Abstract
It has been well established that trans-acting small RNAs guide promoter methylation leading to its inactivation and gene silencing at the transcriptional level (TGS). Here we addressed the question of the influence of the locus structure and epigenetic modifications of the target locus on its susceptibility for being paramutated by trans-acting small RNA molecules. Silencing was induced by crossing a 35S promoter silencer locus 271 with two different 35S-driven transgene loci, locus 2 containing a highly expressed single copy gene and locus 1 containing an inverted posttranscriptionally silenced (PTGS) repeat of this gene. Three generations of exposure to RNA signals from the 271 locus were required to complete silencing and methylation of the 35S promoter within locus 2. Segregating methylated locus 2 epialleles were obtained only from the third generation of hybrids, and this methylation was not correlated with silencing. Strikingly, only one generation was required for the PTGS locus 1 to acquire complete TGS and 35S promoter methylation. In this case, paramutated locus 1 epialleles bearing methylated and inactive 35S promoters segregated already from the first generation of hybrids. The results support the hypothesis that PTGS loci containing a palindrome structure and methylation in the coding region are more sensitive to paramutation by small RNAs and exhibit a strong tendency to formation of meiotically transmissible TGS epialleles. These features contrast with a non-methylated single copy transgenic locus that required several generations of contact with RNA silencing molecules to become imprinted in a stable epiallele.
Key words: transcriptional gene silencing, transgene epialleles, DNA methylation, small RNA
Introduction
Paramutation is the transfer of epigenetic information between homologous loci (allelic or non-allelic) that leads to a heritable change in expression of one of these loci (reviewed in ref. 1). This process involves RNA signals2 and epigenetic modification of chromatin.3 Two types of homology dependent silencing mechanisms can be distinguished, transcriptional gene silencing (TGS) and posttranscriptional gene silencing (PTGS).4,5 TGS is frequently correlated with methylation of the promoter6 while PTGS is associated with DNA methylation of the transcribed region.7 The diffusible character of small RNA molecules is precondition for their capacity to methylate homologous sequences in trans. Indeed, small RNA-directed DNA methylation (RdDM) likely accounts for a considerable portion of DNA methylation events and participates in heterochromatin formation.8 Intensive research in the field has led to the identification of critical epigenetic components of RdDM such as DNA methyltransferases (MET1, DRM), RNA dependent RNA polymerases (RdRP), histone deacetylases (HDAC), plant-specific classes of DNA-dependent RNA polymerases (POLV, POLIV) and others (reviewed in ref. 9). Moreover, methylation of the cytosine residues in CG, CHG and CHH sequence contexts are all catalyzed by different enzymatic activities (reviewed in ref. 10).
Cauliflower Mosaic Virus (CaMV) 35S promoter (P35S) has been widely studied in different plant systems because of its frequent usage in transgenic technologies. Despite the fact that it represents one of the strongest promoters known, its activity is frequently modified through epigenetic mechanisms.11–13 The sensitivity of P35S to epigenetic inactivation may vary from line to line and may be influenced by locus structure and target species. The silenced state may be transient or metastable, perturbed by epigenetic deficiency,14,15 cultivation conditions16 or environmental factors.17 In addition, sites binding critical factors for promoter activity have been identified and shown to be methylation-sensitive.18
The frequency of transgene silencing can be influenced by the organization of transgene insertion,19 chromosomal environment,20 expression threshold21 and ploidy level.22 While hairpin and inverted repeat structures have been widely recognized as triggers of trans-silencing capacity, the factors influencing the susceptibility of targets to undergo silencing are less clear. Repeat rich regions stimulate trans-silencing while loci located in repeat poor (gene rich) regions are less prone to trans-silencing effects in Arabidopsis transgenic lines.20 In tobacco, distinct chromosomal configurations of transgenes and proximity of intercalary heterochromatin were proposed to favor trans-silencing effects.23 The repeats apparently trigger paramutation in certain endogenous loci of maize24 while in other systems, they do not seem to play a role in silencing.25 There are also conflicting reports as to whether transgene expression is influenced by specific structural DNA features such as MARs.26,27 Anyway, a viroid sequence failed to be methylated by RdDM mechanism when embedded within a highly structured satellite mosaic virus sequence.28
Here, we elaborated a hybrid system comprising a transcriptional silencer locus and a homologous target locus. The transcriptional silencer locus (271) induces silencing of the 35S promoter-linked genes. The two target loci contain neomycin phosphotransferase transgene driven by the same 35S promoter (35S:nptII) but at different positions and in a different epigenetic state. We asked whether 271-induced trans-silencing occurred at both target genes, how efficient it was, and whether trans-TGS and promoter methylation were stable upon 271 segregation.
Results
Description of the transgenic loci and experimental setup.
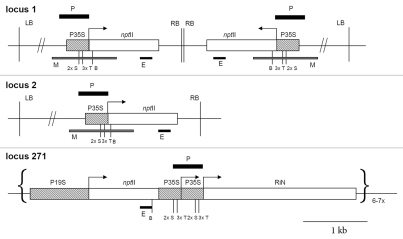
Tobacco (Nicotiana tabacum) transgenic locus 1 (Lo1, Fig. 1), which contains two copies of a T-DNA in an inverted repeat arrangement, was previously described in detail in references 29 and 30. The convergently transcribed neomycin phosphotransferase II (nptII) reporter transgenes driven by 35S promoters (35S:nptII) are posttranscriptionally silenced and methylated in the coding region but not in the promoter. The transgenic locus 2 (Lo2, Fig. 1) consists of a single non-methylated T-DNA copy insertion and is characterized by high and stable expression of reporter transgene nptII.29,31 Transgenic locus 271 consists of the complex insertion of six to seven copies of the tobacco nitrite reductase (NiR) sequence in antisense orientation driven by two 35S promoters and an nptII transgene driven by a CaMV 19S promoter (Fig. 1). This locus was shown to be able to effectively silence in trans all transgenes driven by 19S and 35S promoters at the transcriptional level and silence the endogenous nitrite reductase gene at the posttranscriptional level.32,33 These homologous interactions were associated with the presence of small RNA molecules.34
Figure 1.
Genomic organization of transgenic loci. Transgenic locus 1 contains two copies of the T-DNA organized as an inverted repeat; transgenic locus 2 encompasses one T-DNA copy. The distance between right (RB) and left (LB) border is ∼5 kb in locus 1 and locus 2. There is a hygromycine resistance gene between the LB and the nptII gene. Restriction sites used for methylation analysis were TaiI (T), Sau96I (S) and BclI (B). A thick P bar indicates region of probe hybridization. M and E bars correspond to PCR fragments used for bisulfite and expression analysis, respectively. Silencing trigger locus 271 comprises several transgene copies, some of them being incomplete. P35S, P19S: promoters from the cauliflower mosaic virus; nptII: bacterial neomycin phosphotransferase II gene; RiN: inverted copy of the tobacco nitrite reductase gene.
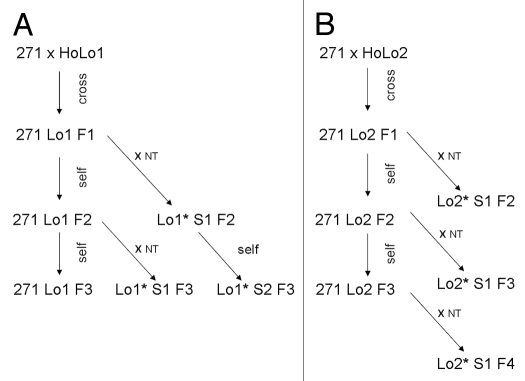
The experimental setup and scheme of crosses is schematically depicted in Figure 2. Lines carrying silencer locus 271 and silencing targets (locus 1 and locus 2) were crossed to obtain F1 hybrids. These hybrids (called 271 Lo1 and 271 Lo2) were selfed for two more generations (F2, F3). In each generation the individuals were genotyped and analyzed for nptII transcript levels and 35S promoter methylation. From the 271 Lo1 hybrid, progeny plants were also selected that contained possibly imprinted Lo1 (termed Lo1*), but that did not inherit the 271 silencer locus. The first generation of segregants (Lo1*S1 F2) was selfed to obtain an additional generation (Lo1*S2 F3) without silencer locus. Also 271 Lo2 hybrids of the F1–F3 generations were crossed with non-transgenic tobacco plants to obtain plants (Lo2*S1 F2, Lo2*S1 F3, Lo2*S1 F4) carrying transgenic locus 2 only.
Figure 2.
Scheme of the experimental strategy. Crossing experiments leading to the required hybrids and segregants encompassing the target locus 1 are schematically represented. The same setup was applied to obtain hybrids and segregants with the locus 2 target. 271: tobacco plant line homozygous for the transgenic locus 271; HoLo1: tobacco plant line homozygous for the transgenic locus 1; 271 Lo1: hybrid plant consisting of the silencing trigger locus 271 and the locus 1 target; Lo1*S1 and Lo1*S2: segregated progenies of the first and second generation, respectively, having inherited the transgenic locus 1; F1, F2 and F3: first, second and third generation, respectively, after crossing the target with the silencing trigger; NT, non-transgenic tobacco plant; self, self-pollination.
Progressive transcriptional gene silencing of target loci in hybrid plants.
The nptII reporter transgene expression was analyzed in parental plant lines and derived hybrids. We used quantitative RT-PCR and northern blot analysis to determine the nptII specific mRNA levels, while the NPTII protein accumulation levels were analyzed by NPTII ELISA.
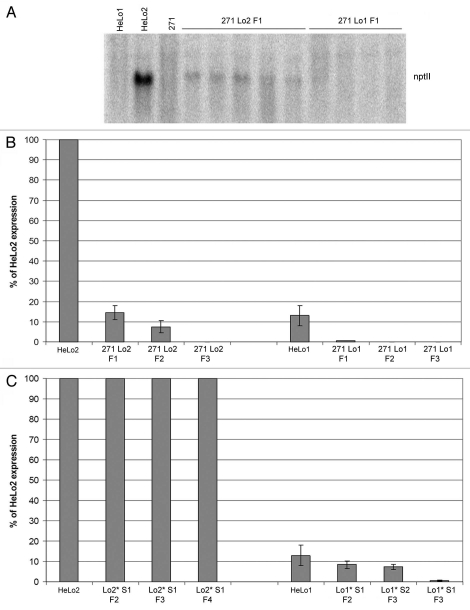
Locus 2 within the parental HeLo2 line (Fig. 3A) synthesizes high levels of transgene-specific transcripts and proteins.29 In contrast, the level of nptII transcripts in the 271 parental line was beyond the sensitivity limit of both northern and quantitative RT PCR assays. In leaves of the 271 Lo2 F1 hybrids, the nptII RNA levels were reduced to ∼15% of the levels in parental HeLo2 leaves, and they were gradually decreasing to ∼8% and <0.1% (beyond the sensitivity limit) in F2 and F3 hybrids, respectively (Fig. 3B). The NPTII protein accumulation levels were reduced >700 fold in all generations of 271 Lo2 hybrids as compared to those in the parental plants (Sup. Fig. 1A). Thus, similarly as upon posttranscriptional silencing, the transcriptional silencing was (much) more pronounced at the protein level than at the RNA level.
Figure 3.
Analysis of the nptII expression in the 271 Lo2 and 271 Lo1 hybrid plants and the segregating progenies. (A) Northern blot analysis of the nptII reporter transgene expression in F1 hybrids. About five micrograms of total RNA were loaded per line and hybridized against the nptII DNA probe. (B) Analysis of the nptII expression by quantitative RT-PCR in the 271 Lo2 and 271 Lo1 hybrids over three generations (F1–F3). The nptII RNA levels were normalized to tobacco actin mRNA. The expression level of the non-silenced HeLo2 parental line has been arbitrary chosen as 100%. Three to five plants of each line were analyzed in three technical replicates. (C) Analysis of the nptII expression by quantitative RT-PCR in the progenies that inherited locus 2 (left) and locus 1 (right) targets without the 271 silencer. S1, S2: first generation and second generation of segregants, respectively.
In the parental HeLo1 line, expression of the nptII gene is silenced by a posttranscriptional mechanism (Fig. 3A). The nptII-specific RNA accumulation levels were about 13% of those of the non-silenced HeLo2 line (Fig. 3B).35 At the protein level (Sup. Fig. 1B), the difference was much more pronounced and NPTII proteins accumulated in Helo1 plants to less than 1% of those in Helo2 plants (>100 fold) consistent with previous observations.36 In 271 Lo1 hybrids, the nptII transcripts were barely detectable already in F1 individuals and thus were reduced further to 1% or less compared to the non-silenced line (Fig. 3B). The 271-induced silencing was stable also in the F2 and F3 generations thereafter.
Progressive silencing is correlated with enhanced promoter methylation.
To determine the methylation status of the 35S promoters, we carried out Southern blot analysis using methylation-sensitive restriction endonucleases (Fig. 4) and bisulfite sequencing (Fig. 5).
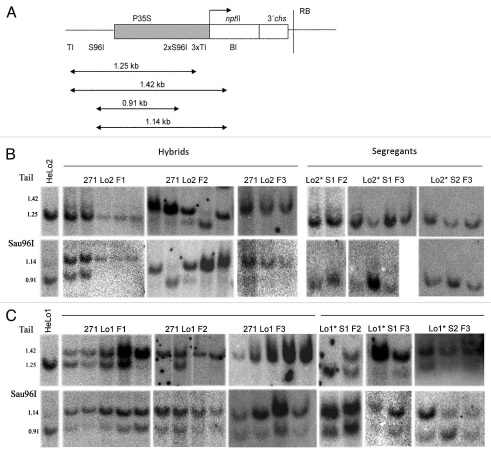
Figure 4.
Analysis of the 35S promoter methylation in the 271 Lo2 and 271 Lo1 hybrid plants and segregating progenies using methylation-sensitive restriction endonucleases. (A) Restriction maps of the P35S in the target loci. BclI (BI) enzyme was used for the dissection of the particular regions of T-DNA in hybrid lines. TaiI (TI) restriction endonuclease was used to determine the methylation of cytosines in CG motif, Sau96I (S96I) enzyme to analyze the methylation of cytosines in non-symmetrical sequence context. Arrows schematically indicate the lengths of the digested fragments. Abbreviations are the same as in Figure 1. (B) Southern blot analysis of 35S promoter methylation in the 271 Lo2 hybrids and locus 2* segregants. DNAs were predigested by BclI restriction enzyme and then digested with methylation-sensitive TaiI (top part) and Sau96I (bottom part). The 1.42 kb BclI/TaiI band indicates the complete methylation of all three in the promoter located TaiI sites; the 1.14 kb BclI/Sau96I band reveals methylation of two Sau96I sites located in the promoter. The 1.25 kb and 0.91 kb bands evidenced the non-methylated cytosine(s) in TaiI and Sau96I restriction sites, respectively. Blot cuts are shown; the examples of full blots are given in Supplemental Figure S2. (C) Southern blot analysis of 35S promoter methylation in the 271 Lo1 hybrids and locus 1* segregants. The experimental strategy is the same as described in (B).
Figure 5.
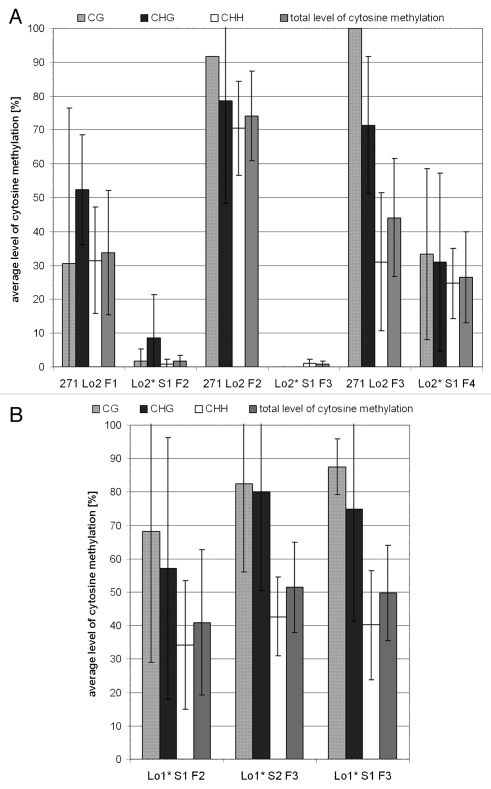
Analysis of the 35S promoter methylation in the parental hybrids and derived segregants by bisulfite genomic sequencing. (A) Summary of cytosine methylation levels in 271 Lo2 hybrids (F1–F3) and derived segregants with the “-/Lo2” genotype. The analyzed 344 bp region comprise both core promoter (−1/−46) and enhancer sequences at −46/−105.51 The diagram is organized to show the methylation pattern in the parent the first followed by that of the segregant (S1). [BR2]The data are averaged values from 4–6 clones per sample. Variation in methylation density between the clones is indicated by error bars (standard deviation). (B) Summary of cytosine methylation levels in segregating progenies from the 271 Lo1 line (“-/Lo1” genotype).
More specifically, the methylation status was analyzed at restriction sites TaiI or Sau96I by double digestion with the methylation-insensitive BclI enzyme to separate target and trigger loci from each other. The three TaiI restriction sites within the 35S promoter are located at −79, −119 and −130 with respect to the transcription start site (Fig. 4A) and are not cleaved when the cytosine in the CG sequence context is methylated (AmCGT). Thus, a 1.25 kb fragment is generated if at least one TaiI site is free of methylation, while a 1.42 kb fragment is generated when cytosines in all three TaiI sites in promoter are methylated. The non-symmetrical methylation in the promoter at positions −163 and −175 was analyzed by Sau96I which is sensitive to methylation of both cytosines (GGNmCmC). Thus, a 1.14 kb fragment is indicative for methylation of both sites, while a 0.91 kb band is generated when at least one Sau96I site is non-methylated (Fig. 4A).
In parental HeLo1 and HeLo2 lines the P35S probe hybridized to the 1.25 kb TaiI and 0.91 kb Sau96I fragments indicating that promoters of both target loci 1 and 2 were non-methylated (Fig. 4B and C). In contrast, only high molecular weight bands were visualized in DNA from locus 271 (Sup. Fig. 2), which is consistent with heavy methylation of P35S copies in this locus.33
DNA from 271 Lo2 F1 hybrids was partially methylated at cytosines in non symmetrical context as indicated by partial resistance to Sau96I digestion while no methylation was observed at CG sites as shown by fully digested TaiI fragments. In the F2 and F3 hybrids, increased resistance to TaiI appeared, revealing increased CG methylation while the level of CHH methylation remained unchanged (Fig. 4B).
In the 271 Lo1 F1 hybrids, hybridization fragments corresponding to partial methylation of both TaiI and Sau96I sites were observed. In F2 and F3, complete resistance of DNA to TaiI digestion appeared to indicate full methylation of CG sites. Similarly, the Sau96I hybridization profiles were indicative for complete methylation of the cytosines in a non-symmetrical context [shift from a 0.91 kb fragment towards a 1.14 kb fragment (Fig. 4C)].
Bisulfite sequencing analysis was carried out to reveal the methylation profile of the 35S promoters within locus 2 in the parental line and derived hybrids (Fig. 5A and Sup. Fig. 1A). The primers amplified 35S:nptII junctions that do not occur in locus 271, hence enabling to selectively analyze the target promoters in the hybrids (Fig. 1). While the promoter was completely unmethylated in the parental HeLo2, methylation appeared in F1 hybrids (34% of all cytosines) and further increased to about 43% of all cytosines in F3 hybrids, in particular at the CG sites, in agreement with the Southern blot results. The non-symmetrical methylation showed some fluctuation across generations. Significant methylation was also observed downstream from the transcription start site in the F3 individual (not shown).
Meiotic stability of epialleles generated by trans-TGS.
The meiotic stability of epigenetically modified 35S promoters was analyzed in the locus 2 segregants by comparing the expression level and the methylation status to the hybrid from which they were derived. In all analyzed samples, the nptII transcript and protein levels were comparable to those in the parental HeLo2 plant (Figs. 3C and Sup. Fig. 1A) indicating full restoration of transcription and translation activity. Thus, in the case of locus 2 target, silencing was entirely dependent on the presence of locus 271 and small RNA signals. Also, the stability of the methylation imprint was analyzed. The TaiI and Sau96I restriction sites in locus 2 promoter were non-methylated in F2 and F3 segregants (Lo2*S1 F2 and Lo2*S1 F3) (Fig. 4B), and also no significant methylation was revealed by bisulfite sequencing (Fig. 5A). However, notwithstanding that expression was fully restored, Lo2*S1 F4 segregant retained significant DNA methylation of cytosines in the promoter region in both CG and non-CG sequence contexts (Fig. 5A).
The meiotic stability of the transcriptional silencing and the methylation status of the 35S promoters in locus 1 were different. In the F2 segregants (Lo1*S1 F2), nptII RNA levels were about 60% of the levels in the parental HeLo1 line, and they slightly decreased in a further generation (Lo1*S2 F3). Lo1*S1 F3 segregants showed barely detectable transcripts, corresponding to 0–4% of the parental HeLo1 transcript levels (Fig. 3C and Sup. Fig. 3B). Moreover, no expression reappeared in subsequent generations (not shown). Thus, once established, the TGS became meiotically stable in the paramutated epiallele of locus 1. In accordance with this, methylation appeared to be inherited in both TaiI and Sau96I sites (Fig. 4C) and bisulfite sequencing confirmed inheritance of cytosine methylation in CG and non-CG motifs in all analyzed locus 1 segregants (Fig. 5B). In a Lo1*S2 F3, obtained by selfing a Lo1*S1 F2 individual (Fig. 2), there was a significant increase of methylation, indicating deepening of methylation even in the absence of the 271 silencing inducer.
In short, TGS silenced and methylated epialleles from an inverted repeat locus were stable and fixed after being target for one generation of a transcriptional silencing locus, while partially silenced and methylated epialleles of a single copy gene were unstable and immediately re-expressed, losing all methylation (Fig. 6). However, increased stability of locus 2 methylation patterns was observed upon the prolonged exposure to RNA signals, even when expression was near to parental non-methylated locus 2 (Fig. 5A).
Figure 6.
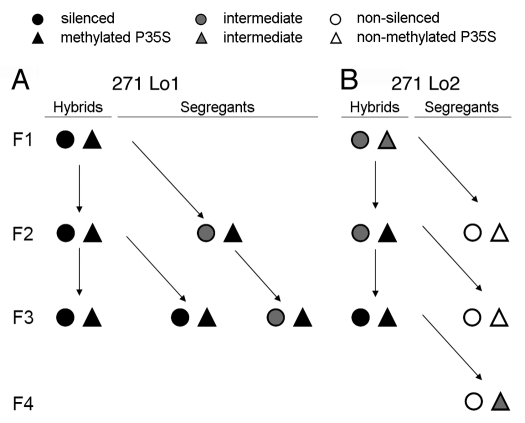
Inheritance of silencing and methylation. Epigenetic patterns of targets in line 271 Lo1 (A), 271 Lo2 (B) and derived segregants. Symbols: circle: expression status of target (filled: fully silenced; empty: non-silenced; and grey: intermediate); triangle: P35S methylation (filled: >40% Cs methylated; empty: non-methylated; grey: <40% methylation). The expression data were taken from quantitative PCR experiments (Fig. 3); the methylation data were taken from Southern blot (Fig. 4) and bisulfite (Fig. 5) analysis.
Promoter derived small RNA molecules are strictly correlated with the presence of the 271 locus.
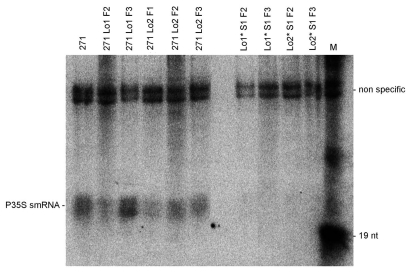
The silencer locus 271 mediates in trans-silencing and methylation of homologous 35S promoters via small RNA (smRNA) molecules.34 To elucidate whether segregants with a methylated promoter would have acquired the capacity to generate themselves smRNAs, we carried out northern blots to analyze the presence of P35S-specific smRNAs in three generations of 271 Lo2 and 271 Lo1 hybrids and segregating progenies. The 35S sense RNA probe hybridized to a doublet of bands migrating in the 20–25 nt region in both types of hybrids (Fig. 7) evidencing the role of smRNAs during the installation of TGS at the target promoter. However, the smRNA signal was not detected in any of the segregants without the silencer locus 271.
Based on these results, we conclude that smRNA molecules are required for the establishment of RNA-directed methylation patterns but they do not seem to be necessary for their maintenance in the absence of silencer locus.
Discussion
Progression of small RNA-mediated transcriptional silencing over generations.
In this work, we addressed the question how quickly a 35S promoter in different contexts becomes transcriptionally silenced and methylated by trans-acting RNA molecules generated by homologous silencer locus 271. The single copy target (locus 2) exposed to RNA silencing signals showed clear evidence of silencing progression over three generations of hybrids: in the F1 generation (271 Lo2 F1), the accumulation level of the nptII-specific RNA was reduced about 7 fold compared to hemizygous locus 2 plants, and gradually decreased in the F2 and F3 generation (Fig. 6). In F3, we were unable to detect nptII-specific RNA by a sensitive quantitative RT-PCR assay, demonstrating that expression was reduced by more than 100 fold compared to the parental line. While methylation occurred in both CG and non-CG contexts already in the first F1 generation, mainly CG methylation increased in the F2 and F3 generations (Fig. 5A) suggesting involvement of a DNA methyltransferase maintenance activity in silencing progression. Significantly, completely inactive 35S promoters showed full methylation of the CG sites at the −83 and −63 positions, known to bind critical activation factors for the 35S promoter.18 Methylation also occurred in a short segment of the transcribed region in accordance with the hypothesis that not only sites upstream but also downstream of TSS may be important for transcriptional silencing.37,38 In a previous study, trans-silencing elicited by locus 271 was relatively weak in young seedlings compared to fully developed plants33 suggesting developmental regulation. Based on our findings it is possible that the RNA directed TGS is gradually established at the target locus throughout mitotic and meiotic cycles starting from a primary zygote. In addition, clone to clone methylation variability (Sup. Fig. 3A) may account for variegated phenotypes in early generations.
At the protein level the differences between generations were not so pronounced and complete silencing appeared already in F1 (Sup. Fig. 1). While similar discrepancy between RNA and protein levels has been observed during PTGS,36 for TGS this has not been reported. There can be several explanations: (1) the NPTII protein might be less stable than its corresponding RNA. However, numerous studies demonstrated already the stability of the NPTII protein in plant cells. (2) The nptII-specific mRNAs may be degraded in hybrids. However, northern blot hybridization failed to show nptII-specific small RNAs in both 271 Lo1 and 271 Lo2 hybrids (not shown). (3) Finally, RNA transcripts resulting from partially methylated promoters may be epigenetically modified and poorly translated. Further studies are needed to clarify these issues.
Increased sensitivity of posttranscriptional silenced loci to transcriptional silencing.
Previous studies already indicated that a partially methylated transgenic locus (H2) was more sensitive to trans-silencing by locus 271 than transgenic loci which were not-methylated.32 Here we show that a PTGS locus 1 having no methylation of the promoter but dense methylation of linked coding region acquires TGS more efficiently than the single copy non-methylated locus 2 (Fig. 6). In a comprehensive study, involving several transgenic lines of Arabidopsis, correlation was found between chromosomal environment, such as close-by repeats and locus susceptibility to undergo RNA-directed TGS.20 However, locus 1 is probably located in euchromatin since (1) T-DNA methylation is restricted to the inverted repeat centre;39 (2) there is an active hygromycin gene located close to the T-DNA/plant junction;29 (3) restriction sites in the flanking sequence of T-DNA are non-methylated.30 Therefore, enhanced sensitivity to TGS should be explained in the context of locus 1 itself rather than in the context of the genomic environment. We do not know whether the PTGS process in its own, the inverted repeat structure or the dense methylation of the nptII transcribed region is responsible for the enhanced sensitivity of locus 1 to undergo TGS. The behavior of locus 1 was in some way similar to that of another locus (H2), sensitive to trans-inactivation by locus 271.32 Both loci produce RNA signals although targeting different sequences—while locus 1 silences the nptII targets, locus H2 inactivates nopaline synthase promoter. In a previous study we showed that stable locus 1 epialleles were frequently established from regenerating plants as a result of cell culture induced epigenetic variation.35 We propose that active promoters of linked posttranscriptionally silenced genes may be sensitive to inactivation either in cis or by in trans mechanisms. We favor the hypothesis that the dense methylation of the coding region might attract DNA methyltransferases and perhaps other silencing factors (such as polymerase IV, V) to the chromosomal locus, switching PTGS into TGS.39
Stability of epialleles generated by trans-TGS.
Mitotic and meiotic inheritance is one of the fundamental features of epigenetic marks. However, its efficiency, transgeneration inheritance and ability to communicate silent state to other alleles vary among systems and species. In this work we provide evidence that the heritability of small RNA-induced transgene epialleles depends on the exposure of targets to RNA signals. This is evidenced by segregation experiments in which a single copy locus 2 line exposed to promoter-specific smRNAs lost the silencer (Fig. 6). All individuals with locus 2 genotype segregating from F1-F2 generations of 271 Lo2 hybrids returned to the original unmethylated state. However, a segregant from the F3 generation retained considerable methylation. This may be explained by increased CG methylation of the target in the parental 271 Lo2 F3 individual. However, methylation occurred not only in CG but also at non CG motifs arguing that maintenance activity may influence both types of methylation.10 The locus 2 epiallelic patterns differ from those induced by a PTGS mechanism on the same locus.40 In those experiments, the coding region lost all non-CG but retained most of CG methylation. Perhaps, the non-CG methylation is actively removed from the transcribed region while it is retained in non-transcribed regions. Promoter methylation corresponding to 26% of the cytosines in the locus 2* epiallele did not prevent expression of the nptII gene. Though we cannot exclude the possibility that silencing might have occurred in other individuals that had not been analyzed (only one plant with locus 2* genotype was recovered from the 271 Lo2 F3 cross) it is likely that the low level methylation is compatible with 35S promoter activity. In support of this, 35S promoters bearing comparable (low) level of methylation were shown to be active also in other systems.32 On the other hand, epialleles generated from 271 Lo1 hybrids show substantial reduction of promoter activity and increased overall methylation (∼40%). Remarkably, the second generation of segregants in the absence of silencer even showed enhanced methylation as compared to parental S1 F2 generation (Figs. 4 and 5), suggesting that the epiallelic state can be fixed in the absence of silencer in some sensitive loci. Intriguingly, relatively dense non-symmetrical methylation was maintained in all TGS epialleles of locus 1 in the absence of methylation-inducing RNAs (this work and reviewed in ref. 30) suggesting that the maintenance of non symmetrical methylation can be directed by other signals, such as chromatin structure.13,41,42 The secondary small RNAs derived from a paramutated epiallele may also be needed to transfer silenced state to homologous promoters (termed secondary paramutation).43 Secondary paramutation has not been observed in locus 1* (not shown) and locus H*32 epialleles, both generated by the contact with the 271 silencer. We can conclude that (1) meiotic stability of epialleles, generated by a RdDM process is increasing with the number of generations exposed to a paramutagenic locus; (2) PTGS loci tend to frequently convert into stable TGS epialleles.
Materials and Methods
Generation of the transgenic plants, cultivation of plants.
All transgenic tobacco (Nicotiana tabacum, var. SR1) plants were generated by Agrobacterium-mediated transformation.45 The plants hemizygous for the PTGS locus 1 (HeLo1, Fig. 1) were obtained by crossing a plant homozygous for locus 1 (HoLo1) with an untransformed SR1 tobacco.31 The line hemizygous for locus 2 (HeLo2, Fig. 1) was obtained by crossing a plant homozygous for locus 2 (HoLo2) with an untransformed SR1 tobacco.31 Seeds of the tobacco transgenic plants homozygous for the transgenic locus 271 (Fig. 1) were obtained from INRA Versailles (a gift of Prof. Hervé Vaucheret). Crosses were performed by emasculating flowers manually before they were opened and applying pollen to the stamen. F1 hybrids were obtained by crosses of HoLo1 and HoLo2, respectively, to 271; the experimental strategy to obtain hybrids and relevant segregants is depicted in the Figure 2. Hybrid plants and segregants were genotyped by Southern blot hybridization.
RNA isolation, northern blot hybridization and real time RT-PCR analysis.
Total RNA was isolated from young leaves with the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) and treated by DNaseI (TURBO DNA-free, Applied Biosystems/Life Technologies, Carlsbad, CA). After electrophoresis on a 1.2% (w/v) formaldehyde-agarose gel, the gel was washed for 10 min in sterile water to remove the formaldehyde. The RNA was denatured in 0.05 M NaOH and blotted onto a Hybond-XL nylon membrane (GE-Healthcare, Little Chalfont, UK) in 20× SSC (1× SSC = 150 mM NaCl, 15 mM Na3-citrate, pH 7.0). 32P labeled nptII DNA probes (DekaLabel kit; MBI, Fermentas, Vilnius, Lithuania) were hybridized in ULTRAhyb buffer (Applied Biosystems/Life Technologies) for 24 h at 35°C. After washing under low-stringency conditions (twice for 15 min in 2× SSC + 0.1% (w/v) SDS at 50°C), the hybridization bands were visualized with a PhosphorImager STORM (GE-Healthcare).
The cDNAs were prepared by reverse transcription of RNAs using the Superscript II Reverse Trancriptase (Invitrogen/Life Technologies, Carlsbad, CA) and Random nonamers (Sigma, Saint Louis, MO). Quantification of the nptII level related to the actin transcripts was done using the Fast Start SYBR Green Master (Roche, Mannheim, Germany) by the Rotorgene 6000 (Qiagen). nptII was amplified with the forward primer 5′-CGT TAC AAG AGA GAA ATC GCC-3′ and the reverse primer 5′-TTC TAT CGC CTT CTT GAC GAG-3′; actin was amplified with the forward primer 5′-CTG GAT TTG CTG GTG ATG AT-3′ and the reverse primer 5′-CYC TCT TGG ATT GAG CTT-3′ in the same PCR cycle (initial denaturation at 94°C for 10 min followed by 35 cycles of 20 s at 94°C, 20 s at 56°C and 30 s at 72°C). The amount of nptII transcript was determined for three to five plants of each line in three technical replicates.
DNA isolation and Southern blot hybridization.
Total genomic DNA was isolated from fresh tobacco leaf by a cetyltrimethylammonium bromide method.46 DNA methylation was analyzed with methylation-sensitive restriction endonucleases. Approximately 20 µg of genomic DNA was digested with an enzyme excess (5 units per 1 µg DNA). After digestion, the DNA was separated by electrophoresis on a 1% (w/v) agarose gel. The gels were alkali-blotted onto a Hybond-XL membrane (GE-Healthcare) and hybridized against 32P-labeled DNA probes (DekaLabel kit, Fermentas) for at least 16 h at 65°C. The 35S promoter probe was prepared from the approximately 980 bp insert of the GSJ290 plasmid.36 After washing under high-stringency conditions (twice for 5 min in 2× SSC + 0.1% (w/v) sodium dodecyl sulfate (SDS) and twice for 20 min in 0.2× SSC + 0.1% (w/v) SDS at 65°C), the hybridization bands were visualized with a PhosphorImager STORM and the data were processed with the ImageQuant software (GE-Healthcare).
Bisulfite genomic sequencing.
Bisulfite treatments were carried out on purified genomic DNA using the EpiTect Bisulfite Kit (Qiagen). The primers for the amplification of the 35S promoter and 5′nptII region are as follows: forward primer 5′-CAT TAC ATC ACC CAT AAT AAA TAC TTT CTC-3′, the first reverse primer 5′-GAA TAG AGA GAA AGA TAT ATT TTT TAA GAT-3′ and the second reverse primer 5′-GTA ATA GAG ATT GGA GTT TTT AAG AAA GTA G-3′. The PCR program consisted of 2 min of initial denaturation at 94°C followed by 35 cycles of 0.5 min at 94°C, 1.5 min at 45°C and 1 min at 72°C. The program was ended with an extension step for 10 min at 72°C. The PCR products were cloned into a TA vector (pDrive, Qiagene) and between 5–10 clones from each sample were sequenced (Eurofins MWG Operon, Germany). The data were processed and methylation density calculated using CyMATE software.47
Analysis of small RNA molecules.
Fractions of smRNA molecules were isolated as previously described in reference 48, with minor modifications.30 The smRNAs were separated by electrophoresis (15% polyacrylamide, 7 M urea in 0.5x TBE; 1x TBE = 90 mM Tris-borate, 2 mM EDTA, pH 8.0), blotted onto a nylon Hybond-XL membrane (GE-Healthcare) with a semi-dry blot instrument (Semi Dry Transfer Blotter, Hoefer, Holliston, MA) in 0.5x TBE, and fixed by carbodiimide (SIGMA) crosslinking49 and baking (2 h at 80°C). To estimate the size of the RNA bands, a 19 nt P35S-derived oligonucleotide was used as a marker. The single-stranded RNA probe was transcribed in sense orientation from a linearized plasmid with 32P-UTP using the RNAMaxx™ High Yield Transcription Kit (Stratagene, La Jolla, CA). Hybridization was performed as described in references 48 and 50, in ULTRAhyb buffer (Applied Biosystems/Life Technologies) at 35°C for 48 h. The membrane was washed twice in 2x SSC + 0.1% (w/v) SDS for 30 min at 35°C and in 20 mM Tris-HCl, 5 mM EDTA (pH 8.0), 60 mM NaCl, 10 µgml−1 RNaseA (for 1 h at 37°C) to remove unspecific background and exposed to the PhosphorImager screen.
Conclusions
In the present work we describe a progressive transgenerational inactivation of promoter activity by trans-acting small RNA. The posttranscriptionally silenced locus 1 with a coding region methylation acquired transcriptional gene silencing and promoter methylation more readily than a non-methylated actively expressed locus 2. Thus, the PTGS loci might be inherently more sensitive to undergo transcriptional silencing elicited either in cis39 or by in trans (this study) factors. The PTGS locus also showed increased tendency to paramutation and formation of stable TGS epialleles following segregation of a RNA silencing signal. The observed heritability of in trans imposed epigenetic changes underline their importance for the establishment of evolutionary relevant traits. Spontaneous transcriptionally silenced epialleles of a flowering cycloidiea gene44 stably inherited over many generations serve as a classical example. We propose that progressive RNA-mediated transcriptional gene silencing plays an important role also in natural situations by harmonizing gene expression during allopolyploidy and stress.
Figure 7.
Detection of P35S-derived smRNA molecules in parental plants, silenced hybrids and segregating progenies that lack the 271 locus. The fraction of RNA enriched for low-molecular weight fragments (about 50 µg per line) was hybridized against the sense-specific P35S riboprobe. Promoter oligonucleotides (19 nt long) were used as size markers (M). Non-specific signals at the top of the gel were taken as indicators of balanced loading.
Acknowledgments
We thank Prof. Hervé Vaucheret (INRA, Versailles, France) for providing us with a tobacco 271 line. We thank Jana Kaiserlichova for the maintenance of plants and excellent technical assistance. This work was supported by the Czech Science Foundation (projects P501/11/P667, 204/09/H002 and P501-10-0208), Academy of Science (AVOZ50040507, AVOZ50040702 and MSMT LC0604) and FWO (Belgium) short term fellowships to M.F. and A.K.
Supplementary Material
References
- 1.Chandler VL. Paramutation's properties and puzzles. Science. 2010;330:628–629. doi: 10.1126/science.1191044. [DOI] [PubMed] [Google Scholar]
- 2.Arteaga-Vazquez M, Sidorenko L, Rabanal FA, Shrivistava R, Nobuta K, Green PJ, et al. RNA-mediated trans-communication can establish paramutation at the b1 locus in maize. Proc Natl Acad Sci USA. 2010;107:12986–12991. doi: 10.1073/pnas.1007972107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haring M, Bader R, Louwers M, Schwabe A, van Driel R, Stam M. The role of DNA methylation, nucleosome occupancy and histone modifications in paramutation. Plant J. 2010;63:366–378. doi: 10.1111/j.1365-313X.2010.04245.x. [DOI] [PubMed] [Google Scholar]
- 4.Vaucheret H, Beclin C, Fagard M. Post-transcriptional gene silencing in plants. J Cell Sci. 2001;114:3083–3091. doi: 10.1242/jcs.114.17.3083. [DOI] [PubMed] [Google Scholar]
- 5.Vaucheret H, Fagard M. Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet. 2001;17:29–35. doi: 10.1016/s0168-9525(00)02166-1. [DOI] [PubMed] [Google Scholar]
- 6.Kooter JM, Matzke MA, Meyer P. Listening to the silent genes: transgene silencing, gene regulation and pathogen control. Trends Plant Sci. 1999;4:340–347. doi: 10.1016/s1360-1385(99)01467-3. [DOI] [PubMed] [Google Scholar]
- 7.Jones L, Hamilton AJ, Voinnet O, Thomas CL, Maule AJ, Baulcombe DC. RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell. 1999;11:2291–2301. doi: 10.1105/tpc.11.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wassenegger M. The role of the RNAi machinery in heterochromatin formation. Cell. 2005;122:13–16. doi: 10.1016/j.cell.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 9.Matzke M, Aufsatz W, Kanno T, Daxinger L, Papp I, Mette MF, Matzke AJ. Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim Biophys Acta. 2004;1677:129–141. doi: 10.1016/j.bbaexp.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 10.Meyer P. DNA methylation systems and targets in plants. Febs Lett. 2010;1 doi: 10.1016/j.febslet.2010.08.017. In press. [DOI] [PubMed] [Google Scholar]
- 11.Meyer P, Niedenhof I, ten Lohuis M. Evidence for cytosine methylation of non-symmetrical sequences in transgenic Petunia hybrida. EMBO J. 1994;13:2084–2088. doi: 10.1002/j.1460-2075.1994.tb06483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dieguez MJ, Vaucheret H, Paszkowski J, Mittelsten Scheid O. Cytosine methylation at CG and CNG sites is not a prerequisite for the initiation of transcriptional gene silencing in plants, but it is required for its maintenance. Mol Gen Genet. 1998;259:207–215. doi: 10.1007/s004380050806. [DOI] [PubMed] [Google Scholar]
- 13.Mishiba K, Nishihara M, Nakatsuka T, Abe Y, Hirano H, Yokoi T, Kikuchi A, et al. Consistent transcriptional silencing of 35S-driven transgenes in gentian. Plant J. 2005;44:541–556. doi: 10.1111/j.1365-313X.2005.02556.x. [DOI] [PubMed] [Google Scholar]
- 14.Pecinka A, Dinh HQ, Baubec T, Rosa M, Lettner N, Mittelsten Scheid O. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell. 2010;22:3118–3129. doi: 10.1105/tpc.110.078493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumpatla SP, Hall TC. Longevity of 5-azacytidine-mediated gene expression and re-establishment of silencing in transgenic rice. Plant Mol Biol. 1998;38:1113–1122. doi: 10.1023/a:1006071018039. [DOI] [PubMed] [Google Scholar]
- 16.Nocarova E, Opatrny Z, Fischer L. Successive silencing of tandem reporter genes in potato (Solanum tuberosum) over 5 years of vegetative propagation. Ann Bot. 2010;106:565–572. doi: 10.1093/aob/mcq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer P, Linn F, Heidmann I, Meyer H, Niedenhof I, Saedler H. Endogenous and environmental factors influence 35S promoter methylation of a maize A1 gene construct in transgenic petunia and its colour phenotype. Mol Gen Genet. 1992;231:345–352. doi: 10.1007/BF00292701. [DOI] [PubMed] [Google Scholar]
- 18.Kanazawa A, O'Dell M, Hellens RP. The binding of nuclear factors to the as-1 element in the CaMV 35S promoter is affected by cytosine methylation in vitro. Plant Biol. 2007;9:435–441. doi: 10.1055/s-2006-924633. [DOI] [PubMed] [Google Scholar]
- 19.De Buck S, Windels P, De Loose M, Depicker A. Single-copy T-DNAs integrated at different positions in the Arabidopsis genome display uniform and comparable beta-glucuronidase accumulation levels. Cell Mol Life Sci. 2004;61:2632–2645. doi: 10.1007/s00018-004-4284-8. [DOI] [PubMed] [Google Scholar]
- 20.Fischer U, Kuhlmann M, Pecinka A, Schmidt R, Mette MF. Local DNA features affect RNA-directed transcriptional gene silencing and DNA methylation. Plant J. 2008;53:1–10. doi: 10.1111/j.1365-313X.2007.03311.x. [DOI] [PubMed] [Google Scholar]
- 21.Schubert D, Lechtenberg B, Forsbach A, Gils M, Bahadur S, Schmidt R. Silencing in Arabidopsis T-DNA transformants: The predominant role of a gene-specific RNA sensing mechanism versus position effects. Plant Cell. 2004;16:2561–2572. doi: 10.1105/tpc.104.024547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mittelsten Scheid O, Afsar K, Paszkowski J. Formation of stable epialleles and their paramutation-like interaction in tetraploid Arabidopsis thaliana. Nat Genet. 2003;34:450–454. doi: 10.1038/ng1210. [DOI] [PubMed] [Google Scholar]
- 23.Jakowitsch J, Papp I, Moscone EA, van der Winden J, Matzke M, Matzke AJM. Molecular and cytogenetic characterization of a transgene locus that induces silencing and methylation of homologous promoters in trans. Plant J. 1999;17:131–140. doi: 10.1046/j.1365-313x.1999.00357.x. [DOI] [PubMed] [Google Scholar]
- 24.Haring M, Bader R, Louwers M, Schwabe A, van Driel R, Stam M. The role of DNA methylation, nucleosome occupancy and histone modifications in paramutation. Plant J. 2010;63:366–378. doi: 10.1111/j.1365-313X.2010.04245.x. [DOI] [PubMed] [Google Scholar]
- 25.Kunz C, Narangajavana J, Jakowitsch J, Park YD, Delon TR, Kovarik A, et al. Studies on the effects of a flanking repetitive sequence on the expression of single-copy transgenes in Nicotiana sylvestris and in N-sylvestris-N-tomentosiformis hybrids. Plant Mol Biol. 2003;52:203–215. doi: 10.1023/a:1023937006311. [DOI] [PubMed] [Google Scholar]
- 26.De Bolle MFC, Butaye KMJ, Goderis IJWM, Wouters PFJ, Jacobs A, Delaure SL, et al. The influence of matrix attachment regions on transgene expression in Arabidopsis thaliana wild type and gene silencing mutants. Plant Mol Biol. 2007;63:533–543. doi: 10.1007/s11103-006-9107-x. [DOI] [PubMed] [Google Scholar]
- 27.Mlynarova L, Hricova A, Loonen A, Nap JP. The presence of a chromatin boundary appears to shield a transgene in tobacco from RNA silencing. Plant Cell. 2003;15:2203–2217. doi: 10.1105/tpc.012070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dalakouras A, Moser M, Krczal G, Wassenegger M. A chimeric satellite transgene sequence is inefficiently targeted by viroid-induced DNA methylation in tobacco. Plant Mol Biol. 2010;73:439–447. doi: 10.1007/s11103-010-9631-6. [DOI] [PubMed] [Google Scholar]
- 29.Van Houdt H, Kovarik A, Van Montagu M, Depicker A. Cross-talk between posttranscriptionally silenced neomycin phosphotransferase II transgenes. Febs Lett. 2000;467:41–46. doi: 10.1016/s0014-5793(00)01076-0. [DOI] [PubMed] [Google Scholar]
- 30.Fojtova M, Bleys A, Bedrichova J, Van Houdt H, Krizova K, Depicker A, Kovarik A. The trans-silencing capacity of invertedly repeated transgenes depends on their epigenetic state in tobacco. Nucl Acids Res. 2006;34:2280–2293. doi: 10.1093/nar/gkl180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovarik A, Van Houdt H, Holy A, Depicker A. Drug-induced hypomethylation of a posttranscriptionally silenced transgene locus of tobacco leads to partial release of silencing. Febs Lett. 2000;467:47–51. doi: 10.1016/s0014-5793(00)01077-2. [DOI] [PubMed] [Google Scholar]
- 32.Park YD, Papp I, Moscone EA, Iglesias VA, Vaucheret H, Matzke AJ, Matzke MA. Gene silencing mediated by promoter homology occurs at the level of transcription and results in meiotically heritable alterations in methylation and gene activity. Plant J. 1996;9:183–194. doi: 10.1046/j.1365-313x.1996.09020183.x. [DOI] [PubMed] [Google Scholar]
- 33.Vaucheret H. Promoter-Dependent Trans-Inactivation in Transgenic Tobacco Plants—Kinetic Aspects of Gene Silencing and Gene Reactivation. Cr Acad Sci Iii-Vie. 1994;317:310–323. [Google Scholar]
- 34.Mourrain P, van Blokland R, Kooter JM, Vaucheret H. A single transgene locus triggers both transcriptional and post-transcriptional silencing through double-stranded RNA production. Planta. 2007;225:365–379. doi: 10.1007/s00425-006-0366-1. [DOI] [PubMed] [Google Scholar]
- 35.Krizova K, Fojtova M, Depicker A, Kovarik A. Cell culture-induced gradual and frequent epigenetic reprogramming of invertedly repeated tobacco transgene epialleles. Plant Physiol. 2009;149:1493–1504. doi: 10.1104/pp.108.133165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Houdt H, Ingelbrecht I, Van Montagu M, Depicker A. Post-transcriptional silencing of a neomycin phosphotransferase II transgene correlates with the accumulation of unproductive RNAs and with increased cytosine methylation of 3′ flanking regions. Plant J. 1997;12:379–392. [Google Scholar]
- 37.Dieguez MJ, Bellotto M, Afsar K, Mittelsten Scheid O, Paszkowski J. Methylation of cytosines in nonconventional methylation acceptor sites can contribute to reduced gene expression. Mol Gen Genet. 1997;253:581–588. doi: 10.1007/s004380050360. [DOI] [PubMed] [Google Scholar]
- 38.Fu X, Kohli A, Twyman RM, Christou P. Alternative silencing effects involve distinct types of non-spreading cytosine methylation at a three-gene, single-copy transgenic locus in rice. Mol Gen Genet. 2000;263:106–118. doi: 10.1007/pl00008669. [DOI] [PubMed] [Google Scholar]
- 39.Fojtova M, Van Houdt H, Depicker A, Kovarik A. Epigenetic switch from posttranscriptional to transcriptional silencing is correlated with promoter hypermethylation. Plant Physiol. 2003;133:1240–1250. doi: 10.1104/pp.103.023796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lunerova-Bedrichova J, Bleys A, Fojtova M, Khaitova L, Depicker A, Kovarik A. Trans-generation inheritance of methylation patterns in a tobacco transgene following a post-transcriptional silencing event. Plant J. 2008;54:1049–1062. doi: 10.1111/j.1365-313X.2008.03475.x. [DOI] [PubMed] [Google Scholar]
- 41.Kanazawa A, O'Dell M, Hellens RP. Epigenetic inactivation of chalcone synthase-A transgene transcription in petunia leads to a reversion of the post-transcriptional gene silencing phenotype. Plant Cell Physiol. 2007;48:638–647. doi: 10.1093/pcp/pcm028. [DOI] [PubMed] [Google Scholar]
- 42.Gambino G, Perrone I, Carra A, Chitarra W, Boccacci P, Torello Marinoni D, et al. Transgene silencing in grapevines transformed with GFLV resistance genes: analysis of variable expression of transgene, siRNAs production and cytosine methylation. Transgenic Res. 2010;19:17–27. doi: 10.1007/s11248-009-9289-5. [DOI] [PubMed] [Google Scholar]
- 43.Stam M, Mittelsten Scheid O. Paramutation: an encounter leaving a lasting impression. Trends Plant Sci. 2005;10:283–290. doi: 10.1016/j.tplants.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 44.Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401:157–161. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- 45.Ingelbrecht I, Van Houdt H, Van Montagu M, Depicker A. Posttranscriptional silencing of reporter transgenes in tobacco correlates with DNA methylation. Proc Natl Acad Sci USA. 1994;91:10502–10516. doi: 10.1073/pnas.91.22.10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fojtova M, Kovarik A, Matyasek R. Cytosine methylation of plastid genome in higher plants. Fact or artefact? Plant Sci. 2001;160:585–593. doi: 10.1016/s0168-9452(00)00411-8. [DOI] [PubMed] [Google Scholar]
- 47.Hetzl J, Foerster AM, Raidl G, Mittelsten Scheid O. CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 2007;51:526–536. doi: 10.1111/j.1365-313X.2007.03152.x. [DOI] [PubMed] [Google Scholar]
- 48.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 49.Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007;35:60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matzke MA, Aufsatz W, Kanno T, Mette MF, Matzke AJM. Homology-dependent gene silencing and host defense in plants. In: Wu C, Dunlap J, editors. Homology Effects. 2002. pp. 243–275. [DOI] [PubMed] [Google Scholar]
- 51.Odell JT, Nagy F, Chua NH. Identification of DNA-sequences required for activity of the cauliflower mosaic virus-35s promoter. Nature. 1985;313:810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.