Abstract
IκBα is a crucial regulator of NFκB transcription. NFκB-mediated gene activation is robust because levels of free IκBα are kept extremely low by rapid, ubiquitin-independent degradation of newly synthesized IκBα. IκBα has a weakly folded ankyrin repeat 5–6 (AR5–6) region that is critical in establishing its short intracellular half-life. The AR5–6 region of IκBα folds upon binding to NFκB. The NFκB-bound IκBα has a long half-life and requires ubiquitin-targeted degradation. We present single molecule FRET evidence that the native state of IκBα transiently populates an intrinsically disordered state characterized by a more extended structure and fluctuations on the millisecond time scale. Binding to NFκB or introduction of stabilizing mutations in AR 6 suppressed the fluctuations, whereas higher temperature or small amounts of urea increased them. The results reveal that intrinsically disordered protein regions transition between collapsed and extended conformations under native conditions.
Keywords: intrinsically disordered protein, NFkappaB, transcription factor, protein dynamics
Intrinsically disordered proteins (IDPs) contain unfolded or weakly folded regions that play central roles in signaling, protein function regulation, ligand scavenging, and assembly of supramolecular complexes (1–3). Among IDPs, IκBα stands out as an example of a protein whose functions depend critically on its disordered regions (4). IκBα is the crucial regulator of the NFκB transcription factor, which activates hundreds of genes (5). NFκB gene activation is rapid and robust because the level of free IκBα is kept extremely low by rapid, ubiquitin-independent degradation of any newly synthesized IκBα (6). The disordered ankyrin repeat 5–6 (AR5–6) region in IκBα has been shown to be critical in establishing the short intracellular half-life of free IκBα. Whereas stabilization of the AR5–6 region of IκBα lengthened the intracellular half-life, stabilization outside the AR5–6 region had no effect (7, 8). Like many other IDPs, the disordered AR5–6 region folds on binding to NFκB, and binding stabilizes the disordered region and switches the degradation mechanism to one requiring phosphorylation and ubiquitination (6).
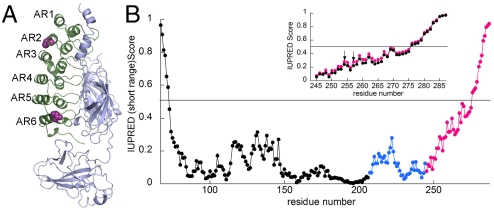
The gene encoding IκBα is highly NFκB-responsive (9–11), resulting in strong postinduction repression of NFκB signaling (12, 13). We recently showed that IκBα dramatically enhances the dissociation of NFκB from transcription sites, and this function, too, depends on the disordered region (14). Because so many critical functions depend on the disordered AR5–6 region of IκBα, structural information on this region is highly sought-after, but so far IκBα has resisted crystallization except in the NFκB-bound state (15, 16)(Fig. 1A).
Fig. 1.
(A) Crystal structure of IκBα (green) bound to NFκB (gray) (PDB ID code 1NFI). Amino acids 70–282 of IκBα are observed to be ordered in this structure (16). After replacing all wild-type cysteines with serines, two cysteines were introduced at positions 128 (AR 2) and 262 (AR 6) (purple spheres) for labeling with Alexa 555 and 647. (B) The IUPRED analysis of IκBα(67–287) is plotted for ARs 1–4 (black), AR5 (blue), and AR6 (magenta). The inset shows a comparison of the IUPRED results for the wild-type sequence of IκBα (magenta) vs. the Y254L/T257A sequence (black) expanded to show only AR6. The arrows indicate the Y254L and T257A mutation sites.
IUPRED analysis (17, 18) of the AR domain (ARD) of IκBα predicts a monotonic increase in intrinsic disorder through the AR5–6 region to the end of the protein (Fig. 1B). Other predictors including FoldIndex (19), PONDR (20), and Disopred2 (21) gave similar results. Experimentally, all the amides in the AR5–6 region rapidly exchange in free IκBα (22, 23) and nearly all of the cross peaks for these residues are broadened and missing in the NMR spectra, indicating that free IκBα is molten globular or weakly folded. On the other hand, the NFκB-bound IκBα could be fully characterized (24). In addition, the AR5–6 region is not part of the cooperatively folding ARD (25). A “prefolded” mutant IκBα, in which two AR 6 residues (mutations Y254L and T257A) are returned to the consensus sequence for stable ARs, has a more folded AR 6 than wild-type IκBα, as judged by H/D exchange and NMR (7). The IUPRED score for this mutant shows slightly less disorder throughout the AR5–6 region (Fig. 1B, Inset). This mutant has a longer half-life in cells (7) and is much less efficient at accelerating dissociation of NFκB off the DNA (14).
Despite the many biophysical approaches used to characterize IκBα, previous strategies have failed to provide structural or dynamic information about the intrinsically disordered AR5–6 region. Single molecule fluorescence resonance energy transfer (smFRET) has been a powerful technique for studying the expanded structures of IDPs, including α-synuclein (26, 27) and p53 (28). We present smFRET results showing that like these other IDPs, IκBα can adopt an expanded structure. In addition, by following the molecules over longer periods of time, we are able to observe that IκBα interconverts between a more compact, folded conformation and an expanded IDP structure in real time.
Results
To investigate the structure and dynamics of AR5–6 by smFRET, we introduced cysteine residues into a cysteine-free construct of IκBα in place of residues E128 (AR 2) and S262 (AR 6), which are away from the NFκB binding face (Fig. 1A). The resulting E128C/S262C IκBα (hereafter referred to as IκBα) retained NFκB binding ability, as demonstrated by SPR (Fig. S1). IκBα was labeled with a mixture of Alexa Fluor 555 and Alexa Fluor 647 fluorophores and examined by ensemble FRET. Bulk FRET measurements failed to reveal a significant difference between the free IκBα and NFκB-bound IκBα, suggesting a minimal structural rearrangement of AR5–6 from the weakly folded free IκBα to the compact NFκB-bound protein. Given the flexibility observed previously in AR5–6 for free IκBα (23, 24), we suspected that the ensemble experiment only reflected a major well-folded protein population. Thus, we used smFRET to look for possible rare dynamic states of IκBα.
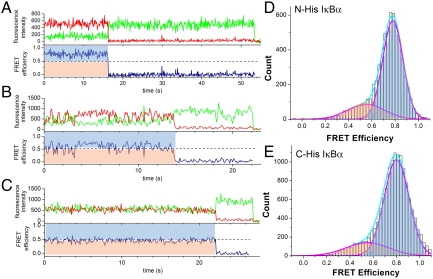
In order to study the individual molecular motions of IκBα for significant periods of time, we used a prism-based total internal reflection fluorescence (TIRF) microscope (29) to observe surface-immobilized IκBα. We introduced a His6 tag at either the N- (before amino acid 67) or C-terminal (after amino acid 287) extensions beyond the IκBα ARD, and the protein was bound to the microscope slide by way of a biotinylated anti-His5 antibody that was attached to neutravidin bound to the sparsely biotinylated PEG surface of the microscope slide (Fig. 2). In this way, we measured the FRET of hundreds of single molecules with 100 ms time resolution for up to 70 s. smFRET traces were collected and only those that showed a single photobleaching event for both fluorophores were used. Traces from at least 200 such bona fide single molecules were analyzed for each experimental condition. The majority of the smFRET traces showed stable, high FRET (Fig. 3A), but occasionally fluctuating (Fig. 3B) or mid-FRET (Fig. 3C) traces were observed. To analyze the aggregate smFRET signal and to compare the different experimental conditions, we prepared histograms by binning the FRET signal by 100 ms. Using this approach, the N-His and the C-His immobilized IκBα were seen to give identical results; the majority of the signal was high-FRET (E = 0.76 - 0.8) but a minor broad mid-FRET signal was also observed (E = ∼ 0.5) (Fig. 3 D and E). Control experiments omitting anti-His5 antibody showed that His6-IκBα immobilization was through the His6 tag (less than 1% of molecules were observed to bind nonspecifically). Because both the N-His and C-His IκBα provided the same results, the rest of the experiments were performed with N-His IκBα in order to avoid crowding effects at the C terminus during NFκB binding.
Fig. 2.
Schematic diagram of the TIRF setup used for the smFRET measurements (only one of two possible ways the protein could be labeled with the Alexa dyes is depicted).
Fig. 3.
(A) Sample trace of an IκBα molecule showing stable high FRET. Traces similar to this one represented approximately 80% of the molecules at room temperature. (B) Sample trace of an IκBα molecule showing fluctuating FRET. Traces similar to this one represented 14–18% of the molecules at room temperature. (C) Sample trace of an IκBα molecule showing stable mid FRET. Traces similar to this one represented 2.5–7% of the molecules at room temperature. In A, B, and C, a dashed line at E = 0.5 marks the boundary for high vs low FRET and shaded blue (> 0.5 FRET efficiencies) and peach (< 0.5 FRET efficiencies) areas in the FRET efficiency traces correspond respectively to the blue and peach areas in the histograms shown in D and E. (D) smFRET histogram of N-terminally His6-tagged IκBα in the absence of NFκB at 21 °C. The histogram revealed a major high-FRET curve centered at 0.76 and a broad shoulder centered at 0.49. (E) smFRET histogram of C-terminally His6-tagged IκBα in the absence of NFκB at 21 °C. All histograms were fitted with up to two Gaussian peaks.
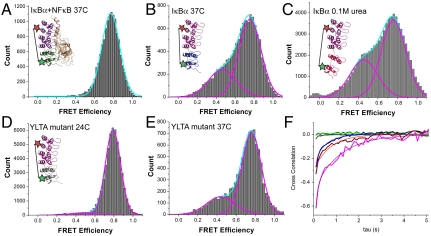
The stable high-FRET signal observed in most of the traces (Fig. 3A) was consistent with the distance between the Alexa 555 and 647 expected for the fully structured ARD observed in the crystal structure (Fig. 1A) (15, 16). To confirm the assignment of the high-FRET signal to the folded IκBα ARD, we also analyzed NFκB-bound IκBα. At both 21 °C (Fig. S2) and 37 °C (Fig. 4A), traces from the NFκB-bound IκBα nearly all resembled the trace shown in Fig. 3A, and the resulting histogram showed only the high-FRET (E = 0.76 - 0.8) population (Fig. 4A).
Fig. 4.
(A) smFRET histogram of the IκBα–NFκB complex at 37 °C demonstrating that binding NFκB to IκBα completely suppressed the low- to mid-FRET populations observed for free IκBα, even at the higher temperature. The inset illustrates the IκBα–NFκB complex (ARs 5–6 in green) and the placement of the fluorophores, which are represented by stars (only one of two possible ways the protein could be labeled with two different dyes is depicted). (B) In the absence of NFκB, the IκBα fluctuations, and hence the low-mid-FRET efficiencies, increased at 37 °C. The inset depicts an extended IκBα in the absence of NFκB at 37 °C, with ARs 5–6 in blue. (C) smFRET histogram of free IκBα in 0.1 M urea shows a marked increase in the mid-FRET population. The inset represents a schematic of an extended IκBα in the presence of 0.1 M urea, with ARs 5–6 in red. (D) smFRET histogram of N-terminally His-tagged YLTA mutant IκBα at 24 °C. The lower-FRET population is greatly suppressed compared to the wild-type, despite the higher temperature. The inset illustrates the stabilized YLTA mutant with ARs 5–6 in gray. (E) At 37 °C, the YLTA construct begins to show a lower-FRET population. (F) Cross-correlation plot averaged over the traces in each condition quantifies the degree of fluctuation. Smooth curves represent single-exponential fitting. The green and gray curves correspond to NFκB-bound and YLTA mutant IκBα; the blue curve corresponds to native IκBα at 37 °C; and the red and magenta curves correspond to IκBα in 0.1 and 0.5 M urea, respectively. The insets in A–D depict the molecules in the same color scheme.
About 20% of the smFRET traces for free IκBα showed anticorrelated fluctuations of donor and acceptor signals (Fig. 3B) or stable mid-FRET (Fig. 3C) consistent with the IκBα ARD occasionally visiting extended conformations. The fluctuation characteristics, however, appeared heterogeneous between molecules, as some were rapid, others slow, some fluctuated to low FRET, and others only to mid FRET, indicating that the molecules were in many different states, all of which were more expanded than the high-FRET, folded state (Fig. S3).
To understand the structural origin of the observed fluctuating FRET signal, we probed the response of IκBα upon thermal or denaturant challenge. At 37 °C, a substantial increase in the number of fluctuating molecules was observed (Table 1). The resulting histogram also showed an increase in the broad, mid-FRET signal, but the mean FRET values did not change significantly (Fig. 4B). We previously showed that when the helical circular dichroism signal is monitored during urea titration, the intrinsically disordered AR5–6 region follows a noncooperative transition, whereas ARs 1–4 cooperatively unfold above 3 M urea (25). These previous results suggested that we could perturb the AR5–6 region with very low urea concentrations without unfolding the AR1–4 region. Indeed, even urea concentrations of 0.1 M markedly increased the number of fluctuating molecules (Table 1) and the amount of lower FRET signal observed (Fig. 4C).
Table 1.
Number of molecules with stable or fluctuating smFRET
| Classification of smFRET traces | |||||
| Sample | Molecule Count and Percentage | ||||
| Protein | T(°C) | n | Stable High | Stable Mid | Fluctuating |
| N-His IκBα | 21 | 207 | 163 (79%) | 15 (7%) | 29 (14%) |
| C-His IκBα | 21 | 202 | 157 (78%) | 5 (2%) | 40 (20%) |
| N-His IκBα | 24 | 213 | 122 (57%) | 34 (16%) | 57 (27%) |
| N-His IκBα | 37 | 226 | 113 (50%) | 36 (16%) | 77 (34%) |
| N-His IκBα + NFκB | 37 | 207 | 176 (85%) | 19 (9%) | 12 (6%) |
| YLTA IκBα | 24 | 215 | 186 (86%) | 12 (6%) | 17 (8%) |
| N-His IκBα + 0.1 M urea | 24 | 221 | 104 (47%) | 26 (12%) | 91 (41%) |
| N-His IκBα + 0.5 M urea | 24 | 200 | 98 (49%) | 25 (12%) | 77 (39%) |
| Number of transitioning molecules | ||||||
| Protein | T(°C) | n | Stable High | Stable Mid | Fluctuating | Transitioning* |
| N-His IκBα | 21 | 210 | 183 (87%) | 5 (2%) | 18 (9%) | 4 (2%) |
| N-His IκBα | 37 | 201 | 109 (54%) | 21 (10%) | 60 (30%) | 11 (6%) |
*Molecules were classified as transitioning if they spent more than 5 s in one state and then transitioned to another state in which they also spent more than 5 s.
We previously showed that introduction of two mutations (Y254L/T257A) in AR6 that restored these residues to the consensus residues for stable ARDs resulted in a more folded AR6 (7). Very few fluctuating molecules were observed for this YLTA mutant IκBα and overall, the mid-FRET population was barely observed at 24 °C (Fig. 4D and Table 1). At 37 °C, the YLTA mutant showed an increase of mid-FRET population and in the number of fluctuating molecules approaching the number seen for the wild-type protein at 24 °C (Fig. 4E).
In order to quantify the amount and time scale of fluctuations, we calculated the cross-correlation of donor and acceptor intensity time traces averaged over 200 molecules each (Fig. 4F). The amplitude of the cross-correlation increased with urea and high temperatures, consistent with the population analysis presented in Table 1. Incubation of wild type and the YLTA mutant IκBα in 0.1 M and 0.5 M urea markedly increased the low-FRET population with a concomitant increase in the number of fluctuating molecules in a urea-dependent manner (Fig. 4E and Table 1 and Fig. S4). This analysis revealed that the number of molecules accessing the fluctuating state increases with urea and higher temperatures. The time scale of fluctuations, calculated as the average time obtained from the exponential fits of the cross-correlation curves ranged from 0.1 to 2 s. This broad distribution of the time scale of the fluctuations is consistent with the heterogeneous time scales observed in the smFRET traces (Fig. S3). The heterogeneity of the fluctuations was similar whether the molecules were tethered via the N terminus (Fig. S3 A–E) or the C terminus (Fig. S3 F–J).
Examination of all traces revealed that most molecules were either in the high-FRET state or in the mid-FRET/fluctuating state. Molecules were only occasionally observed to transition from the stable high-FRET state to the fluctuating state or vice versa. Traces showing such transitions resembled those shown in Fig. 5 A and B. Out of 201 molecules of free IκBα at 37 °C, 54% had stable, high-FRET; 40% were either fluctuating or in a stable mid-FRET state; and 6% (11 molecules) were observed to transition between a fluctuating or mid-FRET state to a stable high-FRET state (as assessed by average E ∼ 0.75 for more than 5 sec) (Table 1). In a sense, then, the native state of IκBα is a very slowly converting “two-state” system that transitions between a high-FRET state, that most likely resembles the folded state seen in the crystal structure of the NFκB-IκBα complex, and an intrinsically disordered state that comprises a number of different conformations that interconvert at various rates (Fig. 6). The equilibrium between these two states can be altered by temperature, denaturant, and stabilizing mutations.
Fig. 5.
(A) Representative smFRET trace of a molecule that switched from fluctuating to a stable high-FRET state during data collection. (B) Representative smFRET trace of a molecule that switched from mid-FRET to a stable high-FRET state during data collection. A dashed line marks the boundary between high- and low-FRET regions of each trace.
Fig. 6.
Schematic free energy profile showing the native state and the broad, heterogeneous intrinsically disordered state. Higher temperatures or addition of small amounts of urea increase the population of the intrinsically disordered state. Below the graph are schematic drawings of possible structures in each state. The colors for ARs 5–6 follow the same scheme as Fig. 4F (magenta: IκBα in 0.5 M urea; blue: IκBα at 37 °C; gray: YLTA mutant at 24 °C; green: IκBα-NFκB complex at 37 °C).
Discussion
Based on the predicted and experimental evidence of disorder in the C-terminal region of IκBα, we naively expected to observe an increase in the bulk FRET signal of AR2–AR6 labeled IκBα upon NFκB binding, but the bulk FRET showed a very small difference between the bound and free proteins. Despite the fact that the AR5–6 region has no protected amides and NMR results are consistent with a molten globular structure, stable high FRET consistent with the distance expected for the completely folded state seen in the NFκB-bound structure was observed for most of the unbound IκBα molecules. Although a small peak of mid-FRET signal was observed, calculation of the average FRET efficiency across the whole histogram gave a value of 0.73 for the free IκBα and 0.76 for the NFκB-bound IκBα. These very similar averages help explain why we did not observe a large difference in the bulk FRET. The observation that the majority of the molecules were actually folded even in the free state was a surprise. Intrinsic disorder is expected to be manifested in an expanded, random coil structure (1), but the fluctuating traces reveal that individual IκBα molecules visit the expanded structure only occasionally. One explanation for this behavior is that the disordered region of IκBα is adjacent to a folded region. Such smaller intrinsically disordered regions adjacent to folded domains are actually predicted to be more common than completely disordered proteins (30), but no other studies have examined the behavior of such adjacent disordered regions.
TIRF microscopy on immobilized molecules has, in most cases, been superseded by confocal approaches mainly out of concern regarding artifacts arising from immobilization (31, 32). Nevertheless, we chose the TIRF approach in order to observe single molecules for long periods of time, which is not possible using the confocal approach. Our immobilization approach used neutravidin binding to a sparsely biotinylated PEGylated microscope slide. A biotinylated anti-His5 antibody was then bound to the neutravidin and the His6-tagged IκBα was finally bound to the antibody. Over 50% of the molecules tethered in this way showed single bleaching events for both fluorophores, and we could be confident we were observing single molecules. In addition, the fact that we observed similar smFRET signals over hundreds of molecules when the IκBα was tethered at either the N or C terminus strongly argues that this tethering approach is free from artifacts.
By observing the IκBα molecules over many seconds, we observed that most had stable high-FRET consistent with the expected folded structure observed in the crystal structure of the NFκB-bound form (15, 16), but 20% appeared to be more disordered. Although many factors need to be considered in computing exact distances from smFRET efficiencies, the average FRET efficiency of 0.76 for the high-FRET molecules matches well with the expected distance between the fluorophores based on the structure of IκBα (40.5 Å) (15, 16). In the disordered molecules, the FRET efficiency fluctuated down to values of 0.2 and, although widely distributed, the average FRET efficiency of the disordered state was 0.5, suggesting a distance between the fluorophores of approximately 51 Å. This distance is consistent with one or more of the ARs adopting a more extended conformation. MD simulations are in progress, which should provide a clearer understanding of the partially folded structures that correspond to the extended conformations.
The most remarkable observation of the disordered state was the instability of the FRET signal. Although a few molecules appeared to have stable mid FRET, the majority showed FRET signals that were anticorrelated and fluctuated between high and low FRET on time scales ranging from the limit of our measurements (100 ms) to seconds (some example traces are shown in Fig. S3). Importantly, the observed heterogeneity of the fluctuations was similar for the IκBα molecules that were tethered by way of an N-terminal His6 tag (Fig. S3 A–E) or a C-terminal His6 tag (Fig. S3 F–J), strongly suggesting that the fluctuations are not due to proximity to the surface. The stable mid-FRET molecules may correspond to a longer-lived partially unfolded state. It is also possible that the mid-FRET state that appears not to be fluctuating may merely have been fluctuating too fast to be detected by our method. The broad range of different fluctuating behaviors observed in the smFRET traces made it impossible to assign defined structural states. Instead, we simply assign all of the lower FRET molecules to a broad ensemble of intrinsically disordered states that appear to undergo slow fluctuations between collapsed and extended conformations (Fig. 6).
Binding of NFκB markedly decreased the number of fluctuating molecules, consistent with previous studies showing that the disordered AR5–6 region folds on binding (23). Introduction of just two mutations (Y254L/T257A) that stabilize AR6 also markedly decreased the number of fluctuating molecules, consistent with the previous observation that amide exchange in the disordered AR5–6 region of this mutant is reduced indicative of a more folded structure (7). The fact that minor perturbations in temperature and/or introduction of a few mutations dramatically altered the fluctuating behavior provides strong evidence that the fluctuations are a property of the intrinsic disorder in native IκBα and not an artifact of the experimental design. The observation that the YLTA mutant doesn’t fluctuate as much as wild type correlates with the dramatically reduced ability of the YLTA mutant to accelerate dissociation of NFκB from the DNA (14) and suggests that the enhancement of NFκB dissociation may be functionally linked to the fluctuations of the AR5–6 region. The fluctuations of the intrinsically disordered state of IκBα probably enhance the ability of IκBα to get in between the DNA and NFκB during thermal fluctuations, resulting in accelerated dissociation. The YLTA mutant is also degraded more slowly in vivo, suggesting that the fluctuations may also be functionally linked to the ubiquitin-independent degradation of free IκBα (7).
Our results reveal previously unreported behaviors of intrinsically disordered protein segments: (i) The AR5–6 region of IκBα is predicted to be just on the edge of being intrinsically disordered and the smFRET measurement shows that a majority of the molecules have stable high FRET, consistent with a folded structure (Fig. 6). The intrinsically disordered state appears to only occasionally be visited and it appears to be a broad manifold of disordered, expanded, interconverting structures. The ability of the weakly folded AR5–6 region of IκBα to expand and contract may be a feature of its topology. AR domains rely solely on local interactions for fold stabilization, and as we have demonstrated, a few mutations can dramatically alter the stability of a local region. Force-induced unfolding experiments have demonstrated that well-folded ARDs behave like nanosprings, generating a force upon refolding (33). The behavior of IκBα shows what happens when the local interactions are not strong enough to achieve a stable fold and the spring-like topology fluctuates in the native state in the absence of force. (ii) The intrinsically disordered state often undergoes large magnitude, low frequency fluctuations for which the time scale varies widely but can often be very slow, on the order of hundreds of milliseconds. Others have measured smFRET of intrinsically disordered proteins, but the experimental setup did not allow observation for the long times required to observe the slow events we report here (26, 27). It will be interesting to see whether proteins of other topologies that contain intrinsically disordered regions undergo such local unfolding events and on what time scales.
Materials and Methods
IκBα Isolation and Labeling for FRET.
All the IκBα wild-type cysteines were replaced with serines by site-directed mutagenesis. Then, positions 128 (AR 2) and 262 (AR 6) were mutated to cysteines. Expression of IκBα followed the described procedure (22) except for protein induction using 0.2 mM IPTG at 18 °C. IκBα isolation used standard nickel affinity purification protocols with Ni-NTA resin (Bio-Rad). IκBα was purified further by size exclusion chromatography using labeling buffer (25 mM Tris, pH 7.2; 150 mM NaCl; 1 mM EDTA; 10 mM TCEP) and concentrated to 150 μM before adding an equimolar mixture of the maleimide-conjugated Alexa 555/647 pair (Invitrogen) in a 10-fold excess (1.5 mM for each fluorophore). The reaction mixture was protected from light and incubated at room temperature for 1 h, followed by 4 °C overnight. The labeled protein was isolated by gel filtration in a Sephadex G-25 resin (Sigma) using isolation buffer (25 mM Tris, pH 7.2; 150 mM NaCl; 1 mM EDTA; 25 mM CHAPS; 25 mM tryptophan; 10% DMSO) to reduce hydrophobic binding of the fluorophores to IκBα. A final size exclusion step into IκBα buffer (25 mM Tris, pH 7.2; 150 mM NaCl; 1 mM EDTA; 1 mM DTT) removed traces of free fluorophores and possible protein aggregates. Labeling efficiencies were determined by measuring the absorbance of the sample at 280, 555, and 650 nm, and using Beer’s Law to independently determine the concentration of IκBα (ϵ = 12,950 cm-1 M-1), Alexa Fluor 555 (ϵ = 150,000 cm-1 M-1), and Alexa Fluor 647 (ϵ = 239,000 cm-1 M-1) in the sample (1 cm light path). The IκBα concentration was corrected in Beer’s equation by subtracting the absorbance of both Alexa Fluor 555 (8% of the absorbance at 555 nm) and Alexa Fluor 647 (3% of the absorbance at 650 nm) from the IκBα absorbance at 280 nm. Overall labeling efficiencies ranged from 30 to 60%, as calculated from the equation:
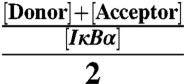 |
smFRET Measurements and Analysis.
We used total internal reflection fluorescence microscopy for imaging as described previously (34). A clean quartz surface was coated with polyethylene glycol (PEG) and biotinylated PEG (100∶1). The His6-tagged IκBα was immobilized via an anti-His5-tag antibody (Qiagen, product number 34440) attached through neutravidins to the PEG surface. This immobilization strategy has been successfully used for other proteins, preserving their functions (35, 36). The imaging solution contained 1 mg/mL glucose oxidase, 0.04 mg/mL catalase, 0.8% dextrose, and saturated trolox (approximately 3 mM), in addition to the IκBα buffer. From the fluorescence intensity of each molecule measured by an EMCCD camera, we calculated FRET efficiency defined with leakage correction as EFRET = (IA-0.08 ∗ ID)/(ID + IA) where ID and IA are the detected emission intensities of donor and acceptor, respectively. We selected molecules exhibiting single photobleaching steps of both dyes. In the FRET histograms, signal acquired for 100 ms represents one count.
Supplementary Material
ACKNOWLEDGMENTS
E.A.K. and J.A.L. acknowledge support from National Institutes of Health (NIH) Grant P01-GM071862. H.K., K.S.L., and T.H. were supported by US National Science Foundation Physics Frontier Center Grant 0822613 and by NIH Grant R21 RR025341. T.H. is an employee of Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102226108/-/DCSupplemental.
References
- 1.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turoverov KK, Kuznetsova IM, Uversky VN. The protein kingdom extended: Ordered and intrinsically disordered proteins, their folding, supramolecular complex formation, and aggregation. Prog Biophys Mol Biol. 2010;102:73–84. doi: 10.1016/j.pbiomolbio.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 4.Ferreiro DU, Komives EA. Molecular mechanisms of system control of NF-kappaB signaling by IkappaBalpha. Biochemistry. 2010;49:1560–1567. doi: 10.1021/bi901948j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pahl HL. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 6.O’Dea EL, et al. A homeostatic model of IkappaB metabolism to control constitutive NF-kappaB activity. Mol Syst Biol. 2007;3:111. doi: 10.1038/msb4100148. (1–7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Truhlar SM, Mathes E, Cervantes CF, Ghosh G, Komives EA. Pre-folding IkappaBalpha alters control of NF-kappaB signaling. J Mol Biol. 2008;380:67–82. doi: 10.1016/j.jmb.2008.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathes E, O’Dea EL, Hoffmann A, Ghosh G. NF-kappaB dictates the degradation pathway of IkappaBalpha. EMBO J. 2008;27:1357–1367. doi: 10.1038/emboj.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown K, Park S, Kanno T, Franzoso G, Siebenlist U. Mutual regulation of the transcriptional activator NF-kappa B and its inhibitor, I kappa B-alpha. Proc Natl Acad Sci USA. 1993;90:2532–2536. doi: 10.1073/pnas.90.6.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott ML, Fujita T, Liou HC, Nolan GP, Baltimore D. The p65 subunit of NF-kappa B regulates I kappa B by two distinct mechanisms. Genes Dev. 1993;7:1266–1276. doi: 10.1101/gad.7.7a.1266. [DOI] [PubMed] [Google Scholar]
- 11.Sun SC, Ganchi PA, Ballard DW, Greene WC. NF-kappa B controls expression of inhibitor I kappa B alpha: Evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 12.Arenzana-Seisdedos F, et al. Nuclear localization of IkBa promotes active transport of NF-kB from the nucleus to the cytoplasm. J Cell Sci. 1997;110:369–378. doi: 10.1242/jcs.110.3.369. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: Temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 14.Bergqvist S, et al. Kinetic enhancement of NF-kappaB•DNA dissociation by IkappaBalpha. Proc Natl Acad Sci USA. 2009;106:19328–19333. doi: 10.1073/pnas.0908797106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huxford T, Huang DB, Malek S, Ghosh G. The crystal structure of the IkappaBalpha/NF-kappaB complex reveals mechanisms of NF-kappaB inactivation. Cell. 1998;95:759–770. doi: 10.1016/s0092-8674(00)81699-2. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs MD, Harrison SC. Structure of an IkappaBalpha/NF-kappaB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 17.Dosztányi Z, Csizmók V, Tompa P, I S. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 18.Dosztányi Z, Csizmók V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 19.Prilusky J, et al. FoldIndex: A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 20.Obradovic Z, Peng K, Vucetic S, Radivojac P, Dunker AK. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins. 2005;61:176–182. doi: 10.1002/prot.20735. [DOI] [PubMed] [Google Scholar]
- 21.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Croy CH, Bergqvist S, Huxford T, Ghosh G, Komives EA. Biophysical characterization of the free IkappaBalpha ankyrin repeat domain in solution. Protein Sci. 2004;13:1767–1777. doi: 10.1110/ps.04731004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Truhlar SM, Torpey JW, Komives EA. Regions of IkappaBalpha that are critical for its inhibition of NF-kappaB. DNA interaction fold upon binding to NF-kappaB. Proc Natl Acad Sci USA. 2006;103:18951–18956. doi: 10.1073/pnas.0605794103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sue SC, Cervantes C, Komives EA, Dyson HJ. Transfer of flexibility between ankyrin repeats in IkappaB* upon formation of the NF-kappaB complex. J Mol Biol. 2008;380:917–931. doi: 10.1016/j.jmb.2008.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferreiro DU, et al. Stabilizing IkappaBalpha by “consensus” design. J Mol Biol. 2007;365:1201–1216. doi: 10.1016/j.jmb.2006.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferreon AC, Gambin Y, Lemke EA, Deniz AA. Interplay of alpha-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc Natl Acad Sci USA. 2009;106:5645–5650. doi: 10.1073/pnas.0809232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trexler AJ, Rhoades E. Single molecule characterization of α-synuclein in aggregation-prone states. Biophys J. 2010;99:3048–3055. doi: 10.1016/j.bpj.2010.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang F, et al. Multiple conformations of full-length p53 detected with single-molecule fluorescence resonance energy transfer. Proc Natl Acad Sci USA. 2009;106:20758–20763. doi: 10.1073/pnas.0909644106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Gall T, Romero PR, Cortese MS, Uversky VN, Dunker AK. Intrinsic disorder in the Protein Data Bank. J Biomol Struct Dyn. 2007;24:325–342. doi: 10.1080/07391102.2007.10507123. [DOI] [PubMed] [Google Scholar]
- 31.Deniz AA, et al. Ratiometric single-molecule studies of freely diffusing biomolecules. Annu Rev Phys Chem. 2001;52:233–253. doi: 10.1146/annurev.physchem.52.1.233. [DOI] [PubMed] [Google Scholar]
- 32.Friedel M, Baumketner A, Shea JE. Effects of surface tethering on protein folding mechanisms. Proc Natl Acad Sci USA. 2006;103:8396–8401. doi: 10.1073/pnas.0601210103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee G, et al. Nanospring behaviour of ankyrin repeats. Nature. 2006;440:246–249. doi: 10.1038/nature04437. [DOI] [PubMed] [Google Scholar]
- 34.Ha T. Single Molecule Fluorescence Resonance Transfer. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 35.Perkins TT, Dalal RV, Mitsis PG, Block SM. Sequence-dependent pausing of single lambda exonuclease molecules. Science. 2003;301:1914–1918. doi: 10.1126/science.1088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park J, et al. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.