Abstract
HetR is an essential regulator of heterocyst development in cyanobacteria. HetR binds to a DNA palindrome upstream of the hetP gene. We report the crystal structure of HetR from Fischerella at 3.0 Å. The protein is a dimer comprised of a central DNA-binding unit containing the N-terminal regions of the two subunits organized with two helix-turn-helix motifs; two globular flaps extending in opposite directions; and a hood over the central core formed from the C-terminal subdomains. The flaps and hood have no structural precedent in the protein database, therefore representing new folds. The structural assignments are supported by site-directed mutagenesis and DNA-binding studies. We suggest that HetR serves as a scaffold for assembly of transcription components critical for heterocyst development.
Keywords: Anabaena/Fischerella, HTH motifs, palindrome binding, patterned development
The cyanobacterium Anabaena can carry out oxygenic photosynthesis and the fixation of nitrogen at the same time. Anabaena grows as a filament that executes these two incompatible processes simultaneously by differentiating specialized cells for nitrogen fixation within each string. The specialized cells, called heterocysts, are spaced at regular intervals along the 100–200-cell filaments. In the strain we studied, Anabaena sp. strain PCC 7120 (Anabaena hereafter), approximately one in ten vegetative cells differentiates into a heterocyst when a culture is deprived of fixed nitrogen (1, 2).
The time required for differentiation of a vegetative cell into a heterocyst is approximately the same as the division time of the vegetative cells. Heterocysts are the source of inhibitory signals that diffuse along the filament, resulting in a gradient whose minimum occurs halfway between two heterocysts. At that site, a vegetative cell differentiates, maintaining the spacing pattern (1, 2).
The diffusing inhibitors, initially thought to be the products of nitrogen fixation (glutamine or arginine), are now considered more likely to be a peptide (PatS or a derivative thereof) and a protein (HetN or a derivative), both of which contain the sequence RGSGR (3–5). A peptide with that sequence prevents binding of HetR to DNA and promotes HetR destruction. HetR is a positive factor essential for heterocyst differentiation, initiating a cascade that ultimately is responsible for the activation of more than a thousand genes (6). HetR normally turns over rapidly in vivo. It appears that both the PatS peptide and HetN promote the turnover of HetR such that a gradient of these proteins away from the heterocyst is responsible for a gradient of HetR. Thus the concentration of HetR is highest midway between two heterocysts and that is where the next heterocyst will be generated by differentiation of a vegetative cell (7).
HetR binds to a specific DNA sequence (8). One binding site has been defined precisely, a 17-base pair palindromic sequence located within the upstream promoter region of hetP, another gene whose product is involved in differentiation (9). The identical palindrome is found at a corresponding position in another strain, Anabaena variabilis ATCC 29413, but in both genomes it is the only occurrence of this motif, making the mechanism of how HetR activates thousands of genes truly enigmatic.
HetR is one of the genes discovered when Anabaena mutants incapable of nitrogen fixation were first isolated and complemented by using cosmid libraries of wild-type DNA to identify individual genes needed for differentiation (6). In that work, HetR was found to be required for the earliest steps in differentiation. When wild-type Anabaena was provided with extra copies of the hetR gene, the frequency of heterocysts increased (6). Most curiously, the amino acid sequence of the HetR protein provided no clue to its function, a mystery that persists to this day. The original hetR mutant was found to contain a serine 179 → asparagine replacement. Later, the wild-type HetR protein expressed in Escherichia coli was seen to be sensitive to proteolysis (or autoproteolysis), leading to its classification as a serine protease (Peptidase S48) (10). Nevertheless, the mutant phenotype of failing to express any of the genes required for differentiation suggested that, directly or indirectly, HetR is a transcriptional regulator.
Results and Discussion
We made many attempts to crystallize the Anabaena 7120 HetR protein. Although some crystals were obtained, they did not diffract well enough for structure determination. We then decided to attempt the cloning, purification, and crystallization of HetR from several cyanobacteria. The HetR sequence is highly conserved among heterocystous cyanobacteria (Fig. S1). This conservation extends to Trichodesmium erythraeum, a filamentous cyanobacterium that does not differentiate heterocysts but nevertheless can fix nitrogen in air (11). Fig. S1 shows the amino acid sequences of the HetR proteins from several cyanobacterial species including Anabaena (6), Nostoc punctiforme (12), Fischerella MV11 (13), and Trichodesmium erythraeum (11). Fischerella grows at elevated temperatures, up to 60 °C (13), so we thought that its HetR might provide better crystals for structure determination.
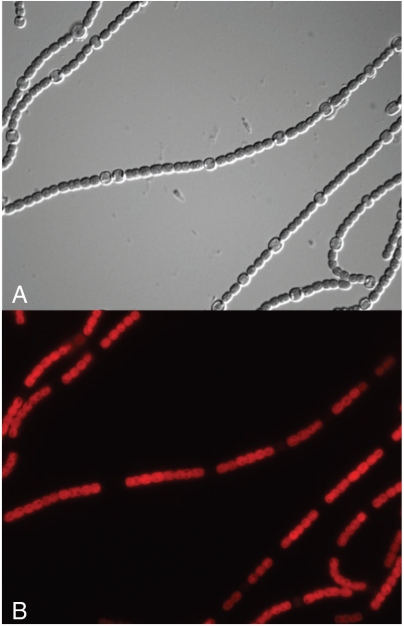
The PCR-amplified hetR genes from each of these organisms were cloned into plasmid p505 and transferred by conjugation from E. coli into a hetR mutant of Anabaena 7120 (6). The purpose of this complementation experiment was to verify that each of the HetR proteins was capable of interacting properly with the PatS peptide or with HetN to produce the normal pattern of heterocyst differentiation. This outcome was observed with all of the heterologous HetR proteins, including that of Trichodesmium. The resulting complemented strain with the Fischerella HetR protein is shown in Fig. 1. The heterocyst spacing is similar to that of the same strain complemented with the Anabaena wild-type HetR, as is the periodic pattern of chlorophyll fluorescence emission, with the heterocysts dimmed due to loss of the phycobiliproteins of photosystem II.
Fig. 1.
Complementation of Anabaena sp. 7120 HetR mutant with the hetR gene from Fischerella sp. MV11. (A) Differential Interference Contrast image showing the heterocyst spacing of the complemented strain grown in N-free medium. (B) The same field viewed with fluorescent excitation at 480 nm and emission at 650 nm, showing energy transfer from phycobilisomes to chlorophyll. The heterocysts are dim due to the destruction of the phycobilisomes during their differentiation.
All of the heterologous HetR proteins could be crystallized but the best diffraction was observed with the protein from Fischerella. We report here the 3.0 Å crystal structure of HetR from Fischerella MV11. The protein represents a unique protein fold.
The HetR Structure.
The crystal structure of HetR from Fischerella MV11 was determined first at 3.38 Å resolution using the single-wavelength anomalous diffraction (SAD) approach applied to seleno-methionine (SeMet)-labeled protein. The structure was of reasonable quality (Table 1) but a unique crystal form was obtained subsequently that diffracted to higher resolution. The structure of HetR described below is based on the 3.00 Å model of native HetR determined using molecular replacement with the 3.38 Å structure as a search model. The final model exhibited good crystallographic and geometric statistics (Table 1). HetR is very loosely packed; a number of regions in the structure are poorly ordered and are missing from the final model (see Methods). Overall, subunit B is better defined (based on 1.2σ of the 2Fo-Fc electron density map) than subunit A. HetR is mostly α-helical with several connecting loops and β-strands (see SI Text).
Table 1.
Summary of HetR crystallographic data
| Data collection statistics | HetR 1 | HetR 2 |
| Space group | P31 | P65 |
| Unit cell (Å) | a = 92.93 b = 92.93 c = 97.65 | a = 123.38 b = 123.38 c = 109.6 |
| Wavelength (Å) | 0.9791 | 0.9793 |
| Highest resolution bin (Å) | 3.05–3.00 | 3.46–3.25 |
| Number of observed reflections | 18,807 (905) | 23,425 (671) |
| Rmerge(%) * | 15.7 (70.9) † | 14.0 (77.0) † |
| Completeness (%) | 100 (100) | 99.9 (100) |
| I/σI | 5.3 (2.0) † | 9.3 (3.0)† |
| Phasing and refinement | MR | SAD |
| Search model: chain B of HetR 2 | Phasing power/FOM: 1.29/0.24 | |
| Phasing resolution range (Å) | 46.5–3.00 | 40.4–3.38 |
| Number of SeMet | 0 | 11 |
| Refinement resolution range (Å) | 46.5–3.00 | 40–3.38 |
| Rcrystz (%) | 18.3 | 20.7 |
| Rfree (%) | 24.7 | 26.7 |
| Number of protein residues | 598 | 598 |
| Number of DNA residues | 0 | 0 |
| Solvent molecules | 1 | 4 |
| Bond lengths (Å) | 0.009 | 0.017 |
| Bond angles (deg) | 1.498 | 1.896 |
| B-factors (Å2) | 52.9 | 112.3 |
| Protein main chain | 55.75 | 110.5 |
| Protein side chain | 51.30 | 113.34 |
| Solvent (water) | 28.1 | 83.3 |
| Wilson B-factor (Å2) | 55.44 | 94.22 |
| Twinning ‡ | ||
| Operator | −k,−h,−l | |
| Fraction | 0.500 | |
| Ramachandran plot (%) § | ||
| Preferred | 92.8 | 91.5 |
| Generously allowed | 7.5 | 12.7 |
| Disallowed | 0 | 1.4 |
| PDB ID | 3QOD | 3QOE |
*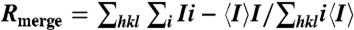 , where Ii is the intensity for the ith measurement of an equivalent reflection with indices h, k, and l.
, where Ii is the intensity for the ith measurement of an equivalent reflection with indices h, k, and l.
†Numbers in parentheses are values for the highest-resolution bin.
‡Refined using Phenix.
§Defined in COOT.
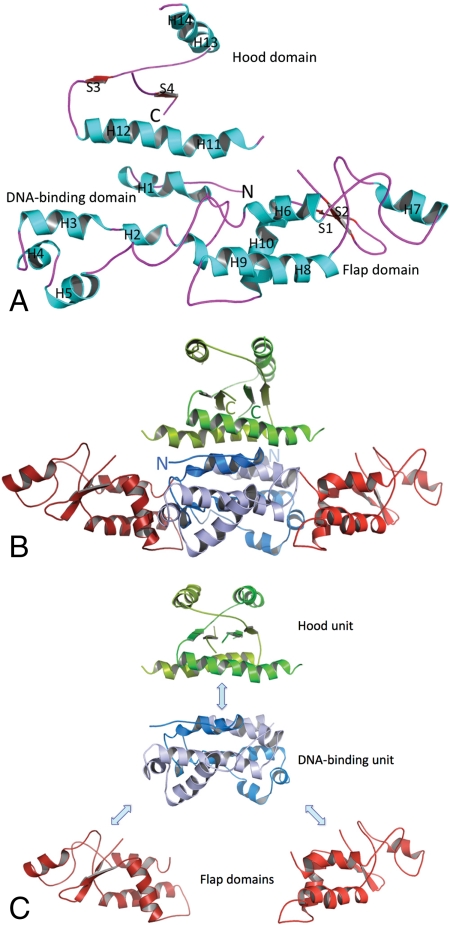
The HetR monomer is composed of three distinct domains: the N-terminal domain (residues 1–98), the middle domain (residues 99–213) designated the “flap”, and a slightly smaller C-terminal domain (residues 214–296), designated the “hood” (Fig. 2). The structure of the monomer is quite extended and the only globular region, the flap domain, is in the middle of the sequence. This domain is composed of four α-helices (H6–H9), one 310-helix, and a β-hairpin (Figs. S1 and S2). Both the N-terminal DNA binding and C-terminal hood regions are in open conformations and are very unlikely to exist alone as such. While the (helix-turn-helix) HTH-like domain shows similarity to proteins with known structures, as a whole HetR represents a unique fold. It appears that the functional hetR gene was formed by combining two new domains (the flap and hood) with a well-defined DNA-binding module.
Fig. 2.
HetR structure. (A) The HetR subunit B fold. α-Helices (H) are in aqua; β-strands (S) are red, and loops are purple. N and C termini are labeled. (B) Crystal structure of the HetR dimer. A ribbon representation of the HetR structure. Secondary structure elements are indicated by H (helices) and S (β-strands). Subunits A and B are shown in lighter colors (light red, blue, and green) and darker colors (dark red, blue, and green), respectively. The DNA-binding unit is shown in blue, flap domains in red, and the hood in green. Protein N and C termini are labeled. (C) Domain organization of the HetR dimer.
HetR Forms a Dimer.
HetR monomers interact (Fig. 2B) to form a highly intertwined, compact dimer with three distinct structural units (Fig. 2C). We determined two crystal structures of HetR. Both the trigonal form obtained at low ionic strength and the hexagonal form, obtained at high ionic strength, contain one dimer of HetR in the asymmetric unit, with the same dimer arrangement, but with somewhat different domain orientations and alterations of secondary structure elements (Fig. S2). The dimeric state is stabilized by interactions between the α-helical and loop regions of the N-terminal and C-terminal domains of each subunit (Fig. 2B). The dimer interface is very extensive; it buries a large, solvent-accessible surface area of 5,100 Å2 per monomer with ΔG = -80 kcal/mol. This arrangement leads to the formation of a structured α-helical central core stabilized by hydrophobic and van der Waals interactions and several intermonomer hydrogen bonds [S14 with carbonyl of L95′ (and carboxylate of D17), Y22 with D240′ and carbonyl of R236′, H66 with E184′ and carbonyl of A181′], and one intersubunit salt bridge formed between R29 and E56′ (also involving interaction with Q57′ and the carbonyl of T53′) and their corresponding symmetry mates. The stability of the dimer interface seems critical to HetR function as suggested by the point mutation D17E, which interferes with DNA binding and heterocyst formation in Anabaena (14). It has been suggested that HetR forms functional homodimers (8) and that the dimers are stabilized by a disulfide bond involving a single conserved cysteine residue (C49). The HetR structure shows that a disulfide bond is not possible between the two chains; the cysteines are 22 Å apart. Although the cysteines are found on the interface between the two subunits, they are buried.
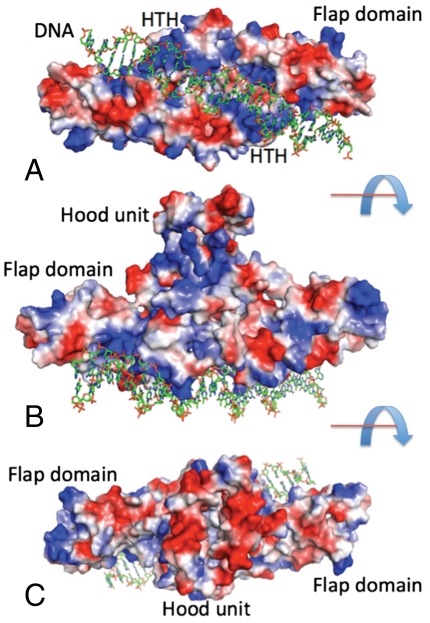
The central core is formed by the N-terminal regions assembled into an all α-helical DNA-binding unit (H1–H5 and H1′–H5′) with two classic HTH motifs separated by ∼36 Å (H4/H5 and H4′/H5′). It is expected that the DNA duplex would bind across the DNA-binding unit with the HTH motifs interacting with the major groove of B-DNA (Fig. 3). The DNA-binding unit of the dimer would span the approximately 16–17 base pairs of the palindromic element of B-DNA identified as the HetR-binding site in the Anabaena hetP promoter (9). The C-terminal regions of the monomers form an appendage, the hood, positioned on top of the DNA-binding unit. This unit forms a three-layer α/β/α sandwich with the top and bottom layers having four α-helices (H11H12/H11′H12′—bottom layer; H13H14/H13′H14′—top-layer) and can be understood only as an integral component of the dimer structure. These helices sandwich a four-stranded β-sheet (S3↑, S4′↓, S4↑, and S3′↓). Helices from the top layer run parallel to the β-strands while helices from the bottom layer are perpendicular to the β-strands (Fig. 3). The hood unit is also stabilized by an intersubunit hydrogen bond (T248 with carbonyl of F294′). However, most of this unit is packed rather loosely and shows a high degree of flexibility (Fig. S2). At the top there is a large negatively-charged cavity on which other components of a transcription complex might dock (Fig. 3B). The two flap domains protrude away from the central N-terminal-C-terminal core structure. These domains are also in position to contact DNA, perhaps at the exterior phosphates, which could enhance the interaction with DNA throughout the length of HetR (Fig. 3C).
Fig. 3.
A model for DNA binding. 27 base-pair B-DNA containing the 17-bp palindrome identified in (9) was modeled by manually fitting to the DNA-binding unit of HetR. The protein is shown in surface rendering, using PyMol (26) to map the local electrostatic potential of HetR. Three views are shown, B and C rotated 90° with respect to A: down the DNA-binding unit (A), sideways (B), and down the hood unit (C). Blue and red represent the positive and negative charge potential at the + and -70 kT e-1 scale, respectively.
The overall structure of the dimer is quite unusual: the multilayered central core is assembled by the DNA-binding unit and the hood appendage unit with two auxiliary flap domains projecting in opposite directions from the central core (Figs. 2 and 3). Although many structural elements of the dimer are related by a 2-fold axis, the dimer shows considerable asymmetry in the positioning of the flap and hood domains. Also, a number of small segments of the secondary structure elements show somewhat different conformations between the two subunits. For example, some α-helices are partly unwound, the loops assume different conformations, and some parts of the structure are disordered, which suggests high flexibility, particularly in the absence of DNA (Fig. S2). The average B-factor of 55 Å2 also suggests high mobility of many elements of the structure including loops and domain-connecting regions in the crystal. The packing of domains is loose with large solvent-accessible channels and cavities and with plenty of space for solvent (which at current resolution can not be modeled), peptides, and ions to access the interior of the protein and for DNA and other proteins to interact with the HetR surface. The HetR dimeric state in solution has been confirmed by size exclusion chromatography (Fig. S3). The structure suggests that this protein can serve as a scaffold for initiating the assembly of complexes with many partners.
HetR Sequence Family.
HetR is a member of a protein family found exclusively in cyanobacteria (PF03574). In this family, approximately 60% of the residues are strictly conserved, spanning almost the entire length of the protein and distributed among all three domains (Fig. S1). The conservation is so high that only 28 out of 299 residues in the sequence of HetR are variable (Fig. S1), suggesting very strong evolutionary pressure to preserve not only the protein structure but also its amino acid sequence. This conservation is remarkable, considering the very long evolutionary history of cyanobacteria. The highly conserved residues comprise the dimerization core and the HTH motifs of the DNA-binding domain. In addition, the flap domain contains three and the hood domain two conserved sequence motifs, respectively. The functional role of these motifs is unknown at present, but the flap and hood domains probably provide sites for binding other components of the transcription complex.
Structural Homologs of HetR.
Structural comparison of HetR with proteins deposited in the Protein Data Bank (PDB) using the ProFunc and DALI servers (15, 16) revealed only one structural homolog. The E. coli Fis protein (PDB code 3FIS, Z-score 2.79, rmsd 2.3 Å, sequence identity 12.3%, covering ∼80 residues) maps to the DNA-binding unit of HetR involving helices H2 and H3 as well as the HTH motif H4/H5. Fis is known as a “nucleoid-associated” or “histone-like” protein (17, 18). Fis was originally identified as a factor required for inversion of the Hin and Gin elements by site-specific DNA recombinases of Salmonella and phage Mu, respectively. This small protein bends DNA upon binding and functions in many cellular processes including phage lambda site-specific recombination, transcriptional activation of rRNA and tRNA operons, repression of its own synthesis, and oriC-directed DNA replication.
Several other factors affecting transcription show structural similarity to the HetR HTH motif (H4/H4′; residues 62–88). These factors include a multiple antibiotic resistance repressor, MarR (PDB code 1JGS, RMSD 1.15 Å, 26 residues overlap) (19), diphtheria toxin repressor (PDB code 2DTR, RMSD 0.98 Å, 25 residues overlap) (20), and catabolite gene activator protein (PDB code 1CGP, RMSD 1.26 Å, 25 residues overlap) (21). A more distant homolog is the E. coli trp operon repressor (PDB code 1JHG, RMSD 1.69 Å, 27 residues overlap) (22).
The other HetR structural units show even lower homology to known protein structures. The flap domain shows very distant homology to one of the domains of edema factor from Bacillus anthracis (PDB code 1XFY, Z-score 0.28, RMSD 4.71 Å, 52 residues overlap) and to melanoma-associated antigen G1 (PDB code 3NW0, Z-score 4.5, RMSD 2.8 Å, 72 residues overlap). No homologs were found for the hood structural unit. We conclude that HetR is comprised of several unique protein folds linked to a familiar DNA-binding module.
DNA Binding.
The structural comparisons suggest that HetR binds a specific DNA target. Such a target has been identified in the Anabaena genome (9). Residues in the N-terminal part of the second helix (H5) (EPKRVK) in the HTH motif may recognize specific base edges in the major groove of the target DNA directly. Considering the nearly 100% residue conservation among HetRs in the C-terminal region of H4 and the N-terminal region of H5, it is likely that the DNA target sequence is equally conserved among different cyanobacterial species. Identical sequences are found in the genomes of two fully sequenced cyanobacteria—A. variabilis and Anabaena sp. PCC 7120 (23). To verify that residues in the HTH motif are involved in the recognition of DNA sequence, we mutated three basic residues from the HTH motif in Anabaena HetR (Arg62 in H4, and Lys73 and Arg74 in H5) to glutamic acid. All of these mutants were found to be deficient in specific binding to a 29-bp palindromic oligonucleotide containing the Anabaena HetR recognition sequence (22, Fig. S4). Another DNA binding deficient mutation at position D17 results in a Het- phenotype in Anabaena (14). D17 is on the dimer interface and its substitution may affect dimer stability. Two other mutations were reported for the DNA-binding unit at G36 and H69. Both of these mutations reduce heterocyst differentiation substantially (14). H69 is in the HTH motif and also at the interface between the DNA binding and flap domains. Mutation of this residue should interfere with both DNA binding and the conformation of the flap domain. G36 is on the dimer interface and is in close contact (4.6 Å) with H69, suggesting that this residue could be important for H69 orientation.
Analysis of the HetR structure also identifies the protein surface that is involved in binding the specific DNA sequence. The 2 N-terminal DNA-binding units, with symmetrically positioned HTH motifs, define a proximal DNA-binding unit that should obey twofold symmetry, consistent with the palindromic nature of the HetR recognition sequence identified experimentally (9). This part of the HetR surface is positively charged (Fig. 3), particularly the HTH motifs. The size of the DNA-binding unit suggests that the DNA target is approximately16–17 bp long. A cavity between the two HTH motifs is clearly large enough to accommodate the minor groove and phosphate units of DNA. The positively charged patch extends beyond the HTH motifs into the flap domains, so the DNA target interacting with HetR may be longer. In that case, the flap domain might provide an additional grip on DNA that is less sequence-specific. Including these domains would extend the size of the DNA target to ∼28 bp. We have modeled the HetR/DNA complex using the HetR dimer and 27-bp B-DNA (Fig. 3). It is clear that DNA can interact with the DNA-binding unit and both flap domains. The flap surface close to the DNA and HTH motifs may also be a good candidate for the binding site of the inhibitory peptide RGSGR because it would interfere with accommodating DNA along this path. The HetR DNA-binding surface shows some curvature (Fig. 3B), suggesting that the bound DNA target might be bent. Fis protein, the closest structural homolog of HetR, bends its target sequence in E. coli (17, 18). The structure of the proposed complex can also be seen in Movie S1 in the SI Text.
Possible Site for RGSGR Peptide Binding.
The peptide RGSGR prevents HetR interaction with its DNA target (8). We have analyzed the structure of the HetR dimer for possible peptide and other ligand binding sites. Nest analysis can identify protein sites capable of accommodating a whole or partial negatively charged atom (15, 24). Such charged atoms could be stabilized by hydrogen-bonding with the main chain NH groups. Three nests were found in the DNA-binding domain (S31G32H33, H68H69L70, and L11G12P13). These nests are in close proximity to each other and two of them are on the interface between the DNA-binding unit and the flap domain. Note that mutation of H69 results in thousand-fold reduction in heterocyst differentiation (14). Two additional nests were found in the flap domain (S193G194T195V196) and the hood domain (E243K244R245). For example, these sites could bind a phosphate group of DNA or a carboxylate of the pentapeptide RGSGR.
The crystal structure of HetR was used to model the RGSGR pentapeptide into the structure. HetR was first analyzed to identify cavities with sufficient accessibility and volume to accommodate a pentapeptide. Comparing the HetR surface to existing protein-peptide binding surfaces in the PDB identified three candidate surfaces (Fig. S5 A–C) (25). The most prominent site involves the cavity formed within the hood structural unit (Fig. S5 A and B, magenta), although binding a peptide to this site would not interfere with DNA binding. The second is found on the interface between the DNA-binding unit and flap domains (Fig. S5 C and D, blue), and the third cuts diagonally across the DNA-binding domain (Fig. S5 E and F, green). Interestingly, the second site includes the nest S193G194T195V196, and modeling suggests that it could be involved in binding the C-terminal carboxylate of the RGSGR pentapeptide. The second and third sites, when occupied by peptide, might interfere with DNA binding. The first and third sites correspond to peptide:HetR stoichiometry of 1∶2, while the second would bind two peptides per HetR dimer.
Summarizing, we have observed two structures of the HetR protein from Fischerella sp.MV11 at high and low ionic strength, respectively. HetR is a member of a highly conserved cyanobacterial protein family that is essential for heterocyst differentiation. The two HetR structures show a protein dimer with considerable local differences in domain orientation and secondary element conformations, suggesting a highly flexible structure. The protein is composed of three segments that constitute well defined structural units: the N-terminal DNA binding and the C-terminal hood units comprised of both chains, with each chain contributing one flap domain. We have confirmed HetR DNA binding and we have shown, by site-directed mutagenesis, that binding involves strictly conserved residues present in the HTH motif. The unique HetR structure reveals an unusual dimer clearly capable of accommodating DNA, other proteins and peptides, and perhaps other molecules. Structural comparison of HetR with other proteins in the PDB reveals that the DNA-binding unit is distantly related to other DNA-binding proteins, such as Fis and several other transcription factors but, as a whole, the HetR structure represents a unique fold.
How does this structure illuminate the role played by HetR in the differentiation of heterocysts? It is known that absence of functional HetR leads to the inability to express many of the genes whose products are required for differentiation and nitrogen fixation. Yet HetR has only a single strong binding site in the Anabaena genome and perhaps several weaker ones. And the structure does not show how it might enhance transcription from that site or nearby ones. Because of the HTH DNA-binding unit dimensions and the DNA recognition site symmetry, HetR specifically recognizes a 17-bp target and could function as a bidirectional activator. However, the DNA symmetry breaks down 10 bp away from the 2-fold axis and the structure shows that HetR can interact with an additional 5–6 bp on each side. As a result, HetR can present to potential partners the same flap domain associated with different DNA sequences, breaking the 2-fold symmetry in the complex. Additional features that may be important are the richness in histidine residues, the presence of the hood unit, and the flexibility of the flaps. We imagine that the latter might accrete RNA polymerase as well as other factors (proteins and RNAs) that direct HetR to additional sites.
Methods
The following methods are described in detail in the SI Text: gene cloning and protein expression, protein purification, DNA-binding assays using gel shifts, determination of the molecular weight of HetR dimers by size exclusion chromatography, crystallization procedures, data collection, structure determination, and modeling the possible binding of the peptide RGSGR to the observed structure of HetR.
Supplementary Material
Acknowledgments.
We thank members of the Structural Biology Center at Argonne National Laboratory for their help in conducting these experiments. This work was supported by National Institutes of Health Grant GM094585 (A.J.), National Science Foundation (NSF) Grant IOS-0919878 (S.M.C.), Federal Ministry of Education and Research Grant 0313921 (W.R.H.) and by the Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357 (A.J.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3QOD and 3QOE).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106840108/-/DCSupplemental.
References
- 1.Meeks JC, Elhai J. Regulation of cellular differentiation in filamentous cyanobacteria in free-living and plant-associated symbiotic growth states. Microbiol Mol Biol R. 2002;66:94–121. doi: 10.1128/MMBR.66.1.94-121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flores E, Herrero A. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat Rev Microbiol. 2010;8:39–50. doi: 10.1038/nrmicro2242. [DOI] [PubMed] [Google Scholar]
- 3.Callahan SM, Buikema WJ. The role of HetN in maintenance of the heterocyst pattern in Anabaena sp. PCC 7120. Mol Microbiol. 2001;40:941–950. doi: 10.1046/j.1365-2958.2001.02437.x. [DOI] [PubMed] [Google Scholar]
- 4.Li B, Huang X, Zhao J. Expression of hetN during heterocyst differentiation and its inhibition of hetR up-regulation in the cyanobacterium Anabaena sp. PCC 7120 FEBS Lett. 2002;517:87–91. doi: 10.1016/s0014-5793(02)02582-6. [DOI] [PubMed] [Google Scholar]
- 5.Yoon HS, Golden JW. Heterocyst pattern formation controlled by a diffusible peptide. Science. 1998;282:935–938. doi: 10.1126/science.282.5390.935. [DOI] [PubMed] [Google Scholar]
- 6.Buikema WJ, Haselkorn R. Characterization of a gene controlling heterocyst differentiation in the cyanobacterium Anabaena 7120. Genes Dev. 1991;5:321–330. doi: 10.1101/gad.5.2.321. [DOI] [PubMed] [Google Scholar]
- 7.Risser DD, Callahan SM. Genetic and cytological evidence that heterocyst patterning is regulated by inhibitor gradients that promote activator decay. Proc Natl Acad Sci USA. 2009;106:19884–19888. doi: 10.1073/pnas.0909152106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang X, Dong Y, Zhao J. HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc Natl Acad Sci USA. 2004;101:4848–4853. doi: 10.1073/pnas.0400429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higa KC, Callahan SM. Ectopic expression of HetP can partially bypass the need for HetR in heterocyst differentiation by Anabaena sp. strain PCC 7120. Mol Microbiol. 2010;77:562–574. doi: 10.1111/j.1365-2958.2010.07257.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhou R, et al. Evidence that HetR protein is an unusual serine-type protease. Proc Natl Acad Sci USA. 1998;95:4959–4963. doi: 10.1073/pnas.95.9.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schiefer W, Schutz K, Hachtel W, Happe T. Molecular cloning and characterization of hetR genes from filamentous cyanobacteria. Biochim Biophys Acta. 2002;1577:139–143. doi: 10.1016/s0167-4781(02)00399-8. [DOI] [PubMed] [Google Scholar]
- 12.Wong FC, Meeks JC. The hetF gene product is essential to heterocyst differentiation and affects HetR function in the cyanobacterium Nostoc punctiforme. J Bacteriol. 2001;183:2654–2661. doi: 10.1128/JB.183.8.2654-2661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finsinger K, et al. Characterization of true-branching cyanobacteria from geothermal sites and hot springs of Costa Rica. Environ Microbiol. 2008;10:460–473. doi: 10.1111/j.1462-2920.2007.01467.x. [DOI] [PubMed] [Google Scholar]
- 14.Risser DD, Callahan SM. Mutagenesis of HetR reveals amino acids necessary for HetR function in the heterocystous cyanobacterium Anabaena sp. strain PCC 7120 33. J Bacteriol. 2007;189:2460–2467. doi: 10.1128/JB.01241-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res 33 (Web Server issue) 2005;33:W89–93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 17.Yuan HS, et al. The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding. Proc Natl Acad Sci USA. 1991;88:9558–9562. doi: 10.1073/pnas.88.21.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kostrewa D, et al. Crystal structure of the factor for inversion stimulation FIS at 2.0 A resolution. J Mol Biol. 1992;226:209–226. doi: 10.1016/0022-2836(92)90134-6. [DOI] [PubMed] [Google Scholar]
- 19.Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat Struct Biol. 2001;8:710–714. doi: 10.1038/90429. [DOI] [PubMed] [Google Scholar]
- 20.Qiu X, Pohl E, Holmes RK, Hol WG. High-resolution structure of the diphtheria toxin repressor complexed with cobalt and manganese reveals an SH3-like third domain and suggests a possible role of phosphate as co-corepressor. Biochemistry. 1996;35:12292–12302. doi: 10.1021/bi960861d. [DOI] [PubMed] [Google Scholar]
- 21.Schultz SC, Shields GC, Steitz TA. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 22.Lawson C. An atomic view of the L-tryptophan binding site of trp repressor. Nat Struct Biol. 1996;3:986–987. doi: 10.1038/nsb1296-986. [DOI] [PubMed] [Google Scholar]
- 23.Rajagopalan R, Callahan SM. Temporal and spatial regulation of the four transcription start sites of hetR from Anabaena sp strain PCC 7120. J Bacteriol. 2010;192:1088–1096. doi: 10.1128/JB.01297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watson JD, Milner-White EJ. A novel main-chain anion-binding site in proteins: the nest. A particular combination of phi, psi values in successive residues gives rise to anion-binding sites that occur commonly and are found often at functionally important regions. J Mol Biol. 2002;315:171–182. doi: 10.1006/jmbi.2001.5227. [DOI] [PubMed] [Google Scholar]
- 25.Binkowski TA, Joachimiak A. Protein functional surfaces: global shape matching and local spatial alignments of ligand binding sites. BMC Struct Biol. 2008;8:45. doi: 10.1186/1472-6807-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLano WL. PyMOL. New York, NY: Schrödinger, LLC; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.