein–protein interaction (Spencer et al., 1993) offers the opportunity to activate the association of two intracellular species. However, this high-affinity interaction is slow to dissociate, and thus the resulting control is poorly reversible (Terrillon and Bouvier, 2004).More recently, inputs have been designed that utilize light-induced conformational changes in naturally light-responsive proteins such as channelrhodopsin (Gunaydin et al., 2010) LOV domains (Strickland et al., 2008), or proteins incorporating photocaged amino acids (Gautier et al., 2010; Lemke et al., 2007), although each has limitations. Applying light-activatable LOV domains to control additional signaling processes requires considerable protein engineering, and LOV domain inactivation occurs spontaneously but cannot be directly controlled by light. Uncaging of photocaged proteins can be performed quickly and selectively, but is an irreversible modification. Finally, light control of channelrhodopsin is fast and reversible but limited to controlling transmembrane cation flux, a specific signaling currency.
Ideally, a controllable intracellular signaling input would be genetically encoded (easily “wired in” to control a variety of proteins), photoreversible, nontoxic to the cell, and high resolution in both time and space. A recently developed light-dependent binding interaction using plant phytochrome proteins satisfies each of these constraints (Levskaya et al., 2009; Ni et al., 1999). This interaction has already proven useful for applying complex temporal and spatial intracellular inputs to live cells in a variety of species (Levskaya et al., 2009; Shimizu-Sato et al., 2002). In principle, this approach could be used for spatiotemporal control over any cellular process that is dependent on association of two proteins. Because induced protein interaction is such a common mechanism for controlling molecular activity, this method has the potential to be highly generalizable.
Despite these advantages, it can be challenging to prepare light-gated genetic constructs, purify the small molecule chromophore required for their function, and assay the function of all these components inside living cells. In this chapter, we address these challenges by presenting a detailed and optimized methodology based on that introduced in Levskaya et al. (2009) for visualizing light-induced binding in mammalian cells. After a brief introduction to the system (Section 3) we present methods for genetically encoding the light-responsive protein domains (Section 4), purifying the small-molecule chromophore that is required for their interaction (Section 5), and validating the light-mediated protein translocation in mammalian cells (Sections 6 and 7).
2. Light-Controlled Phy–PIF Interaction
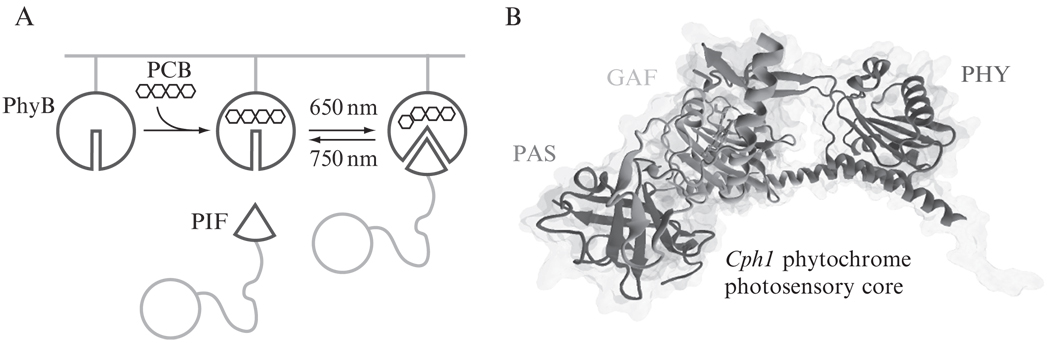
The Phy–PIF system takes advantage of a light-controllable binding interaction between two genetically encoded components: a fragment of Arabidopsis thaliana phytochrome B, referred to here as Phy; and a fragment of phytochrome interaction factor 6, referred to here as PIF. Phy becomes light-responsive following conjugation to the membrane-permeable small molecule chromophore, phycocyanobilin (PCB). Exposure to 650 nm induces association of PIF and Phy, while exposure to 750 nm light induces dissociation of PIF from Phy (Fig. 17.1A). Phy can be reversibly switched between PIF-interacting and -noninteracting states using light within seconds, and switching can be performed for hundreds of cycles without toxicity to the cell or any measurable degradation of the system’s performance (Levskaya et al., 2009).
Figure 17.1.
Schematic of the Phy–PIF interaction. (A) After incorporation of the chromophore PCB, the conformation of Phy can be controlled by exposure to two wavelengths of light (650 and 750 nm). The PIF domain binds only one of these domains with high affinity. By controlling the ratio of 650:750 nm light, the fraction of Phy in a state permissive for binding can be tuned, modulating the total amount of PIF recruitment. (B) Crystal structure of a fragment of the cyanobacterial phytochrome protein Cph1 bound to the small molecule chromophore PCB (shown as licorice) (PDB ID: 2VEA) (Essen et al., 2008). The PAS, GAF, and PHY domains, each required for Phy–PIF interaction, are shown in purple, orange, and red, respectively, while the N terminal 26 amino acids are shown in green. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this chapter.)
How can this system be used to enable light control of a range of cellular activities? Because protein association and dissociation is such a general currency of cell signaling, this light-gated heterodimerization scheme has been applied to a broad range of signaling processes such as transcription, splicing, plasma membrane signaling, and modulating actin assembly in vitro (Leung et al., 2802). It was first used outside of its native context in a 2-hybrid approach, in which the split DNA binding and transcriptional activation domains of Gal4 were fused to Phy and PIF to enable light-gated transcriptional control in yeast (Shimizu-Sato et al., 2002). A similar concept using a split protein (intein) was used to activate splicing in yeast using light (Tyszkiewicz and Muir, 2008). The first application of this system to mammalian cell signaling used Phy and PIF to activate GTPase signaling in mammalian cells by recruiting a constitutively active GEF to its G protein target on the plasma membrane (Levskaya et al., 2009). In this chapter, we focus on a simple, direct assay of light-controllable Phy–PIF binding, in which a fluorescently tagged Phy directed to the plasma membrane where it can act as a light-controlled binding site for cytoplasmic fluorescently tagged PIF. To observe this localization change, we will focus on a typical experimental context for using the Phy–PIF system, in which Phy and PIF constructs are introduced into mammalian cells by retroviral infection and imaged by fluorescence microscopy.
3. Genetic Constructs Utilizing Phy and PIF Components
The Phy and PIF components of the light system can be expressed as fusions to other proteins of interest to elicit binding and activation of a variety of intracellular signaling processes. However, care must be taken to validate Phy and PIF expression and localization for each fusion construct. In this section, we describe some known constraints of the system and elucidate details for establishing successful light-controllable fusion constructs.
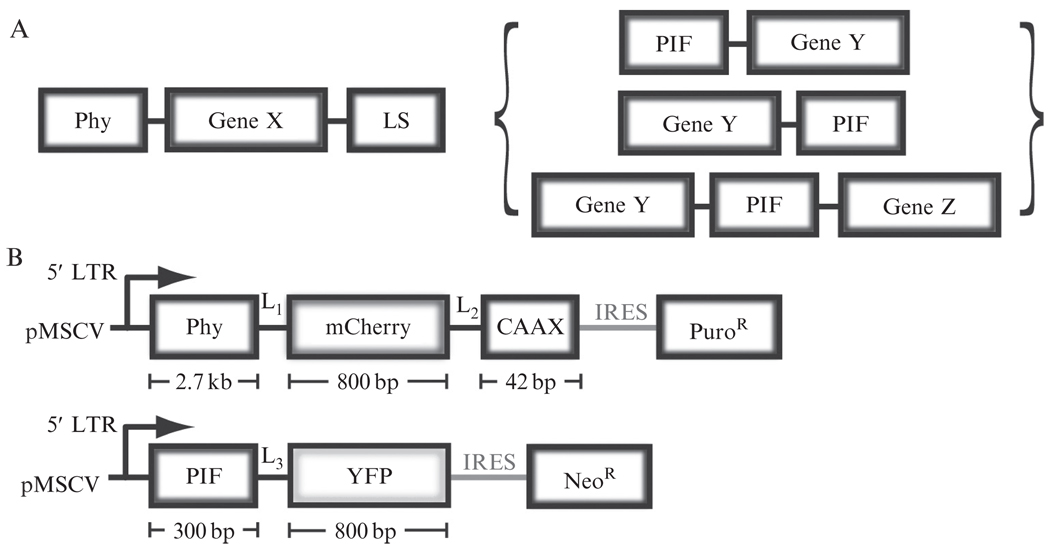
The first of these components is Phy, consisting of residues 1–908 of the A. thaliana PhyB protein (Entrez Gene ID: 816394; see Fig. 17.1B for the structure of a related phytochrome, Cph1) (Essen et al., 2008; Levskaya et al., 2009). Phy has been expressed successfully without codon optimization in Saccharomyces cerevisiae and NIH-3T3 cells; however, we have found codon optimization to facilitate strong Phy expression in some contexts (as for HL-60 cells and Dictyostelium). Whereas PIF tolerates a large range of fusion orientations, we have found that Phy fusion protein expression and function is particularly sensitive to linker lengths and component orientation. Phy appears to work most robustly as an N-terminal fusion component (Fig. 17.2A). While a useful guideline, this is not a strict rule; PIF recruitment has also been observed from some C-terminal and internal Phy fusion constructs. We have had success using a 15 amino acid linker (linker L1: EFDSAGSAGSAGGSS) between the C-terminus of Phy and the N-terminus of downstream fusion constructs; this linker performed better than shorter linkers in some applications.
Figure 17.2.
Genetic Phy and PIF constructs for use in recruitment assays. (A) Schematic diagram showing typical Phy and PIF fusion constructs. The larger Phy protein is usually expressed as a membrane-localized component, fused to a gene of interest (GeneX) as well as a C-terminal membrane localization tag (LS). Phy best tolerates fusion when it encodes the N-terminal component. PIF is typically fused to a freely diffusing cytoplasmic component, and tolerates fusion at either or both termini. (B) Schematic of the specific constructs discussed in this article. Phy is fused to mCherry and a plasma membrane localization tag, the KRas CAAX tail, while PIF is fused to YFP. Both are driven using the MSCV retroviral vector system and are expressed along with puromycin or neomycin selection markers. Gly-Ser linkers L1, L2, and L3 are as described in the text.
How can Phy function be validated in different contexts? One useful, previously characterized Phy single mutant (Y276H) fluoresces at far-red frequencies only in the chromophore-bound state (Su and Lagarias, 2007). We previously demonstrated that NIH-3T3 cells expressing this mutant exhibit bright fluorescence after 30 min of incubation with PCB (Levskaya et al., 2009). Thus, the ability of a Phy fusion construct to bind PCB can be tested directly, independently of the recruitment assay described in Section 6. Phy-Y276H can also be used to test the quality of purified PCB (Section 4).
Phy interacts in a light-dependent fashion with PIF, a second component consisting of residues 1–100 of A. thaliana PIF6 protein. PIF does not exhibit any preference toward N or C terminal fusions and also tolerates fusions on both termini simultaneously (Fig. 17.2A). We have not observed any dependence of PIF–Phy binding on linker length within PIF fusion constructs. Typically, Gly-Ser spacers of 10 amino acids are placed between PIF and its fusion partners.
When using the Phy–PIF system to induce binding between membrane-tethered and cytoplasmic proteins, we typically attach Phy to the membrane component and PIF to the cytoplasmic component (Fig. 17.2A), as the smaller PIF domain is less likely to significantly affect diffusion of its fusion partner. Phy can be tethered to the plasma membrane using a linker followed by the KRas “CAAX tail” plasma membrane localization signal (linker L2: SAGSAGKASG; CAAX tag: KKKKKKSKTKCVIM). This tag has been validated in both yeast and mammalian cells (Clarke et al., 1988). Although this CAAX sequence can be used to induce robust membrane localization in NIH-3T3 cells, phosphorylation at its serine residue by PKC causes dissociation from the membrane in some cell types (Bivona et al., 2006); incorporating a Ser-Ala mutation (unphosphorylatable CAAX tag: KKKKKKAKTKCVIM) can be used to stabilize membrane association in these contexts.
We have demonstrated light-controlled Phy–PIF interaction in a number cell lines, and NIH-3T3 cells (ATCC catalog number CRL-1658) continue to be our gold standard for these studies. Transient transfection of fluorescently tagged Phy and PIF constructs works for some applications, but we find that generating stable cell lines from retroviral constructs greatly enriches the population of cells expressing both Phy and PIF and also facilitates robust recruitment. Furthermore, these stable cell lines maintain expression in long-term culture and thus cells can be sorted by expression level to hone in on the optimal recruiting population for each application.
To establish stable cell lines, we typically clone Phy and PIF constructs into the pMSCV retroviral vector system (Clontech catalogue number 634401), in which each construct is driven from the viral LTR promoter. These constructs also contain an internal ribosomal entry site (IRES), followed by neomycin/G418 or puromycin antibiotic resistance to allow selection for the stably infected population (Fig. 17.2B). To produce virus, we transfect each construct into 293-GPG cells, a standard retrovirus packaging cell line (Ory et al., 1996) using the TransIT 293 transfection kit (Mirus catalog number 2700). NIH-3T3 cells should be infected with Phy and PIF viral constructs at a high enough multiplicity of infection (MOI) to lead to >70% infected cells. There is no need to select using antibiotics before assaying recruitment, although this selection can be performed to enrich for doubly infected cells.
Phy–PIF recruitment is easiest to observe if Phy expression levels are high (see Section 6). For more effective viral transduction, it can be helpful to concentrate Phy-containing retrovirus (Kanbe and Zhang, 2004). Briefly, the collected retrovirus is spun at max speed in 1.5 mL centrifuge tubes in a table top microcentrifuge (Eppendorf Centrifuge model 5415D or similar) at 4 °C for 1 h. After centrifugation, discard all but 100 µL or so of the supernatant, and pool together the “invisible” pellets for infection.
4. Purification of PCB from Spirulina
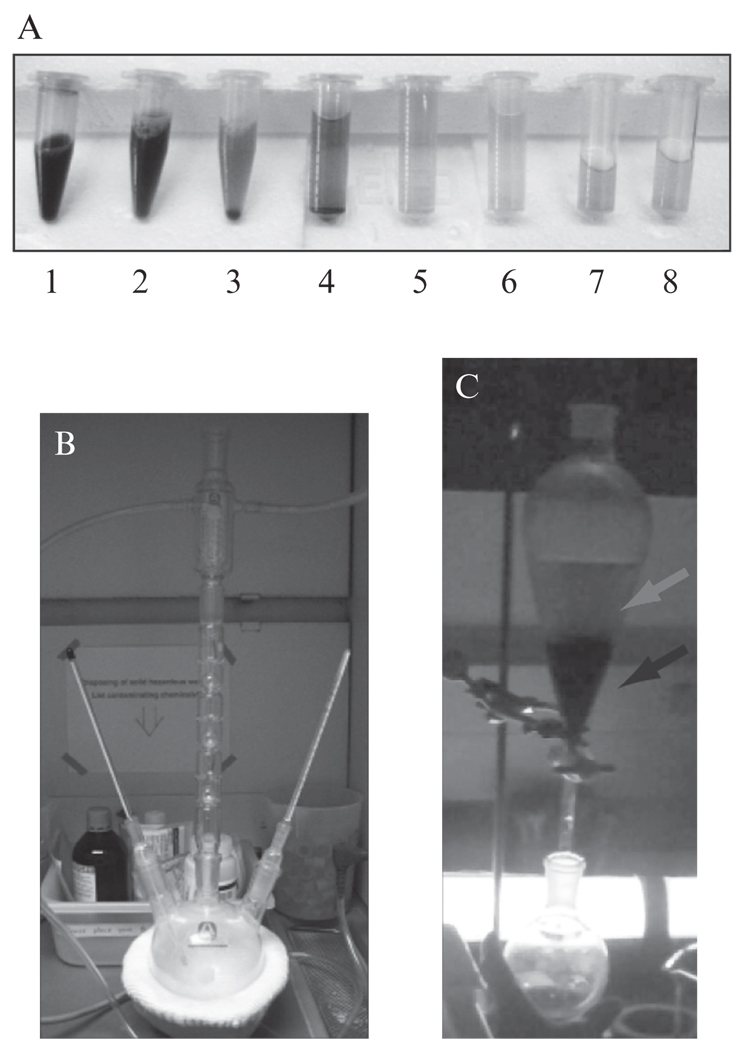
The responsiveness of Phy domains to light depends on their covalent attachment to a small molecule chromophore, PCB. PCB is not synthesized naturally in most nonphotosynthetic cells, but it is easily taken up by all cell types we have tested, penetrating yeast cell walls and freely diffusing through mammalian cell membranes. Thus, free PCB must be obtained and added to cell cultures before light-responsive experiments are conducted. While addition of the PCB chromophore is another required manipulation, it is also an advantage—cells can be handled freely with regard to light exposure until one is ready to perform an experiment, at which point the PCB is added under controlled light conditions. In this section, we provide a detailed step-by-step protocol for PCB purification from Spirulina algae based on the procedure described in Smith and Holmes (1984), adapted from Levskaya et al. (2009) (see Fig. 17.3). This procedure relies on the fact that PCB is the most prevalent protein-bound tetrapyrrole found in Spirulina, so a generic tetrapyrrole purification protocol leads to high enrichment for this compound. Briefly, protein is purified from resuspended Spirulina, an 8 h methanolysis separates tetrapyrroles from their protein binding partners, and chloroform extraction is used to isolate these unbound tetrapyrroles and discard the protein fraction. The resulting purified PCB is stable at − 20 °C for months.
Figure 17.3.
Key steps involved in the purification of PCB. (A) Samples collected at different stages of the PCB preparation procedure. Tubes contain the Spirulina-water mixture (1); the green supernatant after the first spin (2); the TCA protein precipitation, containing supernatant and pellet (3); the supernatant after each of three methanol washes (4–6); and the supernatant after the first and second 8 h methanolysis (7–8). (B) The assembled methanol reflux apparatus. The two thermometers should be placed in the vapor and fluid phases of the PCB-containing solution, respectively. Adjust heating until the temperature in both phases is the same, and the methanol solution is at a low boil. To prevent methanol loss through evaporation, flow cold water through the condenser (middle column of the apparatus). (C) Chloroform separation of PCB from the protein phase, photographed under a green safelight. Separation should result in a white, cloudy aqueous phase (red arrow) above a dark green chloroform phase (black arrow). If the aqueous phase retains a green color, add hydrochloric acid dropwise to acidify the solution, shake vigorously, and allow separation to occur again. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this chapter.)
4.1. Protocol
Resuspend 75 g Spirulina powder (Seltzer Chemical) in 2 L doubly distilled water (~30 mL/g). Stir for 10 min in a 4 L plastic beaker, transfer to 1 L screw-top plastic bottles, then spin at 8000 rpm at 4 °C for 1 h (we use a Sorvall RC5C Plus centrifuge with a Fiberlite F9S-4x1000y rotor).
Collect the supernatant, discarding the dark green pellet. Precipitate soluble protein from the supernatant by adding 20 g TCA (final solution: 1%, w/v, TCA). Wrap the beaker in foil to protect from light and stir solution at 4 °C for 1 h. Centrifuge in 1 L screw-top plastic bottles at 8000 rpm at 4 °C for 10 min (Sorvall RC5C Plus centrifuge; Fiberlite F9S-4x1000y rotor).
-
Resuspend and wash pellets with 100% methanol. Centrifuge in 250 mL screw-top plastic bottles at 6000 rpm for 10 min at 4 °C (Beckman J2-21M centrifuge; Beckman JA-14 rotor), and discard supernatant containing free tetrapyrroles. Repeat washes and centrifugation until supernatant runs clear or is only lightly green; this typically takes three washes. During washes, the pellet will change color, finally achieving a bright cyan color when washing is complete (Fig. 17.3A). These washes can be used to consolidate the material into a smaller volume. We typically perform the first wash in eight 250 mL bottles, the second in four bottles, and the third in two bottles for a final volume of ~500 mL.
Note: during methanolysis and all subsequent steps, you will be working with PCB that is no longer protein-bound. In this state, PCB is light sensitive and the material should be shielded from light using aluminum foil, or by illuminating with a green safelight (Sylvania F40G fluorescent tube wrapped once with a Roscolene 874 sheet and once with a Roscolene 877 sheet to provide illumination at 550 nm) in an otherwise dark room.
Collect the washed cyan pellet and transfer to a 1 L three-neck round-bottomed distillation flask for methanolysis. Add 500 mL methanol; the pellet will not fully resuspend. Add boiling chips to prevent bumping of the methanol during heating. Connect the reflux apparatus, including cold water to recirculate through the condenser, thermometers to measure both the fluid and vapor temperatures, and a heat bath for heating the reaction. Reflux by maintaining at a slow boil with the solution and vapor phase temperatures held at 64.7 °C for 8 h (Fig. 17.3B). Be patient and do not overheat—it may take some time to establish a stable temperature for the reaction.
Transfer the liquid phase of the methanolysis reaction to 500 mL single-necked round-bottomed flask and connect to a rotary evaporator (but do not discard the remaining pellet—see Step 8). Evaporate the methanol to a final volume of 50 mL.
Add 50 mL chloroform and 100 mL water to a separatory funnel, followed by the concentrated PCB solution. Stopper the top of the funnel, and shake vigorously to emulsify the chloroform and aqueous phases. Wait for ~1 min to allow the phases to separate. The aqueous phase should be cloudy and white, while the PCB-containing chloroform phase should be dark green (Fig. 17.3C). If the aqueous phase is green or there is not a distinct color separation between the two phases, it should be taken as an indication that PCB is not well confined to the chloroform phase. In this case, add hydrochloric acid dropwise to acidify the solution. Shake vigorously and repeat until the aqueous phase is colorless.
Separate the chloroform phase (bottom liquid) into a 500 mL single-necked round-bottomed flask, discarding the remaining aqueous phase. Evaporate the chloroform solution using a rotary evaporator to obtain dry PCB. Resuspend the dry PCB in 3 mL DMSO. Aliquot between 20 and 100 µL into dark PCR tubes. PCB can be frozen and thawed tens of times without degradation, but care must be taken not to expose aliquots to excess light.
To obtain the final PCB concentration, dilute the final preparation 1:100 into 1 mL 95:5% MeOH:HCl (37.5%) solution and reading the absorbance at 680 nm. The concentration in mM is calculated as A680 × 2.64; typical final concentrations from this procedure are between 3 and 15 mM.
To increase the yield from this preparation, steps 4–8 can be performed a second time on the pellet remaining after the first 8 h methanolysis reaction. Typically, we perform the two 8-h methanolyses (Step 4) overnight on two consecutive nights, and perform steps 5–8 once on each preparation beginning the next morning, keeping each preparation separate and testing each for its final PCB concentration.
Optional step: The quality of the PCB preparation can be verified independently of Phy–PIF recruitment using cells expressing a Phy mutant (Y276H) that fluoresces upon incorporation of PCB (Levskaya et al., 2009; Su and Lagarias, 2007). Expose cells expressing Phy Y276H to PCB as described in Section 5, and image cells using Cy5 excitation and emission optical configurations.
5. Cell Culture Preparation for Phy–PIF Translocation
After building Phy and PIF recruitment constructs and purifying PCB, it is important to validate the functionality of all components in the experiment of interest. Testing translocation requires two steps: incubating cells with PCB and preparing them for imaging on fibronectin-coated coverslips (the present section), and imaging these cells using confocal microscopy (the next section). Both of these steps require some measure of precision and care, as many variables can affect the quality of observed recruitment.
5.1. Protocol
Coat the glass coverslip on the bottom of a 35 mm MatTek dish (MatTek catalog number P35G-1.5-14C) with 100 µL of 0.2 mg/mL fibronectin diluted in PBS (Sigma catalog number F4759) for at least 1 h at room temperature.
Wash dish twice with 3 mL of Dulbecco’s PBS (D-PBS). Plate cells immediately, or store dishes in PBS at 4 °C for no longer than 2 days prior to use.
Trypsinize and count NIH-3T3 cells expressing Phy and PIF constructs (Section 3) from an existing culture (maintained as described by ATCC). Plate 150,000 cells in 1 mL media on the fibronectin-coated MatTek dish. Place dish in incubator for 30 min to allow cells to adhere.
Note: this step is light-sensitive and care should be taken to minimize PCB’s exposure to light. Perform under low light conditions or under a green safelight (see Step 3). In this step, media and PCB are premixed before adding to cell culture to ensure that cells are not exposed to high concentrations of DMSO. Transfer 100 µL media from the MatTek dish to a 1.5 mL tube. Pipet 4 nmol of PCB (about 1 µL of the 4 mM stock from Step 3) into the tube, and mix well. Add the PCB–media mixture back to dish; swirl to mix. Wrap dish in aluminum foil and place in incubator for at least 30 min. Plates can be maintained in PCB-containing media for a few hours, so multiple plates can be prepared simultaneously and imaged sequentially.
Before imaging, exchange the PCB-containing media for an imaging solution containing 3 mL modified Hank’s balanced salt solution (mHBSS) supplemented with 2% FBS. NIH-3T3 cells should remain healthy at room temperature without supplemental CO2 for at least 6 h under these conditions. For prolonged imaging, replace imaging solution to combat evaporation.
6. Imaging PIF Translocation Using Spinning Disk Confocal Microscopy
After preparing cells with the desired Phy- and PIF-containing constructs and purifying PCB, it is important to verify that binding between Phy and PIF are controllable by light. The following procedure relies on a localization change in one of the components (here, a fluorescent PIF construct) upon binding to the other (membrane-localized Phy). To perform this experiment, the cytoplasmic concentration of PIF is measured by fluorescent imaging using confocal microscopy. Here, we outline the protocol for imaging PIF–YFP and Phy-mCherry-CAAX (mCherry is a fluorescent protein with RFP-like excitation and emission).
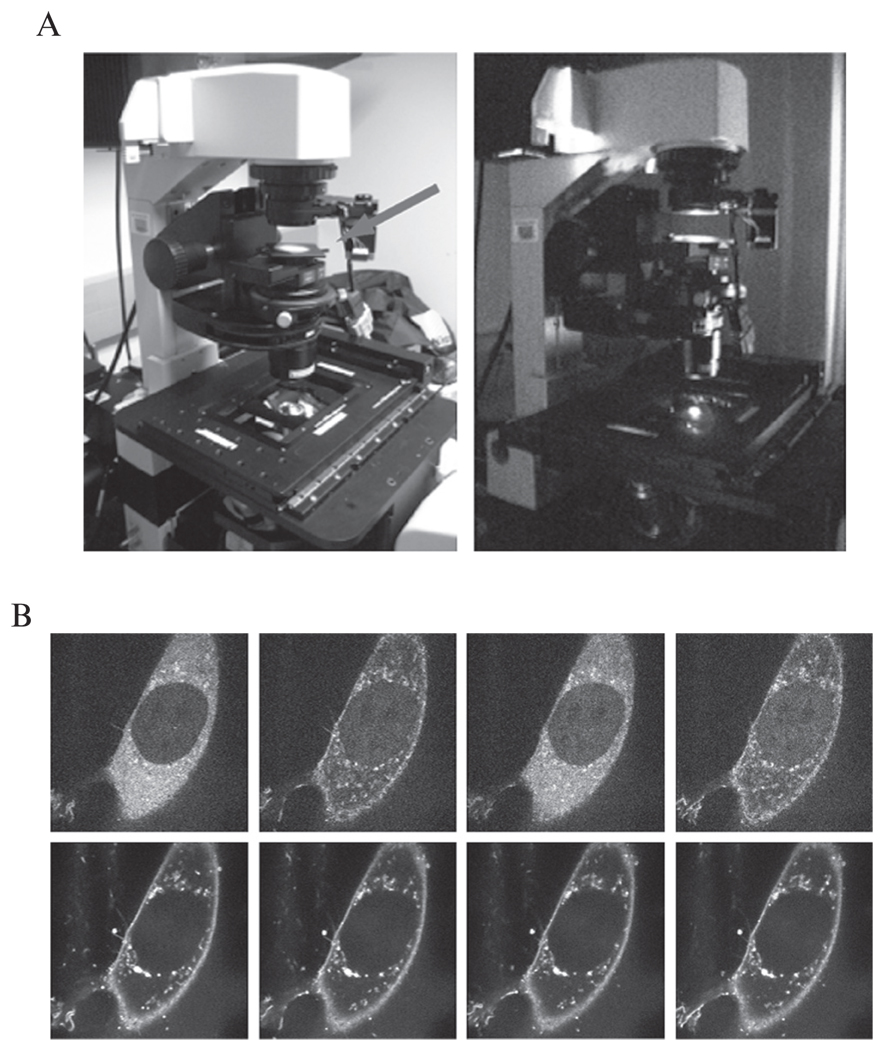
A confocal microscope can be easily used to supply wavelengths required for association and dissociation of Phy–PIF complexes. A red laser emitting at 561 nm (or alternatively, a white light source and 561 nm filter) can be used to maximally activate Phy–PIF association, and these should be available on fluorescent microscopes capable of imaging RFP or similar proteins. However, because the transition to the PIF-binding state is so sensitive, unfiltered brightfield light is sufficient to generate measurable Phy–PIF recruitment, as described in the protocol below. To elicit Phy–PIF dissociation, place a 750 nm long-pass filter (Newport, model FSQ-RG9) on top of the microscope’s condenser (in the brightfield imaging light path) and turn on brightfield illumination (Fig. 17.4A). It is important to ensure that no filters are present in the light path that could interfere with infrared light transmission. It is possible to use a 750 nm filter in conjunction with other light sources (e.g., mercury halide arc lamp; DG4 light source). However, care must be taken to ensure that all optical components are capable of transmitting infrared light (e.g., mirrors, filters, liquid light guides), and that no infrared-blocking filters are present in the light path.
Figure 17.4.
Confocal microscopy to image Phy–PIF translocation. (A) A 750 nm filter can be placed in the brightfield light path to elicit Phy–PIF dissociation. With this filter in place, illuminating with brightfield light leads to Phy–PIF dissociation. Simply removing the filter provides enough activating light to induce Phy–PIF translocation. Alternatively, RFP excitation light (650 nm) can be used to induce association. (B) A montage of confocal images of a NIH-3T3 cell showing PIF–YFP translocation in response to light. Cells were prepared harboring the genetic constructs described in Section 3. The upper panel shows PIF–YFP fluorescence after sequential 30 s exposures of activating (brightfield) and inactivating (750 nm filtered) light. Phy-mCherry levels in corresponding timepoints are shown in the lower panels.
Observing high-quality light-dependent recruitment depends strongly on the expression levels of both Phy and PIF fluorescent fusion proteins. Because this assay relies on observing PIF changing localization from cytoplasm to cell membrane, there must be enough Phy on the membrane to appreciably deplete cytoplasmic PIF levels during exposure to activating light. Thus, it is crucial to select cells with high membrane expression of Phy and low to moderate PIF levels. Cell geometry can also play a crucial role: because small cells have a higher surface area to volume ratio than large cells, smaller cells are further enriched for an excess of Phy molecules compared to PIF, leading to better PIF depletion upon activation.
6.1. Protocol
Select a cell for imaging based on the criteria described above (high Phy-mCherry membrane expression, moderate PIF–YFP expression, and small cell volume).
For the best contrast between recruitment and release, choose an imaging plane in the lower half of the cell, just above the coverslip. Such a focal plane should be close to a large pool of membrane-bound Phy, where PIF cytoplasmic depletion should be maximal. If available, initiate the microscope’s autofocus system to prevent focal plane drift.
Place a 750 nm square filter on top of the microscope condenser, in the brightfield imaging light path (Fig. 17.4A).
Alternate 30 s exposures of brightfield light with and without the 750 nm filter. After each 30 s brightfield exposure, take single PIF–YFP translocation images using the confocal microscope’s YFP imaging mode. A typical series of NIH-3T3 cell images showing recruitment by this technique is shown in Fig. 17.4B.
ACKNOWLEDGMENTS
We thank Anselm Levskaya for advice on PCB purification and light system manipulation. This work was partially supported by the National Institutes of Health grant GM084040, the Cancer Research Institute Postdoctoral Fellowship (J. E. T.), the American Cancer Society Fellowship (D. G.).
REFERENCES
- Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, Miura J, Wiener HH, Wright L, Saba SG, Yim D, Fein A, et al. Mol. Cell. 2006;21:481–493. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Clarke S, Vogel JP, Deschenes RJ, Stock J. Proc. Natl. Acad. Sci. USA. 1988;85:4643–4647. doi: 10.1073/pnas.85.13.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Saidon C, Cohen AA, Sigal A, Liron Y, Alon U. Mol. Cell. 2009;36:885–893. doi: 10.1016/j.molcel.2009.11.025. [DOI] [PubMed] [Google Scholar]
- Essen LO, Mailliet J, Hughes J. Proc. Natl. Acad. Sci. USA. 2008;105:14709–14714. doi: 10.1073/pnas.0806477105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier A, Nguyen DP, Lusic H, An W, Deiters A, Chin JW. J. Am. Chem. Soc. 2010;132:4086–4088. doi: 10.1021/ja910688s. [DOI] [PubMed] [Google Scholar]
- Gunaydin LA, Yizhar O, Berndt A, Sohal VS, Deisseroth K, Hegemann P. Nat. Neurosci. 2010;13:387–392. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- Heim R, Tsien RY. Curr. Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- Ilani T, Vasiliver-Shamis G, Vardhana S, Bretscher A, Dustin ML. Nat. Immunol. 2009;10:531–539. doi: 10.1038/ni.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanbe E, Zhang DE. Blood Cells Mol. Dis. 2004;33:64–67. doi: 10.1016/j.bcmd.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U. Nat. Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- Lemke EA, Summerer D, Geierstanger BH, Brittain SM, Schultz PG. Nat. Chem. Biol. 2007;3:769–772. doi: 10.1038/nchembio.2007.44. [DOI] [PubMed] [Google Scholar]
- Leung DW, Otomo C, Chory J, Rosen MK. Proc. Natl. Acad. Sci. USA. 2008;105(2008):12797–12802. doi: 10.1073/pnas.0801232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A, Weiner OD, Lim WA, Voigt CA. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, Edwards SW, McDowell HP, et al. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. Nature. 1999;400:781–784. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- Ory DS, Neugeboren BA, Mulligan RC. Proc. Natl. Acad. Sci. USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliwal S, Iglesias PA, Campbell K, Hilioti Z, Groisman A, Levchenko A. Nature. 2007;446:46–51. doi: 10.1038/nature05561. [DOI] [PubMed] [Google Scholar]
- Shimizu TS, Tu Y, Berg HC. Molecular Syst. Biol. 2010;6 doi: 10.1038/msb.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. Nat. Biotech. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- Smith H, Holmes MG. Techniques in Photomorphogenesis. Orlando, FL: Academic Press; 1984. [Google Scholar]
- Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- Strickland D, Moffat K, Sosnick TR. Proc. Natl. Acad. Sci. USA. 2008;105:10709–10714. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YS, Lagarias JC. Plant Cell. 2007;19:2124–2139. doi: 10.1105/tpc.107.051516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrillon S, Bouvier M. EMBO J. 2004;23:3950–3961. doi: 10.1038/sj.emboj.7600387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong K, Ikura M. Curr. Opin. Struct. Biol. 2001;11:573–578. doi: 10.1016/s0959-440x(00)00249-9. [DOI] [PubMed] [Google Scholar]
- Tyszkiewicz AB, Muir TW. Nat. Methods. 2008;5:303–305. doi: 10.1038/nmeth.1189. [DOI] [PubMed] [Google Scholar]