Abstract
Prokaryotic clustered regularly interspaced short palindromic repeat (CRISPR)/Cas (CRISPR-associated sequences) systems provide adaptive immunity against viruses when a spacer sequence of small CRISPR RNA (crRNA) matches a protospacer sequence in the viral genome. Viruses that escape CRISPR/Cas resistance carry point mutations in protospacers, though not all protospacer mutations lead to escape. Here, we show that in the case of Escherichia coli subtype CRISPR/Cas system, the requirements for crRNA matching are strict only for a seven-nucleotide seed region of a protospacer immediately following the essential protospacer-adjacent motif. Mutations in the seed region abolish CRISPR/Cas mediated immunity by reducing the binding affinity of the crRNA-guided Cascade complex to protospacer DNA. We propose that the crRNA seed sequence plays a role in the initial scanning of invader DNA for a match, before base pairing of the full-length spacer occurs, which may enhance the protospacer locating efficiency of the E. coli Cascade complex. In agreement with this proposal, single or multiple mutations within the protospacer but outside the seed region do not lead to escape. The relaxed specificity of the CRISPR/Cas system limits escape possibilities and allows a single crRNA to effectively target numerous related viruses.
Keywords: bacteriophage, RNA interference, small RNA
CRISPR (clustered regularly interspaced short palindromic repeats) cassettes are present in virtually every archaeon and in approximately 40% of bacteria (1–3). A CRISPR cassette consists of almost identical direct repeats that are regularly interspersed with spacers (4). In any given cassette, the length of spacers is similar, whereas their sequences vary. CRISPR cassettes are often flanked by a diverse set of CRISPR-associated (cas) genes (2, 5, 6).
CRISPR/Cas (CRISPR-associated sequences) functions as an adaptive immunity system by excluding viruses and other mobile genetic elements that contain sequences matching CRISPR cassette spacers (6–9). Bacterial and archaeal CRISPR/Cas systems generally target DNA (10–13), although one archaeal system has been demonstrated in vitro to interfere at the level of RNA (14). Transcription of a CRISPR cassette, followed by processing with the help of dedicated endoribonucleases, creates small CRISPR RNAs (crRNAs) that guide the Cas machinery to the target, eventually resulting in target cleavage (11, 15–20).
Although a match between a single CRISPR spacer and a foreign DNA sequence called the protospacer can provide immunity to the entry of that DNA into the host, it is not sufficient. Mutations in the conserved protospacer-adjacent motif (PAM, ref. 21) abolish CRISPR-mediated immunity even in the presence of a perfect spacer-protospacer match. Likewise, some point mutations in protospacer that introduce single mismatches with the spacer abolish CRISPR/Cas function even when the PAM is intact (22). Thus, a PAM and a match between a spacer and protospacer are both required for CRISPR/Cas function. Recently, however, instances of point mismatches between a CRISPR spacer and a plasmid protospacer being insufficient for prevention of CRISPR/Cas mediated plasmid exclusion were reported (12, 13). Here, we show that only protospacer positions proximal to the PAM need to perfectly match the CRISPR spacer sequence in Escherichia coli. In contrast, multiple mismatches are tolerated at PAM-distal protospacer positions without affecting the protective function of CRISPR/Cas. The apparent requirement for nucleation of spacer-protospacer recognition at the PAM side revealed by our work is reminiscent of RNAi seed sequences (23, 24) and may point to a fundamental mechanistic similarity of target recognition by RNAi and CRISPR/Cas systems.
Results
Targeting Bacteriophage M13 with Engineered CRISPR Spacer.
Previously, Brouns et al. (17) showed that a plasmid containing an engineered CRISPR cassette carrying at least one spacer matching phage λ DNA rendered the E. coli host resistant to the phage. Here, we engineered a derivative CRISPR cassette plasmid that contains a spacer, which we call g8, targeting the transcribed strand of the double-stranded replicative form of the phage M13 genome but not the DNA strand that is packaged in the M13 virion (see Materials and Methods, Fig. 1A). The spacer was chosen based on the following considerations: (i) the corresponding protospacer in the M13 genome contained an upstream ATG sequence motif, matching in sequence and position the putative E. coli PAM consensus sequence AWG (21), and (ii) the region of the g8 protein encoded by the protospacer is a nonstructured loop connecting two α-helices and was expected to tolerate multiple amino-acid substitutions without affecting the protein function.
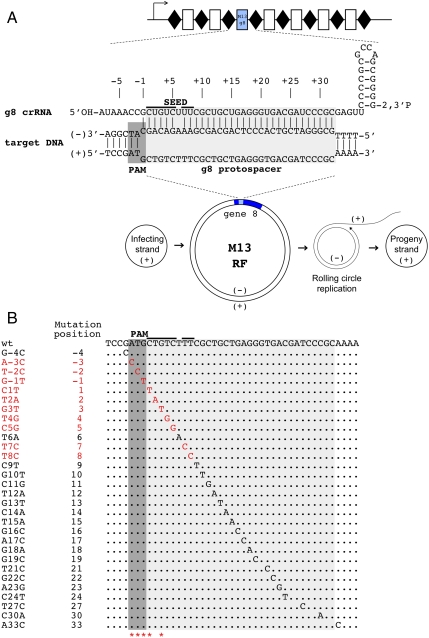
Fig. 1.
CRISPR/Cas mediated restriction of bacteriophage M13 requires intact PAM and PAM-proximal part of the protospacer. (A) An engineered plasmid-borne CRISPR cassette carrying a g8 spacer is schematically shown (Top). Rectangles indicate spacers, rhombi repeats. An arrow indicates the direction of transcription. Bottom) The life cycle of the M13 phage is schematically shown. Phage DNA enters the cell as a circular single-stranded DNA (“infecting” or “(+)” strand DNA). This strand is used as a template to create double-stranded “RF” of the phage genome (shown with gene 8, containing the g8 protospacer, highlighted with blue color). From the RF, the rolling circle replication of the phage genome is initiated, ultimately generating progeny (+) genome strands that are packaged in virions, The g8 protospacer and adjacent M13 sequence and crRNA containing the g8 spacer sequence are expanded above the RF intermediate. The structure shown (Top) forms when crRNA-guided Cascade complex recognizes double-stranded DNA containing the protospacer. (B) The sequence of the g8 protospacer, PAM, and the seed region is shown (Top). Below, point mutations selected as spontaneous escape phages on lawns of cells expressing g8 crRNA and also engineered by site-specific mutagenesis are shown. Mutations indicated with red color, led to escape, whereas mutations shown in black were restricted by CRISPR/Cas (the phage mutants had nonescape phenotype). Asterisks indicate positions of spontaneous escape mutations (see also Figs. S2A and S4). Additional point mutations obtained can be found in Fig. S2B.
Cells containing the engineered M13 targeting CRISPR plasmid, as well as cells carrying a control plasmid in which spacers were inserted that did not target M13 (Fig. S1 and ref. 17), were tested as hosts for M13 phage in a plaque assay (see Materials and Methods). The cells also contained plasmids expressing cas genes (17) and an F-factor encoding pili that are required for M13 adsorption. The efficiency of plaquing (e.o.p.) of wild-type M13 on lawns of cells carrying the g8 plasmid was approximately 105-fold lower than that of control cells lacking the g8 spacer (e.o.p. = 1). Considering the design of our experiment, we conclude that g8 crRNA can efficiently interfere with M13 infection by targeting the (-) strand of M13 DNA. This strand is present only in the dsDNA RF form of the viral genome and in rolling circle replication intermediates of the viral DNA (Fig. 1A).
Selection of Natural M13 Mutants That Escape CRISPR/Cas Mediated Interference.
Despite efficient protection from M13 infection afforded by the g8 CRISPR plasmid, phage plaques were still observed when a sufficient number of phage particles (106 pfu or more) were used in the infection experiment. These “escape” phages appeared to be modified in some way because they had an e.o.p. of 1 on both control and g8 CRISPR plasmid-containing cells. The g8-targeted protospacers of 50 such independent escape plaques, each displaying an e.o.p. of 1 on g8 CRISPR plasmid-containing cells, were sequenced. For clarity, we use the following numbering scheme for protospacer positions: The position immediately downstream of PAM is called 1, with subsequent positions being 2, 3, etc., up to position 32, which is the last position of the protospacer; the PAM positions are referred to as -1, -2, and -3, with -1 the closest to the protospacer. All escape phages carried single nucleotide substitutions, which clustered in the predicted PAM sequence and in positions 1 and 3 of the protospacer (Fig. 1B and Fig. S2A).
Systematic Mutagenesis of M13 Protospacer.
Because individual mutations were obtained several times, the data indicated that we were approaching saturation with respect to the variability of natural escape phages. Given the highly unequal distribution of natural mutations leading to phage escape phenotype, we generated a series of phages with site-specific point mutations in the g8 protospacer to assess the effect of these mutations on M13 ability to escape CRISPR-mediated exclusion. Viable phages were obtained with single nucleotide substitutions in protospacer positions 1–19, 21–24, 27, and 30. Plaque assays showed that only phages with substitutions at positions 1–5, 7, and 8 displayed an escape phenotype (Fig. 1B and Fig. S2B), indicating that perfect base pairing of only this part of crRNA (denoted “seed sequence”) with the corresponding part of the protospacer (denoted “seed region”) as well as an intact PAM sequence are required for interference by the E. coli subtype CRISPR/Cas system. All other mutants were restricted on g8 CRISPR plasmid-containing cell lawns as effectively as the wild-type phage (Fig. 1B).
To demonstrate that the observed lack of stringent requirement for a perfect spacer-protospacer match is a general feature of E. coli subtype CRISPR/Cas system, we performed an independent experiment in which a plasmid was introduced into E. coli cells overexpressing Cascade, Cas3, and an engineered crRNA targeting this plasmid. An approximately 10,000-fold drop in plasmid transformation efficiency (compared to efficiency of transformation into cells lacking crRNA targeting the plasmid) was observed. Plasmids that escaped the CRISPR/Cas-imposed transformation block contained point mutations at positions -3 and -2 in PAM and positions 1, 4, 5, and 7 in the protospacer (see Materials and Methods and Fig. S3). Based on Fig. S3, statistically significant variation of transformation efficiency among the plasmids that escaped the transformation block was observed, possibly indicating that some mutations weakened, but did not completely abolish the CRISPR/Cas function. In contrast, the all-or-none type of effect was observed in phage infection experiments, with only escape mutants forming visible plaques on lawns of g8 crRNA containing cells. Although this matter was not further investigated, we surmise that the quantitative difference in the two assays has to do with the fact that the results of the plasmid transformation assay depend on a single restricting act of the CRISPR/Cas system upon cell entry of a plasmid molecule during transformation, followed by positive selection for plasmid presence in antibiotic-containing medium. The phage assay reports on the formation of a plaque that results from multiple infection events, of which each can be affected by CRISPR/Cas. Even a noncomplete block of individual cycles of phage development by CRISPR/Cas can have a large effect on plaque formation, thus preventing us from detection of gradual effects.
To prove that restriction of M13 phage containing engineered protospacer mutations was indeed due to the function of CRISPR/Cas system, several escape phages were selected on g8-spacer CRISPR plasmid-containing cells and sequenced. The results, summarized in Fig. 1B and Fig. S4, demonstrate that all escape phages contained, in addition to site-specifically introduced protospacer mutations, point substitutions at PAM or protospacer positions 1 and 3. We therefore conclude that most point substitutions introducing single mismatches between a protospacer and crRNA spacer sequence do not prevent CRISPR/Cas mediated interference.
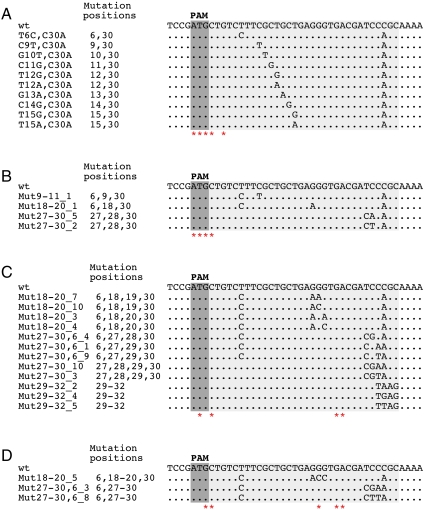
Double mutant phages were generated by combining a substitution at position +30 with several other silent (i.e., having no effect on CRISPR interference) point mutations in the protospacer (Fig. 2A). All mutants were viable, and each of them was restricted on lawns of cells containing the g8 plasmid as efficiently as the wild-type phage. Escape phages selected from individual double mutant phages contained an additional mutation at PAM or at protospacer positions 1 and 3 (Fig. 2A). Thus, a tested double mismatch between a CRISPR spacer and the target protospacer is insufficient to escape CRISPR interference. Phages combining three silent substitutions in the protospacer were constructed and were found to be subject to CRISPR-mediated exclusion (Fig. 2B). Triple mutants that escaped CRISPR/Cas mediated exclusion again contained additional mutations that occurred at PAM and protospacer positions 1 and 3 (Fig. 2B and Fig. S5A).
Fig. 2.
Multiple mutations in the protospacer are tolerated without affecting CRISPR/Cas mediated exclusion. Double (A), triple (B), quadruple (C), and quintuple (D) substitutions in the g8 protospacer that did not affect CRISPR/Cas mediated exclusion are listed below the wild-type protospacer sequence. In each panel, asterisks indicate positions of point mutations identified in escape phages selected on the background of individual restricted phage mutants.
Quadruple and quintuple protospacer phage mutants were created in a similar fashion (Fig. 2 C and D, correspondingly). With the exception of one quadruple and three quintuple mutants (Fig. S6), all were found to be restricted on g8 plasmid-containing cell lawns. Analysis of escape phages selected from phages carrying quadruple and quintuple mutations revealed that in addition to mutations in PAM and PAM-proximal protospacer positions uncovered earlier, substitutions in positions 19, 22, and 23 were also present (Fig. 2 C and D and Fig. S5 B and C). Unlike mutations in the PAM and seed region, these mutations by themselves did not lead to escape. The results thus indicate that when the number of individually silent mismatches between a spacer and a protospacer equals or exceeds 4, substitutions at functionally unimportant protospacer positions start to affect CRISPR/Cas function. In other words, a limit is being approached beyond which no efficient recognition of protospacer target can occur.
It could be argued that the relaxed specificity of CRISPR system revealed in our experimental system is due to nonphysiological levels of expression of CRISPR RNA and/or cas gene products. To exclude this possibility, we engineered an E. coli strain M13g8 containing a g8 spacer in the genomic CRISPR I cassette. The strain also lacked the hns gene, because recent work demonstrated that the histone-like protein (H-NS) of E. coli inhibits transcription initiation from cas promoters (25, 26). Experiments with wild-type and several mutant phages were repeated with the M13g8 strain; the results were identical to those with cells carrying a plasmid-borne CRISPR cassette with the g8 spacer and cas gene plasmids (Fig. S7). Thus, relaxed requirements for CRISPR spacer/target DNA protospacer identity are not due to overexpression of plasmid-borne cas genes and/or CRISPR RNA but rather faithfully reflect a basic mechanistic feature of the CRISPR/Cas system.
Recognition of Mutated M13 Protospacers by the Cascade Complex in Vitro.
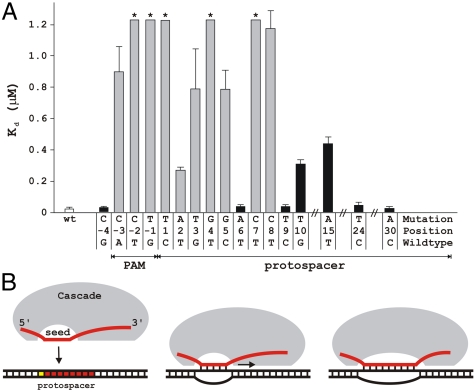
We next determined the consequences of CRISPR spacer–phage protospacer mismatches on target recognition in vitro. A Cascade complex carrying g8 crRNA was prepared, and the affinity of the complex for double-stranded DNA oligonucleotides containing wild-type or mutant PAM, and mutant protospacers was determined using native gel electrophoresis (Fig. S8). The results show that, although the dissociation constant (Kd) for the wild-type DNA probe is 24 nM, Kd of all escape mutant probes is at least 10- to 50-fold higher (Fig. 3). Probes with mutations in PAM positions -2 and -1, and in protospacer positions 1, 4, and 7, could not be bound to saturation (Kd > 1.2 μM). We conclude that compromised target DNA binding by the Cascade complex is in good agreement with the viral escape phenotype observed in vivo. However, lower binding affinities of the Cascade complex do not in all cases result in viral escape, as is suggested by the lowered Kd values of the nonescape mutants with substitutions at positions 10 or 15. This suggests that the presence of an intact PAM accompanied by perfect base pairing of the seven-nucleotide crRNA seed sequence (comprising positions 1–5, 7, and 8) with the seed region of target DNA is actively monitored by the Cascade complex and is a prerequisite for the initiation of CRISPR interference. Mutations at nonseed positions in the protospacer lead to mismatches with the crRNA spacer outside the monitored region and form no obstacle to downstream CRISPR interference processes.
Fig. 3.
Mutations in the PAM sequence and mismatches in the seed region decrease target DNA binding affinity of the g8 crRNA-Cascade. (A) Dissociation constants (Kd) of escape mutant (gray bars) and nonescape mutant DNA (black bars) were determined by electrophoretic mobility shift assay. Asterisks indicate dissociation constants of these dsDNA targets that are greater than 1.2 μM. Error bars indicate standard deviations. (B) A model of R-loop formation between crRNA and double-stranded target DNA. Base pairing between the crRNA spacer and target DNA is initiated at crRNA seed sequence area and propagated in 5′ → 3′ direction over the complete protospacer region.
Discussion
A view of target recognition by E. coli subtype Cascade complex that emerges from our work is schematically depicted in Fig. 3B. In the presence of functional PAM, the seed sequence in the crRNA likely functions as a nucleation point for progressive hybridization of crRNA fragment matching the CRISPR spacer with target DNA, leading to local unwinding of the double-stranded protospacer DNA. The subsequent extension stage of seed-nucleated melting appears to be relatively insensitive to mismatches, as productive target complexes occur even in the presence of mismatching (this study, 12, 13). In contrast to the contiguous six to seven nucleotide seed sequence of eukaryotic interfering RNAs (23), the seven-nucleotide crRNA seed sequence is noncontiguous, with matching at position 6 not required (Figs. 1B and 3). This is consistent with an observation that the nucleotide in this position is unpaired and therefore is likely flipped out of the RNA:DNA hybrid in the Cascade-induced R loop (27). We hypothesize that the crRNA seed sequence plays a crucial role in scanning the invader DNA for a matching protospacer sequence. By an initial rapid check for the presence of a PAM and base pairing of the seven-nucleotide crRNA seed sequence a basal level of specificity of binding may be accomplished. Higher accuracy identification of a protospacer sequence may be achieved at the second stage, when the nonseed sequence of the crRNA spacer is allowed to base pair. This stepwise recognition process may enable the Cascade complex to locate targets with greater efficiency.
The relaxed specificity of the CRISPR/Cas system has important consequences for the understanding of host-virus interactions. The number of mismatches that do not affect CRISPR/Cas function (4, 5 mismatches) provides a lower limit of acceptable mismatches, because certain combinations of point substitutions that exceed the limit may still allow CRISPR-mediated exclusion. In conclusion, the relaxed specificity of the CRISPR/Cas system at nonseed positions in the crRNA spacer limits the possibilities for phages to escape immunity by point mutation and may allow a single spacer to effectively target related phages without increasing the length of the CRISPR cassette.
It should be noted that all phage plaques that appeared on lawns of cells carrying the M13 targeting CRISPR plasmid were found to contain escape mutations. This was true both for the wild-type phage infection and for infections with various mutants, including the quintuple mutants, containing five mismatches outside of the seed region. Thus, the experimentally determined e.o.p. values (approximately 10-5) obtained with these phages report on the mutation rate, rather than the actual efficiency of protection provided by the CRISPR/Cas system. In the case of the wild-type phage, all 50 plaques that appeared on targeting cell lawns and that were analyzed were found to be mutant. It therefore follows that a chance of overriding protection afforded by the CRISPR/Cas by the upcoming phage is no more than 2 × 10-7, which is better than the corresponding values for restriction-modification systems. In fact, the above estimate provides a lower limit of CRISPR/Cas efficiency and it is possible that CRISPR/Cas interference is an all-or-none phenomenon, which can be avoided only by introducing mutations in important regions of PAM, the protospacer, or in the CRISPR/Cas components.
Materials and Methods
Molecular Cloning.
A CRISPR cassette plasmid targeting the M13 phage genome was generated by replacing the EcoRI-BamHI fragment in the nontargeting CRISPR plasmid pWUR477 (17, Table S1) by synthetic DNA carrying a 32-bp fragment of the M13 gene 8 (Fig. S1).
Phage Mutagenesis.
Mutations of the g8 protospacer were introduced into the M13 phage genome by QuickChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s protocol. Primers used in mutagenesis are listed in Table S2. Reaction mixtures, in 20 μL, contained 20 ng of double-stranded M13 replicative form (RF) used as a template and 50 ng each of two complementary oligonucleotides carrying the desired mutations. After amplification and DpnI digestion, 1 μL of modified phage DNA was transformed into NovaBlue Singles competent cells (Novagen) by 1 min heat shock at 42 °C followed by 2 min incubation on ice. Ten to 50 μL of transformation reactions was added to soft agar containing 200 μL of overnight culture of F+ E. coli cells [NovaBlue or NovaBlue(DE3) from Novagen] and plated on LB agar plates. Phage plaques were monitored after an overnight incubation at 37 °C and phage DNA analyzed by sequencing through the g8 protospacer region.
Phage Sensitivity Test.
Cell sensitivity to wild-type and mutant M13 phages was determined by a spot test method. Novablue(DE3) cells carrying CRISPR cassette plasmid (with g8 spacer or nontargeting pWUR477) and two compatible plasmids expressing the entire set of cas genes (pWUR397 and pWUR399) (17, Table S1) were used as a host. The cells were grown in LB medium supplemented with 25 μg/mL Str, 25 μg/mL Kan, and 34 μg/mL Cam until OD600 nm reached 0.5. The cultures were concentrated by centrifugation, cell pellets resuspended in 1/10 volume of 10 mM Tris-HCl, pH 8.0, and 10 mM MgSO4 and used in plaque tests. Rectangular 100 × 150 mm Petri dishes with LB agar supplemented with 25 μg/mL Str, 25 μg/mL Kan, and 34 μg/mL Cam were overlaid with 5 mL of soft agar containing 0.75 mL of plating cells suspension. After solidification for 5 min, 5 μL of 10-serial fold dilutions of phage lysates were spotted on the soft agar surface. Plates were allowed to dry and incubated overnight at 30 °C. Efficiency of plaquing was calculated as a ratio of phage titers observed on cells expressing the g8 spacer CRISPR cassette and on nontargeting CRISPR cells. For each phage mutant, plaque assays were performed at least twice. When phage infection was restricted by cells with the g8 spacer, escape phages were selected and analyzed by sequencing through the g8 protospacer region.
Kd Measurements.
PAGE-purified oligonucleotides (Table S2) were annealed and 5′-labeled with 32P γ-ATP (PerkinElmer) using T4 polynucleotide kinase (Fermentas). Oligonucleotides were purified with Qiaquick nucleotide removal kit (Qiagen) and single-stranded DNA was removed with Exonuclease I (Fermentas). Exonuclease I was removed by extraction with an equal volume of phenol∶chloroform∶isoamylalcohol (25∶24∶1) equilibrated at pH 8.0 (Fluka). Cascade-containing crRNA containing the g8 spacer was expressed from pWUR408, pWUR 514, and pWUR615 (Table S1) and purified as described previously (27). EMSA reactions contained 6-, 12-, 30-, 60-, 120-, 300-, 600-, or 1,200-nM Cascade, 1-nM dsDNA target, 100 mM NaCl, 20 mM Tris-Cl pH 8.0, and were incubated for 30 min at 37 °C prior to electrophoresis on a 5% polyacrylamide gel. The gels were analyzed using phosphor storage screens and a PMI phosphor imager (Bio-Rad).
The signals of unbound and bound probe were quantified using Quantity One software (Bio-Rad). The fraction of bound probe was plotted against the total Cascade concentration, and the data were fitted by nonlinear regression analysis to the following equation: Fraction bound probe = [Cascade]total/(Kd + [Cascade]total). Kd values reported are the average of three independent determinations.
Random Mutagenesis, Plasmid Transformation and Escape Mutant Selection.
A random mutant library of a 350-bp λ phage fragment was generated by PCR using the GeneMorph II Random Mutagenesis Kit (Stratagene). Mutated PCR products were cloned into pUC19 using restriction sites BamHI and HindIII and transformed to E. coli NEB5α (New England Biolabs). Approximately 104 colonies were combined, grown to stationary phase in liquid media, and their plasmids were isolated. This plasmid library was used to transform E. coli KRX cells (Promega) carrying pWUR397, pWUR400, and pWUR630 to produce Cascade, Cas3 and J3 pre-crRNA. These cells were pregrown in LB supplemented with the appropriate antibiotics at 37 °C until an OD600 nm of approximately 0.3. The cells were induced with 1 mM isopropyl β-D-1-thiogalactopyranoside and 0.2 % L-arabinose and grown for 45 min at 37 °C prior to harvesting. Cells were made electrocompetent by washing twice with ice cold water and subsequently twice with ice cold 10% glycerol. The mutant library was transformed and escape colonies were selected for plasmid isolation. The pWUR610 plasmids from 59 of these escape mutants were sequenced at Baseclear using BG2455 and BG2456 primers, resulting in the identification of 48 point mutants of 7 different types. To confirm that the escape phenotype is caused by the mutation in pWUR610, the isolated plasmids were retransformed, and the transformation efficiencies were calculated based on duplicate experiments.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health R01 Grant GM59295, Russian Academy of Sciences Molecular and Cellular Biology grant, Federal Program “Scientific and scientific-pedagogical personnel of innovative Russia 2009-2013” State Contract 02.740.11.0771, Russian Foundation for Basic Research Grant 1-04-01373-a (to K.S.), The Netherlands Organisation for Scientific Research Veni Grant 863.08.014 (to S.J.J.B.) and Vici Grant 865.05.001 (to J.v.d.O.), and National Institutes of Health RC1 Grant GM09047 (to B.L.W.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104144108/-/DCSupplemental.
References
- 1.Mojica FJ, Díez-Villaseñor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, bacteria and mitochondria. Mol Microbiol. 2000;36:244–246. doi: 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- 2.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 3.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 4.Jansen R, van Embden JD, Gaastra W, Schouls LM. Identification of a novel family of sequence repeats among prokaryotes. OMICS. 2002;6:23–33. doi: 10.1089/15362310252780816. [DOI] [PubMed] [Google Scholar]
- 5.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: Computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 8.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 9.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 10.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 12.Gudbergsdottir S, et al. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol Microbiol. 2011;79:35–49. doi: 10.1111/j.1365-2958.2010.07452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manica A, Zebec Z, Teichmann D, Schleper C. In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol Microbiol. 2011;80:481–491. doi: 10.1111/j.1365-2958.2011.07586.x. [DOI] [PubMed] [Google Scholar]
- 14.Hale CR, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang TH, et al. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci USA. 2002;99:7536–7541. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lillestøl RK, Redder P, Garrett RA, Brügger K. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–2188. doi: 10.1261/rna.2230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–2579. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 22.Deveau H, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pougach K, et al. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol Microbiol. 2010;77:1367–1379. doi: 10.1111/j.1365-2958.2010.07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pul U, et al. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol Microbiol. 2010;75:1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- 27.Jore MM, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.