Abstract
Methylazoxymethanol (MAM), the genotoxic metabolite of the cycad azoxyglucoside cycasin, induces genetic alterations in bacteria, yeast, plants, insects and mammalian cells, but adult nerve cells are thought to be unaffected. We show that the brains of adult C57BL6 wild-type mice treated with a single systemic dose of MAM acetate display DNA damage (O 6-methyldeoxyguanosine lesions, O 6-mG) that remains constant up to 7 days post-treatment. By contrast, MAM-treated mice lacking a functional gene encoding the DNA repair enzyme O 6-mG DNA methyltransferase (MGMT) showed elevated O 6-mG DNA damage starting at 48 hours post-treatment. The DNA damage was linked to changes in the expression of genes in cell-signaling pathways associated with cancer, human neurodegenerative disease, and neurodevelopmental disorders. These data are consistent with the established developmental neurotoxic and carcinogenic properties of MAM in rodents. They also support the hypothesis that early-life exposure to MAM-glucoside (cycasin) has an etiological association with a declining, prototypical neurodegenerative disease seen in Guam, Japan, and New Guinea populations that formerly used the neurotoxic cycad plant for food or medicine, or both. These findings suggest environmental genotoxins, specifically MAM, target common pathways involved in neurodegeneration and cancer, the outcome depending on whether the cell can divide (cancer) or not (neurodegeneration). Exposure to MAM-related environmental genotoxins may have relevance to the etiology of related tauopathies, notably, Alzheimer's disease.
Introduction
We describe mouse brain cell-signaling pathways that are perturbed by the aglycone (methylazoxymethanol, MAM) metabolite of a plant genotoxin (MAM-glucoside, cycasin) that is strongly associated with a declining neurodegenerative disease: Western Pacific amyotrophic lateral sclerosis and parkinsonism-dementia complex (ALS-PDC). This disease is clinically related to amyotrophic lateral sclerosis, atypical parkinsonism, and Alzheimer's dementia (AD) [1]–[6]. As with AD and certain other human neurodegenerative disorders, the cellular neuropathology of ALS-PDC is hallmarked by neurofibrillary tangles composed of paired helical filaments containing abnormally hyperphosphorylated forms of the microtubule-stabilizing protein tau [3], [6].
Western Pacific ALS-PDC, a prototypical neurodegenerative disorder apparently of environmental origin, has been highly prevalent in three genetically distinct island populations: (a) Japanese in the Kii Peninsula of Honshu Island, (b) Papuan New Guineans in West Papua, Indonesia, and (c) Chamorros on Guam and Rota in the Mariana Islands, migrants from Guam, and a few North American (Caucasian) and Filipino immigrants to Guam [6]–[8]. All three affected populations used the neurotoxic cycad seed for medicinal purposes [9]–[11]. On Guam, where ALS-PDC has been studied scientifically for over 60 years [4], the disease has been repeatedly linked to ingestion of food derived from cycad seed, with highly significant correlations for the cycasin content of cycad flour and for ALS and PD forms of the disease in both males and females [1], [5], [12], [13]. Diminishing use of cycad seed for food and/or medicine as affected populations adopt modern lifestyles is consistent with the progressive decline in disease prevalence in all three geographic isolates of ALS-PDC [11].
Cycasin and its aglycone methylazoxymethanol (MAM) are established developmental neurotoxins. MAM acetate damages neuronal DNA, modulates brain molecular networks and arrests regional brain development when administered systemically to postnatal day-3 mice [14], [15], but the adult rodent brain has been seen as largely refractory to the genotoxin [16], [17]. The promutagen MAM is also an established hepatotoxin and experimental carcinogen [18]. In fact, rodents that have been chronically treated with the MAM precursor azoxymethane (AOM) are widely used as models for investigating the pathogenesis and chemoprevention of human colon carcinoma [19]. Unfortunately, while recent (1998–2002) cancer data are available for Guam [20], longitudinal trends in cancer prevalence comparable to those available for Guam ALS-PDC are unknown.
We undertook this study of the adult mouse brain to test the hypothesis that the DNA-damaging properties of MAM, which are mutagenic and tumorigenic in cycling cells of the colon epithelium [19], activate molecular networks associated with the degeneration of post-mitotic neurons in neurodegenerative disease. While the relationship between environment-induced DNA damage, mutagenesis and malignancy is well accepted, non-nuclear mechanisms are usually considered to underpin neurodegenerative diseases. However, unlike most organs, the adult human brain has a low or absent capacity to repair alkylation-induced DNA damage [21], [22], with implications for long-term survival and eventual degeneration of nerve cells [23]. We addressed the aforementioned hypothesis by comparing the relationship between MAM-induced DNA damage (O 6-methyldeoxyguanosine, O 6-mG) and gene expression patterns in the brains of adult mice that are functionally proficient (wild type, wt) and deficient (Mgmt−/−) in the repair of O 6-mG, the latter lacking the gene coding for the specific DNA repair enzyme O 6-mG methyltransferase [24]. Two separate laboratories treated groups of wt and Mgmt−/− mice with a single systemic dose of MAM, and the combined data were mined for common brain transcriptional profiles. A third laboratory conducted blinded analyses of brain O 6-mG levels.
We present evidence that signaling pathways associated with human neurodegenerative disease are activated in mature mouse brain as the result of unrepaired MAM-induced DNA damage. These pathways involve receptors for certain neurotransmitters, including ionotropic and metabotropic neuronal receptors for glutamate, an excitatory neurotransmitter with the potential to kill nerve cells. While these findings support a role for MAM in the etiology of ALS-PDC, perhaps acting as a “slow toxin” via persistent DNA damage in nerve cells subject to continuous glutamate neurotransmission, they do not exclude a role for other cycad neurotoxins or genetic factors.
We also demonstrate that MAM-induced DNA damage modulates signaling pathways in the mouse brain that are associated with cancer, as well as neurodegeneration, the two phenotypes possibly representing responses of cycling and non-cycling cells, respectively. Others have proposed links between neurodegeneration/cancer and cell cycle regulation, DNA repair, response to oxidative stress [25], [26], aberrant wingless and proto-oncogene Int-1 (Wnt) signaling [27], glycogen synthase kinase 3 beta (GSK3β) regulation [28], modulation of tumor protein (TP53 or P53) expression [29], and perturbations of tau in AD and prostate cancer [30]. Chronic inflammation is another characteristic feature of both cancer and AD [31].
Results
Organ Response to MAM
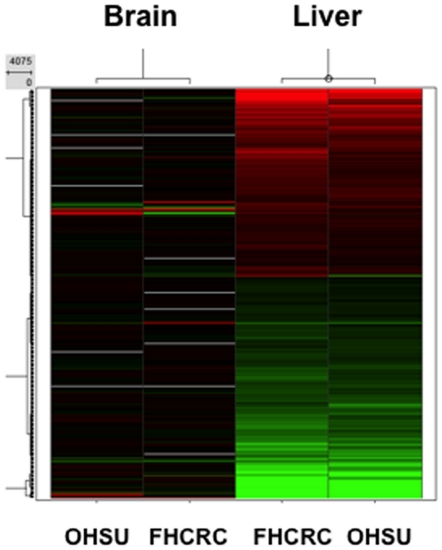
A preliminary study was carried out to determine the immediate transcriptional responses of the mouse wt brain to systemic treatment with MAM relative to that of a non-neural tissue (liver) that is specifically targeted in humans with acute cycad toxicity. Comparable patterns of gene expression were found for the two tissues at the two study sites. Whereas the liver (a primary target of cycad toxicity in humans and rodents) showed a robust response to MAM, relatively few changes were noted in the brain (Fig. 1).
Figure 1. Heat map comparing the post-treatment (6 hr) transcriptional response of brain and liver (positive control) of wt mice to a single intraperitoneal dose of MAM.
The experiment was conducted at two independent laboratories using identical protocols. Green denotes down-regulation and red up-regulation of gene expression. OHSU: Oregon Health & Science University. FHCRC: Fred Hutchinson Cancer Research Center.
DNA Damage
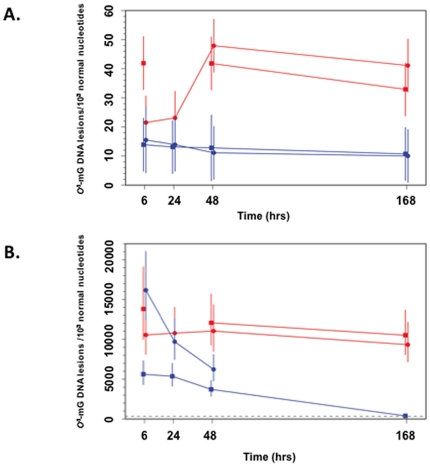
Brain tissue of wt and Mgmt−/− mice showed minimal detectable quantities (MDQ, see Methods) of O6-mG DNA lesions at 6 hr, 24 hr, 48 hr, and 168 hr following treatment with vehicle. After a single dose of MAM, the time-course of DNA damage in brain vs. liver (positive control) yielded fairly consistent data at the two independent study sites (Fig. 2). Levels of O6-mG were three orders of magnitude lower in brain than liver for both wt and Mgmt−/− mice. Significant deviation between the responses of wt and Mgmt−/− brain (p<0.01) and liver (p<0.01) was found at 48 hr, and this was maintained until 168 hr post MAM treatment (Table S1) [32]–[34]. While DNA damage was maintained at low (brain) or declining (liver) levels in the tissues of wt animals, there was persistence of the relatively higher levels of O6-mG DNA lesions in both tissues of Mgmt−/− mice. In sum, O6-mG lesions in both organs were much higher in Mgmt−/− vs. wt mice, and the DNA damage remained elevated in the brains of both wt and Mgmt−/− mice.
Figure 2. Time-course of O6-methylguanosine (O6-mG) DNA damage in the brain (A) and liver (B) of wt (blue) and Mgmt−/− (red) mice following a single intraperitoneal dose of MAM.
Results for the two study sites are shown as separate red and blue lines. The plotting symbols (OHSU: circle; FHCRC: square) denote the estimated medians; lines extend ±2 standard errors from the medians. DNA damage (O6-mG) is three orders of magnitude higher in the liver than in brain, and the significantly elevated O6-mG levels in Mgmt−/− vs. wt tissues at 48 hr are maintained at 168 hr post MAM treatment. Discontinuities in the red and blue lines are attributed to technical errors or where samples were not collected. The dashed gray line in B denotes the maximum observed O6-mG level in the brain (∼330 lesions per 108 normal nucleotides).
Brain Transcriptional Profiles
Analyses were performed on data aggregated from two laboratories. No significant differences in gene expression were noted in the brains of wt mice treated with MAM vs. vehicle. By contrast, significant modulation of gene expression was present in the brains of similarly treated Mgmt−/− mice. Analyses were first performed to determine the transcriptional response to MAM in Mgmt−/− mice and, secondly, to explore the MAM vs. vehicle effect in each genotype and whether the modulation of the effect differed between the genotypes. Subsequently, Ingenuity® pathway analysis (IPA) was used to identify the most significantly enriched biofunctions for each data set by combining significant gene expression changes at all time-points and comparing these data. Gene expression changes were also anchored to O6-mG DNA lesions to determine which genes were modulated by DNA damage. A third analysis, which combined the first two analyses, was used to explore Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways perturbed by MAM relative to vehicle that were either unique to Mgmt−/− or that differed between the two genotypes. The top KEGG pathways were determined using DAVID (the Database for Annotation, Visualization and Integrated Discovery) bioinformatics software [35], [36].
Initial studies were conducted to determine the response of the brain transcriptome to MAM vs. vehicle in DNA repair-deficient mice. The first set revealed 362 genes (of 41,000) that were differentially expressed in the brains of Mgmt−/− mice treated with MAM vs. vehicle (Table 1). Of these 362 genes, 57 were highly correlated (r>0.7) with O 6-mG levels. The four most significant disease biological functions corresponded to Neurological Disease (133 genes), Psychological Disorders (65 genes), Cancer (105 genes) and Genetic Disorder (170 genes). A list of the genes associated with each of these biological functions is provided in the supplemental data (Table S2).
Table 1. Top MAM-sensitive biological functions in Mgmt−/− mouse brains.
| Diseases & Disorders | Mgmt −/− treated with MAM vs. vehicle (N = 362) | Mgmt −/− treated with MAM vs. vehicle (N = 57) anchored to O 6-mG |
| Neurological Disease | 133 | 32 |
| Psychological Disorders | 65 | 19 |
| Cancer | 105 | 40 |
| Genetic Disorder | 170 | 23 |
Pathway analysis showing the four most significant biological functions altered in at least one time point in the brains of MAM-treated vs. vehicle-treated Mgmt−/− mice. The center column shows 362 genes that were significantly modulated by MAM, of which 57 genes individually satisfied criteria for DNA lesion anchoring (right).
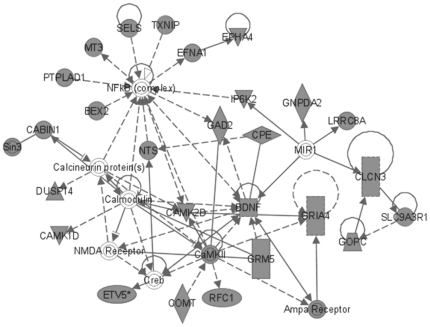
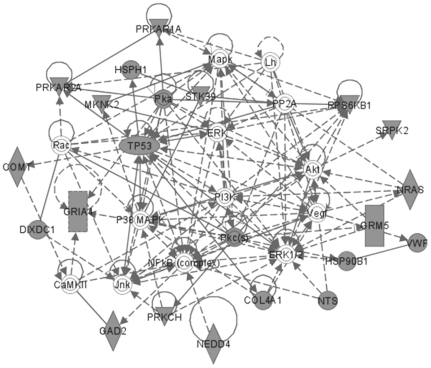
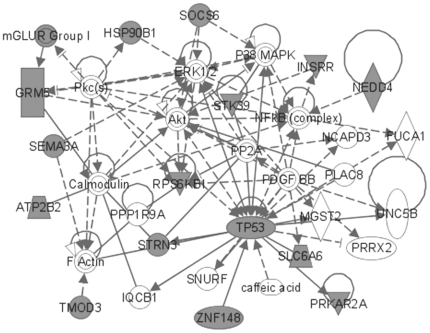
The most significant molecular networks derived from MAM-triggered, differentially expressed genes (362 genes) revealed hubs involving NF-κB (nuclear factor of kappa light polypeptide gene enhancer in B-cells), calcium-binding proteins (i.e., calcineurin, calmodulin), brain-derived neurotrophic factor (BDNF), glutamate receptors N-methyl-D-aspartate (NMDA) and alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA), CREB (cyclic AMP response element-binding), and microRNA1 (MIR1-1) (Fig. 3). When these MAM-induced differentially expressed genes were anchored to O6-mG levels, a subset of 57 genes revealed prominent hubs for NF-κB, extracellular signal-regulated kinase (ERK) and ERK1/2, p38-mitogen-activated protein kinase/c-Jun N-terminal kinases (MAPK/JNK), TP53, and Akt (v-akt murine thymoma viral oncogene homolog) (Fig. 4).
Figure 3. MAM-modulated brain gene products for Mgmt−/−.
Most significant brain expression sub-network modulated by MAM vs. vehicle in Mgmt−/− mice (all time-points combined) composed of 362 differentially expressed genes.
Figure 4. MAM-modulated brain gene products (anchored) for Mgmt−/−.
Most significant brain expression sub-network modulated by MAM vs. vehicle in Mgmt−/− mice (all time-points combined) derived from 57 differentially expressed genes (a sub-set of the 362 genes) that were anchored to O 6-mG levels.
The second analysis explored the effect of MAM relative to vehicle between the two genotypes (wt vs. Mgmt−/−); thus, it determined whether DNA-repair capacity influences the response of the brain transcriptome to MAM. This analysis was sensitive to those genes that may have shown different directions of modulation between the genotypes, even if neither modulation was significant on its own. There were 153 differentially expressed genes reflecting genotype differences between wt and Mgmt−/− in the brain's response to MAM vs. vehicle. Of the 153 genes, 60 genes (∼40%) were anchored to O 6-mG levels. Brains of these animals showed the same four biological functions for disease and disorders as in Table 1, with the single exception that the category Psychological Disorders was not significant in the anchored group (Table 2). A list of the genes associated with each of these biological functions is provided in the supplemental data (Table S3).
Table 2. Top MAM-sensitive biological functions in wt and Mgmt−/− mouse brains.
| Diseases & Disorders | wt vs. Mgmt −/− treated with respect to MAM vs. vehicle (N = 153) | wt vs. Mgmt −/− treated with respect to MAM vs. vehicle (N = 60) anchored to O 6-mG |
| Neurological Disease | 63 | 28 |
| Psychological Disorders | 32 | - |
| Cancer | 22 | 2 |
| Genetic Disorder | 87 | 37 |
Pathway analysis showing the four most significant biological functions altered in the brains of Mgmt−/− vs. wt mice treated with respect to MAM vs. vehicle (all time-points combined).
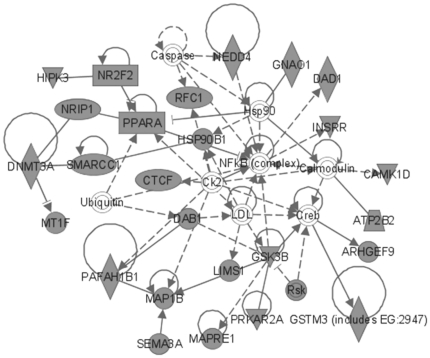
The most significant molecular networks in the 153 gene set contained hubs for NF-κB and CREB, and genes that regulate transcription through epigenetic mechanisms, including DNMT3A (DNA [cytosine-5-]-methyltransferase 3 alpha) and SMARCC1 (SWI/SNF related, Matrix-associated, actin-dependent regulator of chromatin, subfamily c, member 1) and nuclear transcription, namely PPARA (peroxisome proliferator-activated receptor alpha), molecular chaperones (HSP90B1, heat shock protein 90 kDa beta [Grp94], member 1), GSK3β and sema domain, immunoglobulin domain [Ig], short basic domain, secreted, [semaphorin] 3A (SEMA3A), which are implicated in Parkinson's and/or Alzheimer's disease (Fig. 5). As in Figure 3, the anchored set of 60 genes included TP53, ERK1/2 and NF-κB as the most prominent hubs, together with a number of other hubs (Akt, NF-κB, P38-MAPK and calmodulin) (Fig. 6). Glutamate receptors are also represented in this data set.
Figure 5. MAM-modulated brain gene products for Mgmt−/− vs. wild type.
Most significant MAM-modulated expression sub-network in the brain of Mgmt−/− vs. wild type mice (all time-points combined) composed of 153 differentially expressed genes. Note genes involved in epigenetic functions are also modulated: DNMT3A and SMARCC1 regulate chromatin function.
Figure 6. MAM-modulated brain gene products (anchored) for Mgmt−/− vs. wild type.
Most significant MAM-modulated expression sub-network in the brain of Mgmt−/− vs. wild type mice (all time-points combined) composed of 60 differentially expressed genes that were anchored to O 6-mG levels. Note the presence of NF-κB, ERK1, and p38-MAPK hubs, and the involvement of TP53 and glutamate receptors.
The third set of transcriptional data analyses combined the first two data sets (for a total of 443 non-duplicated genes), including the differential response of genotypes to MAM vs. vehicle (Table 2) and the response of Mgmt−/− brains to systemic treatment with MAM vs. vehicle (Table 1). The most significant scoring sub-network of MAM gene products (p<10−46) contained hubs for F-actin, NF-κB, microRNA-1, cofilin, calcium/calmodulin-dependent protein kinase II (CaMKII), glycogen synthase, the AMPA receptor, BDNF, and others. The same four diseases and disorders were the most significant of the biological functions list based on IPA analysis, while Nervous System Development and Function, and Skin and Hair Development and Function, appeared in the list of most significant four perturbed Physiological Systems Development and Functions (Table 3).
Table 3. Top MAM-associated diseases, disorders and physiological functions.
| Mgmt −/− MAM vs. vehicle plus wt vs. Mgmt −/− with respect to MAM vs. vehicle (Total molecules N = 443) | |||
| Diseases and Disorders | Physiological System Development and Function | ||
| Neurological Disease | 159 | Nervous System Development & Function | 64 |
| Psychological Disorders | 75 | Embryonic Development | 22 |
| Cancer | 114 | Organ Development | 14 |
| Genetic Disorder | 212 | Skin and Hair Development and Function | 11 |
Pathway analysis showing the four most significant diseases and disorders linked (left columns) and physiological systems (right columns) altered in the brains of MAM-treated vs. vehicle-treated Mgmt−/− mice plus Mgmt−/− vs. wt mice treated with respect to MAM vs. vehicle. Data were aggregated and all time points were combined for a total of 443 molecules.
Table 4 shows the top KEGG pathways denoting molecular interactions perturbed by MAM in either wild type or Mgmt−/− brains. Pathways involved in cancer, Wnt signaling, and insulin-signaling pathways were among the most significant. Other prominent KEGG pathways included those involved in purine metabolism, MAPK signaling, neurotrophin signaling, chemokine signaling and neuroligand-receptor interaction (Table 4).
Table 4. Top MAM-associated brain KEGG pathways.
| Top KEGG Pathways | Genes | Phenotype |
| Pathways in cancer | 13 | CC |
| Wnt signaling | 10 | AD, CC, skin, bone |
| Insulin signaling | 9 | AD, ALS |
| Purine metabolism | 9 | |
| Prostate cancer | 8 | CC |
| MAPK signaling | 7 | AD, CC |
| Melanogenesis | 6 | PD? CC |
| Neurotrophin signaling | 6 | AD |
| Focal adhesion | 6 | AD, CC |
| Chemokine signaling | 5 | AD |
| Neuroactive ligand-receptor interaction | 5 | AD |
| Calcium signaling pathway | 4 | AD, CC |
Top brain KEGG pathways (derived from 443 MAM-modulated genes, see Table 3) and the number of MAM-modulated genes and human diseases associated with each pathway. Note the prominent involvement of the Wnt, insulin and MAPK signaling pathways, in addition to the presence of pathways involved in inflammatory and other responses. AD: Alzheimer disease. ALS: Amyotrophic lateral sclerosis. CC: Colon cancer. PD: Idiopathic Parkinson's disease.
We also performed a PAINT (Promoter Analysis and Interaction Network Tool) analysis to identify the biologically relevant transcription factor binding sites in the genes that were enriched among the genotypes and differentially expressed genes between MAM and vehicle-treated animals. The 5′-flanking regions of the differentially expressed genes (2000 bp upstream of the transcription start site) were examined for enrichment of commonly expressed transcriptional regulatory elements (TREs). Table 5 shows the TREs among the unique genotype-specific (n = 153) and a subset of anchored (n = 60) genes targeted by MAM. Only TREs that were significantly enriched (p<0.05) in MAM-targeted genes are shown (Fig. 7). The highest scoring TRE was the highly conserved hepatocyte nuclear factor 4 (HNF-4), which binds to the consensus sequence AGGTCAaAGGTCA to activate transcription.
Table 5. MAM-associated enriched transcription factor binding sites.
| Transcriptional Response Element | P Value |
| HNF-4 | <0.001 |
| Lmo-2 | 0.008 |
| Tal-1α/E47 | 0.025 |
| GATA-2 | 0.033 |
| Tal-1β/E47 | 0.042 |
Enriched transcription factor binding sites among genotype-specific differentially expressed genes between MAM and vehicle-treated animals. Data based on 60 differentially expressed genes anchored to O 6-mG levels (see Fig. 6).
Figure 7. Transcription factor binding-site enrichment hierarchy.
Analysis of the promoter regions of the 60-anchored gene sub-set derived from the strain-specific differentially expressed genes between MAM and vehicle-treated animals for transcription regulatory elements. A heat map (interaction matrix) shows the genes (rows) and motifs (columns) that were individually clustered and found in >5% of all promoters. Note the HNF4 binding site is common to 60% of the 60 anchored genes.
Discussion
Brain and Colon: Common Pathways but Different Outcomes
We have shown that the cycad genotoxin MAM induces persistent DNA damage (i.e., O6-mG lesions) and modulates several cell signaling pathways (i.e., TP53, NF-κB, MAPK) in the brains of young adult Mgmt−/− mice. Our data support the hypothesis that MAM-induced O 6-mG DNA lesions alter purine metabolism and modulate cell-signaling pathways associated with both neurodegeneration and cancer. While MAM does not induce brain tumors in singly treated adult mice, the genotoxin consistently triggers tumors in peripheral organs, notably the intestine [37], [38]. The prominent modulation of “cancer genes” in the “tumor-insensitive” brains of MAM-treated adult animals suggests that perturbations of these genes in the brain have consequences other than cancer.
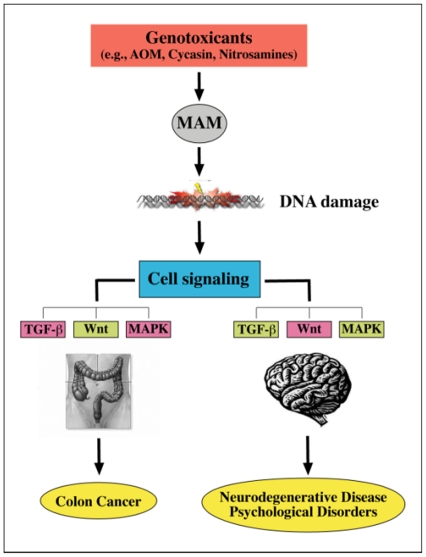
Molecular mechanisms underlying MAM-induced colon cancer have been established in the azoxymethane (AOM) mouse model of colorectal adenocarcinoma in which MAM (the cytochrome P4502E1-mediated metabolite of AOM) is the sole triggering agent [39], [40]. In the AOM mouse model, MAM-induced mutation of K-ras (i.e., transversion from G:C to A:T at codon 12 derived from O6-mG lesions) activates this pathway and the downstream MAPK and Phosphoinositide 3-kinase/Akt (PI3K/Akt) mediators, indicating that MAM perturbs gene expression in these pathways by a DNA damage-dependent mechanism. Mutations in β-catenin blocks its degradation by a GSK-3β-mediated mechanism resulting in its intracellular accumulation and the activation of the Wnt/β-catenin signaling pathway. The nuclear transport of β-catenin leads to the activation of genes that regulate cell proliferation, while expression of pro-apoptotic proteins is inhibited [40], [41]. Such events may explain how MAM modulates the expression of genes with pivotal roles in cell signaling in the brain of young adult mice (Fig. 8).
Figure 8. Proposed relationship between MAM-induced colon cancer and brain disease/disorders.
Genotoxicants that induce O6-methylguanine lesions (DNA damage) (e.g. via methylazoxymethanol, MAM) disturb cell signaling pathways, including transforming growth factor-β (TGF-β), wingless and proto-oncogene Int-1 (Wnt), and mitogen-activated protein kinase (MAPK). In general, the literature supports up-regulation (green) and down-regulation (pink-red) in association with the two distinct phenoptypes. (Modified from Chen and Huang [40]).
Cancer and Neurodegenerative Disease: Two Sides of the Same Coin?
The key finding relevant to ALS-PDC is the presence of MAM-induced transcriptional changes in the brains of young adult mice that lack efficient repair of O 6-methylguanine DNA lesions. This is in contrast to the absence of significant transcriptional changes in the adult brain of MAM-treated wild-type mice. The human brain shows variable amounts of MGMT activity, but most adult brains studied have minimal levels comparable to that of Mgmt−/− mice [21]. If the human brain responds to MAM in a manner comparable to the Mgmt−/− mouse brain, the response would be alterations in cell signaling pathways linked to both neurodegeneration and neuropsychological abnormalities. While the present results are based on short-term studies, the apparent association between the most significant MAM-associated biological functions in the adult mouse brain and cycasin-associated ALS-PDC are obvious. Their corollary provides clear support for further examination of the possible etiologic role of cycasin in the induction of ALS-PDC, one form (dementia) of which shows impressive clinical and neuropathological relationships with AD [42].
Behrens and colleagues [29] have reported an inverse relationship in the incidence of cancer and AD: in a prospective longitudinal study, the risk of developing cancer with time was significantly reduced in participants with AD, while those with a history of cancer had a lower rate of AD. The investigators state: “in cancer, cell regulation mechanisms are disrupted with augmentation of cell survival and/or proliferation, whereas, conversely, AD is associated with increased neuronal death, either caused by, or concomitant with, β-amyloid (Aβ) and tau deposition.” They discuss the putative role of P53 and the Wnt signaling pathway in these inverse disease associations: whereas reduced P53 expression arising from mutations may lead to uncontrolled cell proliferation, as, in colorectal cancer, bone cancer (osteosarcoma), and other tumors, increased P53 expression may activate pathways leading to cell death, such as occurs in AD [29]. The gene coding for TP53 was modulated by MAM in both DNA lesion-anchored sets of brain genes: while TP53 activation is known to occur after DNA damage, continued activation in the Mgmt−/− brain might promote neuronal demise.
Links with Brain Pathology in ALS-PDC and AD
Wnt signaling and insulin signaling are also among the top KEGG pathways perturbed in the brain after systemic MAM treatment. While MAM-induced activation of the Wnt/β-catenin pathway leads to uncontrolled cell proliferation in the AOM model of colon cancer, suppression of this pathway in the brain may promote cell death. Boonen and colleagues [43] propose that disrupting the tightly regulated brain Wnt signaling pathway may constitute a key pathological event in AD. They propose that amyloid-beta (Aβ), a key protein in senile plaques, may down-regulate the Wnt/β-catenin pathway, thereby upregulating GSK3β and its subsequent hyperphosphorylation of tau, linking Aβ and neurofibrillary tangle pathology. Others have shown that inhibition of GSK3β increases mouse brain insulin-like growth factor-1 (IGF-1) [44], which in turn promotes Aβ production [45], [46]. IGF-1 and GSK3β are elevated in the hippocampus and spinal cord of individuals with Guam and Kii ALS [47]. IGF-1 is a potent survival factor for motor neurons in animals, and GSK3β is suspected to play important roles in apoptosis and tau phosphorylation [47].
The involvement of insulin signaling in AD has led to the proposal that this neurodegenerative disorder is a “special form of diabetes mellitus of the brain” [48]. The presence of microRNA1 as a differentially regulated hub in MAM-treated animals is noteworthy because of its ability to regulate insulin signaling (especially the IGF-1 receptor) [49], its association with colon cancer [50], and the key roles of ERK1 and microRNAs in tau phosphorylation and AD [51]. The insulin signaling pathway in diabetes mellitus type 2 is regulated by a number of transcription factors, notably HNFs, especially MODY1 (HNF4α), which regulates a large fraction of the hepatic and pancreatic transcriptomes by binding directly to approximately half of the transcribed genes. Therefore, HNF-4 serves as a “master regulator” of the human genome [52], [53]. There is substantial evidence that HNF-4 has a unique role in glucose-dependent insulin secretory pathways [54], [55], since mutations within the HNF-4α gene are linked to the monogenetic disorder Mature Onset Diabetes of the Young (MODY-1) [56]. HNF-4 was the most over-represented transcription factor-binding site among the promoter regions of brain genes that were modulated by MAM and anchored to DNA lesions. Since HNF-4 binds as a homodimer to a DNA recognition site containing a guanine-rich direct repeat element (AGGTCAaAGGTCA), this transcription factor might be a ‘hotspot’ for MAM-induced O6-mG DNA lesions. Several studies have shown that minor alterations in a nucleobase (e.g., O6-mG, 8-oxoG) at a crucial position within a promoter element can disrupt transcription factor binding and potentially modify gene expression [57]–[61]. Such a mechanism might explain why the HNF-4 consensus sequence was primarily targeted by MAM in the genes that were anchored to O6-mG DNA lesions.
The P38-MAPK signaling pathway is also among the top KEGG pathways perturbed by MAM. This cascade is activated following genotoxic stress [62], involved in the AOM model of colorectal cancer. and is widely believed to contribute to neuroinflammation in AD [63]. P38-MAPK has important roles in brain function, including glutamate (AMPA) receptor trafficking, NMDA-induced outward currents, excitotoxicity, synaptic plasticity and tau phosphorylation [63]–[66]. P38-MAPK hubs, linked to ionotropic and metabotropic glutamate receptors, were prominent in the anchored DNA damage data sets derived from the brains of MAM-treated mice suggesting that MAM-induced genotoxic stress perturbs glutamatergic function via a P38-MAPK-mediated mechanism. Given that MAM modulates glutamate-stimulated neuronal tau mRNA expression in vitro [67], we have proposed elsewhere that continuous MAM activation of glutamate-stimulated tau expression could trigger a slowly progressive neurodegenerative disease (tauopathy) of the type seen in Western Pacific ALS-PDC [68]. This mechanism is consistent with the observation that human exposure to cycad is followed by a clinically silent, long-latent period spanning years or decades [7], [13].
Other links with ALS-PDC
There are other reasons to suspect an etiological relationship between the cycad genotoxins cycasin/MAM and Western Pacific ALS-PDC. First, treatment of postnatal mice with MAM arrests cerebellar development [69] leading to the production of ectopic, multinucleated Purkinje-like cells comparable to those reported in Guam and Kii ALS brains [70]. Second, individuals with ALS-PDC and cycad-exposed animals develop skin and bone changes (summarized by Spencer [71]). The Wnt signaling pathway, which was perturbed by MAM, plays a pivotal role in the regulation of bone mass, with pathway activation in bone regeneration [72], [73]. Wnt proteins (Wnt5a) also regulate epidermal differentiation in adult skin [74]. Animals grazing on cycads have a tendency to lose their horns and hooves, in addition to developing neuromuscular disease, and Guam Chamorros have had a high and familial incidence of benign bony nodules or multiple exostoses (diaphyseal aclasis) [71] and thickened skulls (unpublished data). Cycads speed skin repair in rodents, and the skin of ALS and ALS-PDC patients is uncommonly resistant to bed sores [68]. ALS skin shows increased expression of TDP-43 [75], one of the proteins that accumulates in ALS/PDC brains along with tau, Aβ, α-synuclein and ubiquitin [42].
There are important implications for human health if the association between cycasin/MAM and neurodegeneration is confirmed in longer-term studies. First and foremost, this would radically impact understanding of the etiology of sporadic ALS, AD and other tauopathies, and raise the intriguing possibility of creating animal models of these disorders using environmental agents that perturb genome regulation. Second, in the realm of public health, environmental agents and drugs with MAM-like genotoxic properties (e.g., nitrosamines, hydrazines, streptozotocin) would be suspect etiologic agents for [triggers of] sporadic neurodegenerative disease, particularly in individuals exposed early in life or those with impaired DNA-damage responses. Others have recently advanced the hypothesis that DNA-damaging nitrosamines, previously proposed as risk factors for cancer [76], may increase risk for AD as well as diabetes mellitus [77], [78]. Additionally, injection of the diabetogenic drug streptozotocin directly into the brain results in morphological abnormalities that include hyperphosphorylated tau and Aβ proteins [79]. These conditions, it should be noted, may involve long latent periods prior to clinical expression, consistent with the concept of a “slow toxin” proposed for cycasin [71].
Summary
Given that Western Pacific ALS-PDC has occurred in genetically distinct populations that have used cycad for food and/or medicine, the present findings show that cycasin (the glucoside of MAM) is a plausible etiologic candidate for this disease. Single systemic treatment of adult mice with MAM rapidly damages brain DNA (O6-mG lesions) and modulates signaling pathways and neurotransmitter systems associated with human neurodegenerative disease. This statement is true only for animals with very low brain levels of the specific DNA-repair enzyme MGMT comparable to those in the young adult human brain. Murine brain signal pathways modulated by MAM and linked to human neurodegenerative disease overlap with those associated with MAM-induced colon cancer. The two disease phenotypes, neurodegeneration and cancer, are mechanistically related; their expression may depend on whether (colon epithelium) or not (neuron) MAM-exposed cells are able to undergo mitosis, mutagenesis and uncontrolled cell proliferation.
Our findings are based on the identification of interacting molecular networks, which permits identification of the key hubs for those networks. The hubs and the networks they connect, not the individual genes of each affected network, provide the key information in this study. Brain transcriptional changes linked to MAM-induced DNA damage were assessed within days of systemic treatment with the genotoxin to determine the earliest alterations in cellular function.
Finally, it is important to note that indictment of cycasin/MAM as a potential etiologic agent in ALS-PDC does not exclude a role for other cycad chemicals, including β-N-methylamino-L-alanine [12]; rather, it spurs the search for chemically related compounds that may play a role in sporadic forms of other tauopathies, including AD.
Materials and Methods
Ethics Statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH). The protocol was approved by committees on the ethics of animal experimentation at the participating institutions. The Oregon Health & Science University (OHSU) Institutional Animal Care and Use Committee (IACUC) Protocols are #161 (G.E.K.) and B11100 (P.S.S.). The Fred Hutchinson Cancer Research Center (FHCRC) IACUC Protocol is #1595 (H.Z.).
Mouse treatment with MAM
Methylazoxymethanol acetate (MAM purity ≥96% by GC) was purchased from Midwest Research Institute Chemical Carcinogen Repository (Kansas City, Missouri) and stored at −20°C. Eleven-week-old male C57BL6 wild type and Mgmt −/− mice on a C57BL6 background were acclimated for 5 days to local housing. Mgmt −/− mice were genotyped as previously described [24]. Animals were randomly assigned to two treatment groups (n = 4 per group) and given a single intraperitoneal injection (100 µl adjusted to body weight) with either vehicle (0.05% high-performance liquid chromatography-grade acetic acid in saline) or MAM (20 mg/kg in vehicle). Animals were terminated by guillotine 6 hr, 24 hr, 48 hr or 7 days post-injection, periods during which no signs of evolving neurological or behavioral changes were observed. Brains were rapidly excised and transected longitudinally: the right half was placed in ice-cold RNALater™ (Applied Biosystems/Ambion Inc, Austin, Texas) for transcriptional analysis and the left half flash-frozen for analysis of DNA lesions. This protocol was carried out in two independent laboratories (FHCRC, OHSU).
DNA lesions
A blind assay for DNA damage in liver and individual half-brains was carried out at the Food & Drug Administration National Center for Toxicological Research (Jefferson, Arkansas) using a liquid chromatography tandem mass spectrometry (LC/MS-MS) procedure specific for O6-methyldeoxyguanosine DNA lesions [80]. Genomic DNA was isolated using Qiagen Genomic-tip 100/G columns, as described by the manufacturer (Qiagen, Valencia, California). Briefly, genomic DNA was purified by mechanically disrupting the tissue in lysis buffer with a Potter-Elvehjem homogenizer, the homogenate incubated with proteinase K, then washed twice with 70% ethanol, air-dried, and dissolved in LC-MS buffer (5 mM bis-Tris pH 7.1, 0.1 mM ethylenediaminetetraacetic acid). Purity was checked by 260/280 ratios (range 1.8–2.0), and the concentration determined by the NanoDrop™ method (NanoDrop Technologies, Wilmington, Delaware). An aliquot of genomic DNA (∼10–20 µg) was incubated with 4 units of micrococcal nuclease and 0.5 units of spleen phosphodiesterase overnight at 37°C in hydrolysis buffer (14 mM succinic acid, 8.5 mM calcium chloride, pH 6). Nuclease P1 (1.5 units in 1 mM zinc chloride) was added, the samples incubated at 37°C for 2 h, an internal standard added (15N-O 6-methyldeoxyguanosine) to the digest, and the samples centrifuged before analysis by LC/MS-MS. Reference DNA (calf thymus DNA treated with methyl nitrosourea to give ∼250 lesions in 107 normal nucleotides) was used in each run to control for day-to-day variability. The minimal detectable quantity of DNA lesions in liver and brain tissue was 1–2.6 O6-mG lesions/108 normal nucleotides.
The liver and brain of vehicle-treated wild type and Mgmt−/− mice showed minimal detectable quantities (MDQ = 1–2.6) of O 6-mG DNA lesions/108 normal nucleotides at 6 hr, 24 hr, 48 hr and 168 hr post MAM treatment.
Microarray processing and analysis
RNA was extracted from half-brains using the RNeasy Mini Kit (Qiagen, Valencia, California) and analyzed for concentration and quality with the Agilent Bioanalyzer (Agilent, Andover, Massachusetts). Three of the four RNA samples from each treatment group at each time point were frozen and shipped on dry ice to Icoria, Inc. (Research Triangle Park, North Carolina) where they were hybridized. Total RNA was labeled using the one-cycle target labeling protocol and hybridized to GeneChip® Mouse Genome 430 2.0 Arrays (Affymetrix, Santa Clara, California). Preliminary analysis of brain and liver data was performed by the Rosetta Resolver® system (Rosetta Biosoftware, Seattle, Washington) for gene expression analysis.
Data from raw cell intensity files (.CEL) were pre-processed and normalized using robust multi-array average (RMA). RMA values were calculated with R/Bioconductor's package [81]–[84] using the default arguments (background correction using RMA background correction, quantile normalization, and median polish). Data from each of the four time points were analyzed separately. A linear model was separately fitted to RMA-normalized intensity measures for each of the ∼45,000 probes. Models included terms for the three effects of interest: treatment (MAM vs. vehicle), genotype (wt vs. Mgmt−/−), and the interaction between treatment and genotype. Four additional model terms were included to account for the two laboratories (FHCRC vs. OHSU), and the interaction of laboratory with the three effects of interest. Consistency of these effects was assessed through the interactions involving the laboratory: if p>0.15 for interactions involving the laboratory, the initial model was reduced (by excluding interactions with laboratory) and then refitted and effects of interest estimated; if interactions with the laboratory were significant (p<0.15), then the probe was excluded from further analysis as this indicated effects of genotype, treatment, or the interaction between the two effects varied between laboratories. Probes with consistent effects between laboratories had p-values for treatment, genotype, and the interaction adjusted to control the false discovery rate (FDR) [85]. Probes were declared significant if the FDR-adjusted p-value was <0.05 and the absolute fold change for the particular effect was >1.3.
All data are compliant with the MIAME (Minimum Information About a Microarray Experiment standard. Raw data were deposited in Gene Expression Omnibus (GEO), using the GEO Accession Number GSE26600. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=rxmhhmkgwmiwedq&acc=GSE26600.
Anchoring Microarray Data to DNA lesions
For MAM-treated animals (wt and Mgmt−/−), Spearman's rank correlation coefficient was computed between RMA-normalized signal intensities and the number of O6-mG lesions. Correlations were computed separately for each time point. Correlation coefficients between lesion levels and normalized intensity >0.70 were taken as an indication of “anchoring” between individual gene expression and brain DNA damage. No significant change in brain gene expression was found in wt mice treated with MAM.
Network analysis
Probes that showed significance in the microarray analysis for at least one time point (6 h, 24 h, 48 h or 7 days post-injection) were analyzed for enriched networks using Ingenuity® software (http://www.ingenuity.com/). Networks were developed separately for probe sets where the MAM vs. vehicle effect was modified by genotype and also for probes where the MAM vs. vehicle effect was only significant in the Mgmt−/− mice. Network analysis was further sharpened by restricting the focus to probes anchored to DNA lesion levels.
Transcription Factor Binding-site Analysis
Transcription factor binding-site analysis was performed using PAINT (http://www.dbi.tju.edu/dbi/tools/paint/) [86]. PAINT identifies transcriptional response elements (TRE) in the 5′-untranslated regions of genes and assesses which TREs occur at a frequency different than expected by chance. p<0.05 was considered statistically significant. 5 kb of the 5′-untranslated regions of the genomic sequences were used for the analysis. Default settings were used for all other parameter choices.
Supporting Information
Median number of O 6-mG DNA lesions for wt and Mgmt−/− animals. Analysis of mouse brain DNA adduct levels using robust linear models (RLM, Wilcoxon weights) [32]–[34]. Data obtained from brains 6 hr post-systemic treatment with MAM. Median response (O 6-mG DNA lesions per 108 normal nucleotides) given along with estimated standard error of median in square brackets. Tests compare median number of O 6-mG DNA lesions per 108 normal nucleotides between Mgmt−/− and wild type mice; reported test statistic (TS) is distributed approximately as F(1,28).
(DOCX)
List of genes supporting data in Table 1 . Genes modulated by MAM within each of the four top biological functions (MAM vs. vehicle).
(DOC)
List of genes supporting data in Table 2 . Genes modulated by MAM within each of the four top Biological Functions (wt vs. Mgmt −/−).
(DOC)
Acknowledgments
Rodger Metheny is thanked for facilitating coordination among the centers.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by the National Institutes of Environmental Health Sciences: ES11384 (OHSU), ES11399 (MIT), ES011387 (FHCRC/UW) and ES07033 (UW) (http://www.niehs.nih.gov/research/supported/centers/trc/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zhang ZX, Anderson DW, Mantel N, Roman GC. Motor neuron disease on Guam: geographic and familial occurrence. Acta Neurol Scand. 1996;94:51–9. doi: 10.1111/j.1600-0404.1996.tb00039.x. [DOI] [PubMed] [Google Scholar]
- 2.Roman GC, Zhang Z-X, Ellenberg JA. The neuroepidemiology of Parkinson's disease. In: Ellenberg JA, Koller WC, Langston JW, editors. Etiology of Parkinson's Disease. New York: Marcel Dekker; 1995. pp. 203–43. [Google Scholar]
- 3.Kuzuhara S, Kokubo Y, Sasaki R, Narita Y, Yabana T, et al. Familial amyotrophic lateral sclerosis and parkinsonism-dementia complex of the Kii Peninsula of Japan: clinical and neuropathological study and tau analysis. Ann Neurol. 2001;49:501–11. [PubMed] [Google Scholar]
- 4.Plato CC, Garruto RM, Glasko D, Craig UK, Plato M, et al. Amyotrophic lateral sclerosis and parkinsonism-dementia complex of Guam: changing incidence rates during the past 60 years. Am J Epidemiol. 2003;157:149–57. doi: 10.1093/aje/kwf175. [DOI] [PubMed] [Google Scholar]
- 5.McGeer PL, Steele JC. The ALS/PDC syndrome of Guam: Potential biomarkers for an enigmatic disorder. 2011. Prog Neurobiol April 20 [Epub ahead of print] [DOI] [PubMed]
- 6.Winton J, Joyce S, Zhukareva V, Practico D, Perl DP, et al. Characterization of tau pathologies in gray and white matter of Guam parkinsonism-dementia complex. Acta Neuropathol. 2006;111:401–12. doi: 10.1007/s00401-006-0053-0. [DOI] [PubMed] [Google Scholar]
- 7.Garruto RM, Gajdusek C, Chen KM. Amyotrophic lateral sclerosis among Chamorro migrants from Guam. Ann Neurol. 1980;8:612–9. doi: 10.1002/ana.410080612. [DOI] [PubMed] [Google Scholar]
- 8.Garruto RM, Gajdusek C, Chen KM. Amyotrophic lateral sclerosis and parkinsonism-dementia among Filipino migrants to Guam. Ann Neurol. 1981;10:341–50. doi: 10.1002/ana.410100405. [DOI] [PubMed] [Google Scholar]
- 9.Kurland LT. An appraisal of the neurotoxicity of cycad and the etiology of amyotrophic lateral sclerosis on Guam. Fed Proc. 1972;31:1540–2. [PubMed] [Google Scholar]
- 10.Whiting MG. Toxicity of cycads: implications for neurodegenerative diseases in cancer. Transcripts for four Cycad Conferences. New York: Third World Medical Research Foundation; 1988. [Google Scholar]
- 11.Spencer PS, Palmer, Ludolph AC. On the decline and etiology of high-incidence motor system disease in West Papua (southwest New Guinea). Mov Disord. 2005;20,(Suppl 12):S119–26. doi: 10.1002/mds.20552. [DOI] [PubMed] [Google Scholar]
- 12.Kisby GE, Ellison M, Spencer PS. Content of the neurotoxins cycasin (methylazoxymethanol β-D-glucoside) and BMAA (β-N-methylamino-L-alanine) in cycad flour prepared by Guam Chamorros. Neurology. 1992;42:1336–40. doi: 10.1212/wnl.42.7.1336. [DOI] [PubMed] [Google Scholar]
- 13.Borenstein AR, Mortimer JA, Schofield E, Wu Y, Salmon DP, et al. Cycad exposure and risk of dementia, MCI, and PDC in the Chamorro population of Guam. Neurology. 2007;68:1764–71. doi: 10.1212/01.wnl.0000262027.31623.b2. [DOI] [PubMed] [Google Scholar]
- 14.Kisby GE, Standley K, Lu X, O'Malley J, Lin B, et al. Molecular networks perturbed in a developmental animal model of brain injury. Neurobiol Dis. 2005;19:108–18. doi: 10.1016/j.nbd.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Kisby GE, Olivas A, Park T, Churchwell M, Doerge D, et al. DNA repair modulates the vulnerability of the developing brain to alkylating agents. DNA Repair (Amst) 2009;8:400–12. doi: 10.1016/j.dnarep.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shors TJ, Miesegaes G, Beylin A, Zhao M, Rydel T, Gould E. Neurogenesis in the adult is involved in the formation of trace memories. Nature. 2001;410:372–6. doi: 10.1038/35066584. [DOI] [PubMed] [Google Scholar]
- 17.Shors TJ, Townsend DA, Zhao M, Kozorovitskiy Y, Gould E. Neurogenesis may relate to some but not all types of hippocampal-dependent learning. Hippocampus. 2002;12:578–84. doi: 10.1002/hipo.10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laqueur GL, Mickelsen O, Whiting MG, Kurland LT. Carcinogenic properties of nuts from Cycas circinalis L. indigenous to Guam. J Natl Cancer Inst. 1963;31:919–51. [PubMed] [Google Scholar]
- 19.Rosenberg DW, Giardina C, Tanaka T. Mouse models for the study of colon carcinogenesis. Carcinogenesis. 2009;30:183–96. doi: 10.1093/carcin/bgn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haddock RL, Whippy HJD, Talon RJ, Montano MV. Ethnic disparities in cancer incidence among residents of Guam. Asian Pacific J Cancer Prev. 2009;10:57–62. [PMC free article] [PubMed] [Google Scholar]
- 21.Silber JR, Blank A, Bobola MS, Mueller Lack of the DNA repair enzyme O6-methylguanine DNA methyltransferase in histologically normal brain adjacent to primary human brain tumors. Proc Natl Acad Sci USA. 1996;93:6941–6. doi: 10.1073/pnas.93.14.6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bobola MS, Blank A, Berger MS, Silber JR. O6-Methylguanine-DNA-methyltransferase deficiency in developing brain: implications for brain tumorigenesis. DNA Repair (Amst) 2007;6:1127–33. doi: 10.1016/j.dnarep.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robison SH, Bradley WG. DNA damage and chronic neuronal degenerations. J Neurol Sci. 1984;64:11–20. doi: 10.1016/0022-510x(84)90051-0. [DOI] [PubMed] [Google Scholar]
- 24.Glassner BJ, Weeda G, Allan JM, Broekhof JL, Carls NH, et al. DNA repair methyltransferase (Mgmt) knockout mice are sensitive to the lethal effects of chemotherapeutic alkylating agents. Mutagenesis. 1999;14:339–47. doi: 10.1093/mutage/14.3.339. [DOI] [PubMed] [Google Scholar]
- 25.Staropoli JF. Tumorigenesis and neurodegeneration: two sides of the same coin? Bioessays. 2008;30:719–27. doi: 10.1002/bies.20784. [DOI] [PubMed] [Google Scholar]
- 26.Morris LG, Veeriah S, Chan TA. Genetic determinants at the interface of cancer and neurodegenerative disease. Oncogene. 2010;29:3453–64. doi: 10.1038/onc.2010.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caricasole A, Bakker A, Copani A, Nicoletti F, Gaviraghi G, Terstappen GC. Two sides of the same coin: Wnt signaling and neurodegeneration and neuro-oncology. Biosci Rep. 2005;25:309–27. doi: 10.1007/s10540-005-2893-6. [DOI] [PubMed] [Google Scholar]
- 28.Muyllaert D, Kremer A, Jaworski T, Borghgraef P, Devijver H, et al. Glycogen synthase kinase-3β, or a link between amyloid and tau pathology? Genes Brain Behav. 2008;7(Suppl 1):57–66. doi: 10.1111/j.1601-183X.2007.00376.x. [DOI] [PubMed] [Google Scholar]
- 29.Behrens MI, Lendon C, Roe CM. A common biological mechanism in cancer and Alzheimer's disease. Curr Alzheimer Res. 2009;6:196–204. doi: 10.2174/156720509788486608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Souter S, Lee G. Tubulin-independent tau in Alzheimer's disease and cancer: Implications for disease pathogenesis and treatment. Curr Alzheimer Res. 2010;7:697–707. doi: 10.2174/156720510793611637. [DOI] [PubMed] [Google Scholar]
- 31.Serrano J, Fernández AP, Martínez-Murillo R, Martínez A. High sensitivity to carcinogens in the brain of a mouse model of Alzheimer's disease. Oncogene. 2010;29:2165–71. doi: 10.1038/onc.2009.503. [DOI] [PubMed] [Google Scholar]
- 32.McKean JW. Robust analysis of linear models. Statist Sci. 2001;19:562–570. [Google Scholar]
- 33.Terpstra J, McKean JW. Rank-based analysis of linear models using R. J Statist Software. 2005;14(7) http://www.jstatsoft.org/ [Google Scholar]
- 34.R-code to implement RLM procedure. Weighted Wilcoxon Estimates. http://www.stat.wmich.edu/red5328/WWest/ Accessed March 24, 2011.
- 35.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA. Systemic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 37.Laqueur GL, McDaniel EG, Matsumoto H. Tumor induction in germfree rats with methylazoxymethanol (MAM) and synthetic MAM acetate. J Natl Cancer Inst. 1967;39:355–71. [PubMed] [Google Scholar]
- 38.Suzui M, Sugie S, Morie H, Okuno M, Tanaka T, Moriwaki H. Different mutation status of the β-catenin gene in carcinogen-induced colon, brain, and oral tumors in rats. Mol Carcinog. 2001;32:206–12. doi: 10.1002/mc.10014. [DOI] [PubMed] [Google Scholar]
- 39.Nigro ND. Animal model for colorectal cancer. Prog Clin Biol Res. 1985;186:161–73. [PubMed] [Google Scholar]
- 40.Chen J, Huang XF. The signal pathways in azoxymethane-induced colon cancer and preventive implications. Cancer Biol Therap. 2009;8:1313–7. doi: 10.4161/cbt.8.14.8983. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi M, Wakabayashi L. Gene mutations and altered gene expression in azoxymethane-induced colon carcinogenesis in rodents. Cancer Sci. 2004;95:475–80. doi: 10.1111/j.1349-7006.2004.tb03235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miklossy J, Steele JC, Tu S, McCall S, Sandberg G, et al. Enduring involvement of tau, β-amyloid, α-synuclein, ubiquitin and TDP-43 pathology in the amyotrophic lateral sclerosis/parkinsonism-dementia complex of Guam (ALS/PDC). Acta Neuropathol. 2008;116:625–37. doi: 10.1007/s00401-008-0439-2. [DOI] [PubMed] [Google Scholar]
- 43.Boonen RA, van Tijn P, Zivkovic D. Wnt signaling in Alzheimer's disease: up or down, that is the question. Ageing Res Rev. 2009;8:71–82. doi: 10.1016/j.arr.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 44.Bolos M, Fernandez S, Torres-Aleman I. Oral administration of a GSK3 inhibitor increases brain insulin-like growth factor I levels. J Biol Chem. 2010;285:17693–700. doi: 10.1074/jbc.M109.096594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Araki W, Kume H, Oda A, Tamaoka A, Kametani F. IGF-1 promotes β-amyloid production by a secretase-independent mechanism. Biochem Biophys Res Commun. 2009;380:111–4. doi: 10.1016/j.bbrc.2009.01.044. [DOI] [PubMed] [Google Scholar]
- 46.De la Monte SM. Insulin resistance and Alzheimer's disease. BMB Report. 2009;42:475–81. doi: 10.5483/bmbrep.2009.42.8.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kihira T, Suzuki A, Kondo T, Wakayama I, Yoshida S, et al. Immunohistochemical expression of IGF-1 and GSK in the spinal cord of Kii and Guamanian ALS patients. Neuropathology. 2009;29:548–58. doi: 10.1111/j.1440-1789.2009.01010.x. [DOI] [PubMed] [Google Scholar]
- 48.Steen E, Terry BM, Rivera EJ, Cannon JL, Neeley TR, et al. Impaired insulin and insulin-like growth factor expression and signaling mechanisms in Alzheimer's disease–is this type 3 diabetes? J Alzheimers Dis. 2005;2005;7:63–80. doi: 10.3233/jad-2005-7107. [DOI] [PubMed] [Google Scholar]
- 49.Elia L, Contu R, Quintavalle M, Varrone F, Chimenti C, et al. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation. 2009;120:2377–85. doi: 10.1161/CIRCULATIONAHA.109.879429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarver AL, French J, Borralho PM, Thayanithy V, Oberg AL, et al. Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer. 2010;9:401. doi: 10.1186/1471-2407-9-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hebert SS, Aikaterini SP, Smith P, Galas M-C, Planel E, et al. Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Human Mol Gene. 2010;19:3959–69. doi: 10.1093/hmg/ddq311. [DOI] [PubMed] [Google Scholar]
- 52.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–81. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bolotin E, Liao H, Ta TC, Yang C, Hwang-Verslues W, et al. Integrated approach for the identification of human hepatocyte nuclear factor 4α target genes using protein binding microarrays. Hepatology. 2010;51:642–53. doi: 10.1002/hep.23357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roth U, Curth K, Unterman TG, Kietzmann T. The transcription factors HIF-1 and HNF-4 and the coactivator p300 are involved in insulin-regulated glucokinase gene expression via the phosphatidylinsitol 3-kinase/protein kinase B pathway. J Biol Chem. 2004;279:2623–31. doi: 10.1074/jbc.M308391200. [DOI] [PubMed] [Google Scholar]
- 55.Ganjam GK, Dimova EY, Unterman TG, Kietzmann T. FoxO1 and HNF-4 are involved in regulation of hepatic glucokinase gene expression by resveratrol. J Biol Chem. 2009;284:30783–97. doi: 10.1074/jbc.M109.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ryffel GU. Mutations in the human genes encoding the transcription factors of the hepatocyte nuclear factor (HNF) 1 and HNF4 families: functional and pathological consequences. J Mol Endocrinol. 2001;27:11–29. doi: 10.1677/jme.0.0270011. [DOI] [PubMed] [Google Scholar]
- 57.Bonfanti M, Broggini M, Prontera C, D'Incalci M. O 6-Methylguanine inhibits the binding of transcription factors to DNA. Nucleic Acids Res. 1991;19:5739–42. doi: 10.1093/nar/19.20.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghosh R, Mitchell DL. Effect of oxidative DNA damage in promoter elements on transcription factor binding. Nucleic Acids Res. 1999;27:3213–8. doi: 10.1093/nar/27.15.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parsian AJ, Funk MC, Tao TY, Hunt CR. The effect of DNA damage on the formation of protein/DNA complexes. Mutat Res. 2002;501:105–13. doi: 10.1016/s0027-5107(02)00016-7. [DOI] [PubMed] [Google Scholar]
- 60.Hailer-Morrison MK, Kotler JM, Martin BD, Sugden KD. Oxidized guanine lesions as modulators of gene transcription. Altered p50 binding affinity and repair shielding by 7,8-dihydro-8-oxo-2′-deoxyguanosine lesions in the NF-κB promoter element. Biochemistry. 2003;42:9761–70. doi: 10.1021/bi034546k. [DOI] [PubMed] [Google Scholar]
- 61.Yamini B, Yu X, Dolan ME, Wu MH, Darga TE, et al. Inhibition of nuclear factor κβ activity by temozolomide involves O 6-methylguanine induced inhibition of p65 DNA binding. Cancer Res. 2007;67:6889–98. doi: 10.1158/0008-5472.CAN-06-4496. [DOI] [PubMed] [Google Scholar]
- 62.Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell. 2007;11:175–89. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Munoz L, Ammit AJ. Targeting p38 MAPK pathway for the treatment of Alzheimer's disease. Neuropharmacology. 2010;58:561–8. doi: 10.1016/j.neuropharm.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 64.Chaparro-Huerta V, Flores-Soto ME, Gudino-Cabrera G, Rivera-Cervantes MC, Bitser-Quintero OK, Beas-Zarate C. Role of p38 MAPK and pro-inflammatory cytokines expression in glutamate-induced neuronal death of neonatal rats. Int J Dev Neurosci. 2008;26:487–95. doi: 10.1016/j.ijdevneu.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 65.Park EC, Glodowski DR, Rongo C. The ubiquitin ligase RPM-1 and the p38 MAPK PMK-3 regulate AMPA receptor trafficking. PLoS One. 2009;4:e4284. doi: 10.1371/journal.pone.0004284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang R, Sun L, Hayashi Y, Liu X, Koyama S, et al. Acute p38-mediated inhibition of NMDA-induced outward currents in hippocampal CA1 neurons by interleukin-1β. Neurobiol Dis. 2010;38:68–77. doi: 10.1016/j.nbd.2009.12.028. [DOI] [PubMed] [Google Scholar]
- 67.Esclaire F, Kisby G, Spencer P, Milne J, Lesort M, Hugon J. The Guam cycad toxin methylazoxymethanol damages neuronal DNA and modulates tau mRNA expression and excitotoxicity. Exp Neurol. 1991;155:11–21. doi: 10.1006/exnr.1998.6962. [DOI] [PubMed] [Google Scholar]
- 68.Spencer PS, Kisby GE, Ludolph AC. Slow toxins biologic markers, and long-latency neurodegenerative disease in the western Pacific region. Neurology. 1991;41:62–6. doi: 10.1212/wnl.41.5_suppl_2.62. [DOI] [PubMed] [Google Scholar]
- 69.Jones MZ, Gardner E. Pathogenesis of methylazoxymethanol-induced lesions in the postnatal mouse cerebellum. J Neuropathol Exp Neurol. 1976;35:413–44. doi: 10.1097/00005072-197607000-00004. [DOI] [PubMed] [Google Scholar]
- 70.Shirake H, Yase Y. ALS in Japan. In: Vinken PJ, Bruyn GW, editors. Handbook of Clinical Neurology. Vol. 22. System Disorders and Atrophy Pt 2. American Elsevier, New York; 1975. 353 [Google Scholar]
- 71.Spencer PS. Guam ALS/Parkinsonism-dementia: A long-latency neurotoxic disorder caused by “slow toxin(s)” in food? Can J Neurol Sci. 1987;14:347–357. doi: 10.1017/s0317167100037732. [DOI] [PubMed] [Google Scholar]
- 72.Chen Y, Alman BA. Wnt pathway, an essential role in bone regeneration. J Cell Biochem. 2009;106:353–62. doi: 10.1002/jcb.22020. [DOI] [PubMed] [Google Scholar]
- 73.Michigami T. The genetic basis for skeletal disease. Molecular advances in sclerosing bone disorders. Clin Calcium. 2010;20:1196–201. [PubMed] [Google Scholar]
- 74.Romanowska M, Evans A, Kellock D, Bray SE, McLean K, et al. Wnt5a exhibits layer-specific expression in adult skin, is upregulated in psoriasis, and synergizes with type 1 interferon. PLoS One. 2009;4:e5354. doi: 10.1371/journal.pone.0005354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suzuki M, Mikami H, Watanable T, Yamano T, Yamazaki T, et al. Increased expression of TDP-43 in the skin of amyotrophic lateral sclerosis. Acta Neurol Scand. 2010;122:367–72. doi: 10.1111/j.1600-0404.2010.01321.x. [DOI] [PubMed] [Google Scholar]
- 76.Correa P, Haenszel W, Cuello C, Tannenbaum S, Archer M. A model for gastric cancer epidemiology. Lancet. 1975;2:58–60. doi: 10.1016/s0140-6736(75)90498-5. [DOI] [PubMed] [Google Scholar]
- 77.De la Monte SM, Neusner A, Chu J, Lawton M. Epidemiological trends strongly suggest exposures as etiologic agents in the pathogenesis of sporadic Alzheimer's disease, diabetes mellitus, and non-alcoholic steatohepatitis. J Alzheimers Dis. 2009;17:519–29. doi: 10.3233/JAD-2009-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Salkovic-Petrisic M, Osmanovic J, Grunblatt E, Riederer P, Hoyer S. Modeling sporadic Alzheimer's disease: the insulin resistant brain generates multiple long-term morphobiological abnormalities including hyperphosphorylated tau protein and amyloid-β. J Alzheimers Dis. 2009;18:729–50. doi: 10.3233/JAD-2009-1184. [DOI] [PubMed] [Google Scholar]
- 79.De la Monte SM, Tong M. Mechanisms of nitrosamine-mediated neurodegeneration: potential relevance to sporadic Alzheimer's disease. J Alzheimer's Dis. 2009;17:817–25. doi: 10.3233/JAD-2009-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Churchwell MI, Beland FA, Doerge DR. Quantification of O6-methyl and O6-ethyldeoxyguanosine adducts in C57BL/6N/Tk+/− mice using LC/MS/MS, J. Chromatogr B Analyt. Technol Biomed Life Sci. 2006;844:60–66. doi: 10.1016/j.jchromb.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 81.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bolsta BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 84.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 85.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Royal Stat Soc B. 1995;57:289–300. [Google Scholar]
- 86.Vadigepalli R, Chakravarthula P, Zak DE, Schwaber JS, Gonye GE. PAINT: a promoter analysis and interaction network generation tool for gene regulatory network identification. Omics. 2003;7:235–252. doi: 10.1089/153623103322452378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Median number of O 6-mG DNA lesions for wt and Mgmt−/− animals. Analysis of mouse brain DNA adduct levels using robust linear models (RLM, Wilcoxon weights) [32]–[34]. Data obtained from brains 6 hr post-systemic treatment with MAM. Median response (O 6-mG DNA lesions per 108 normal nucleotides) given along with estimated standard error of median in square brackets. Tests compare median number of O 6-mG DNA lesions per 108 normal nucleotides between Mgmt−/− and wild type mice; reported test statistic (TS) is distributed approximately as F(1,28).
(DOCX)
List of genes supporting data in Table 1 . Genes modulated by MAM within each of the four top biological functions (MAM vs. vehicle).
(DOC)
List of genes supporting data in Table 2 . Genes modulated by MAM within each of the four top Biological Functions (wt vs. Mgmt −/−).
(DOC)