Abstract
Plants have to adjust, grow and establish themselves in various changing environmental conditions. Additionally, the sessile life-style of plants requires the development of response mechanisms for their adaptation in such environmental cues. Under biotic and abiotic stress, plant growth is negatively affected mainly as a result of cell cycle inhibition. The perception of stress involves the activation of signaling cascades that result in a prolonged S-phase and delayed entry into mitosis. Although the molecular interactions that link the cell cycle machinery to perception of stress are not fully understood, recent studies indicated the involvement of Cyclin Dependent Kinases (CDKs) in the plant response machinery. CDKs are core cell cycle regulators but their activity has been implicated in additional diverse cellular processes. Here we review the impact of different types of abiotic stress on plant cell cycle progression and CDK activity, and discuss the contribution of CDK function in the signaling control of stress tolerance.
Key words: abiotic stress, cell cycle, CDK, cyclin
Introduction
As a consequence of their lifestyle, plants are immobile for most of their lifetime and, lacking the mobility of the majority of animals, they have to cope with wide changes in their local environment—they cannot move to a more favourable one. Their successful adaptation to this sessile life-style can be attributed to their ability to adapt and respond to different types of biotic and abiotic stress. Here, the term stress is used to refer to factors that could negatively affect performance and/or physiology. These factors are generally classified into two broad groups, biotic and abiotic. Biotic stress refers to the damaging impact of living organisms, such as bacteria, fungi, viruses, animals and weeds while abiotic stress includes diverse environmental conditions like high temperatures, drought, salinity and UV light (Fig. 1). Exposure to stress can lead to a variety of responses, the extreme manifestation of which is the synthesis of reactive oxygen species (ROS) that can lead to cell death.1
Figure 1.
Types of biotic and abiotic stress that affect plant development.
Stress responses have been extensively studied due to their economic implications, as they can adversely influence the physiology, growth and crop productivity of plants.2 There is now added urgency in understanding stress responses as climate change is likely to lead to environmental deterioration and has raised serious concerns about future food production. As well as breeding more stress tolerant varieties of crops, it is important to understand the genetic and molecular basis of the stress response.3 To this end, stress inducible genes have identified and the complex signalling cascades that mediate stress tolerance are being dissected.4–7 One of the key responses to moderate (i.e., non-lethal) stress is the inhibition of cell proliferation and cell growth. While this response can lead to reduced yield in crops, it has a more general adaptive significance in that the resultant plant is more likely to survive. In this article, we briefly review the molecular response to abiotic stress but focus on the possible roles of Cyclin Dependent Kinases (CDKs), a group of protein kinases that include key cell cycle regulators, with particular emphasis on agronomic importance. Animal CDKs are well known for their role in the cell's response to genotoxic and other cell cycle specific stresses that cause different types of DNA damage.8 In yeast, the Pho85 CDK protein regulates transcriptional response to a variety of stress conditions9,10 and specifically modulates the nuclear localization of Rim101, a key transcription factor in response to alkaline stress.11 There is an increasing body of evidence that plant CDKs also play similar roles in the plant stress response mechanism.
Plant Response Mechanisms to Abiotic Stress: The Role of Protein Kinases
Appropriate perception and rapid response to stressful conditions is thought to contribute to tolerance. The regulatory mechanisms that confer stress tolerance involve the induction of stress-responsive genes. The first group comprises proteins that operate directly in stress tolerance and include late-embryogenesis-abundant (LEA) proteins.12 LEA proteins are also produced during the normal maturation of seeds. They appear to protect proteins and membranes and assist in the renaturation of damaged proteins and, during seed maturation, they may act as a hydration buffer.13 The second group of induced genes includes regulatory molecules, such as protein kinases, involved in signal transduction cascades.12
Ca+2-dependent protein kinases (CDPKs) and sucrose non-fermentation 1 (SNF1)-related kinases (SnRKs) are activated by ABA-dependent and ABA-independent signaling pathways and regulate stress responsive gene expression, including ABA-responsive transcription factors and LEA genes.14–22 Recently, members of the wheat SnRK2 family were implicated in the regulation of enhanced abiotic stress tolerance and growth under normal and stress conditions. Additionally, overexpression of those genes favored root development, benefited water and nutrient uptake and did not retard the growth of transgenic plants, indicating their strong potential in transgenic breeding assays for the improvement of abiotic stress tolerance in crops.23,24
The induction of mitogen-activated protein kinase (MAPK) cascades transduce stress stimuli into cellular responses. MAPK cascades are widespread in eukaryotes and the typical module generally consists of a MAPK kinase kinase (MAPKKK), a MAPK kinase (MAPKK) and a MAPK. For example, a MAPK module (consisting of MEKK1 (MAPKKK), MEK1 (MAPKK) and MPK4/MPK6 (MAPK)) transduces abiotic stress signals to confer tolerance in Arabidopsis by altering the expression of 152 genes that have diverse functions in signal transduction, cellular defense and stress metabolism.25–27 Furthermore, overexpression of the Arabidopsis MPK3 and rice MAPK5 resulted in enhanced biotic and abiotic stress resistance.28,29 Although these cascades are often portrayed as a linear hierarchy, the multiplicity of molecular components at each level within the cascade offers the possibility of crosstalk between pathways and may allow integration and optimization of the organism's response to diverse stimuli. Indeed, the Ca+2-dependent protein kinase CPK3 was recently proposed to mediate stress response at the post translational level through the regulation of membrane-localized target proteins, following the immediate MAPK-dependent transcriptional acclimation to salt stress.30
More recently, a member of the NIMA (never in mitosis A)-related kinases (NEKs) was implicated to osmotic stress response and regulation of plant growth. Animal and fungi NEKs were reported to regulate cell cycle progression.31,32 On the other hand, the function of their plant counterparts was restricted to developmental processes like morphogenesis of epidermal cells, flowering, fruit and shoot development and vascular tissue formation.33–37 Nevertheless, the Arabidopsis AtNEK6 was shown to associate with the novel Absisic Acid (ABA)-signaling component ARIA (Arm repeat protein interacting with ABF), a protein that interacts with ABF-transcription factors and regulates ABA-dependent stress responsive gene expression.38–40 The authors suggested that AtNEK6 participates in ABA and salt stress response during germination, indicating that protein kinases with diverse functions are involved in the plant stress response mechanism (Fig. 2).
Figure 2.
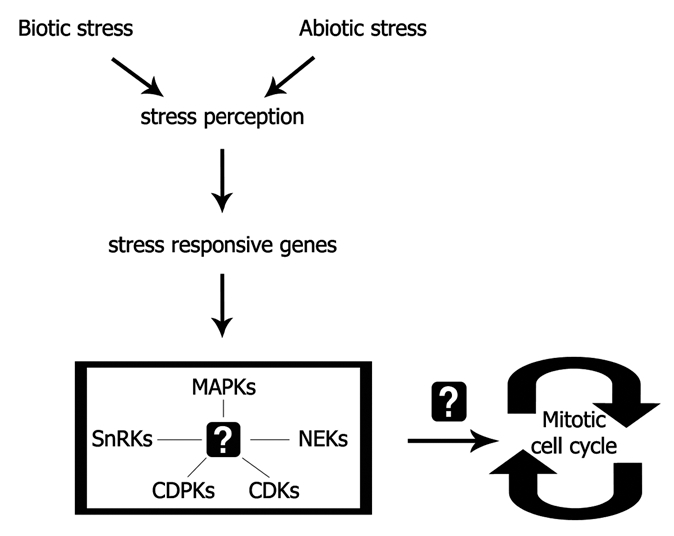
Participation of protein kinases in the plant stress-defense machinery. Crosstalk regulatory pathways among the different kinase modules and integration of their function into cell cycle progression remain to be elucidated.
Several protein kinases involved in stress tolerance are stimulated by ABA, a hormone with a well-documented role in stress response.41 ABA biosynthesis is triggered by stress and in turn activates a variety of stress-inducible genes corresponding to different types of stress. Additionally, ABA may link abiotic stress response with plant development since members of the Arabidopsis SnRK2 family are strongly activated by ABA and regulate growth and reproduction.42,43
Introduction to the CDK Protein Kinase Family
CDKs are a large family of serine/threonine protein kinases with an important role in ensuring that cells progress in an orderly fashion over the different stages of cell division.44,45 Almost all CDKs require binding with a cyclin for their activity. Cyclins, so-called due to their cyclical appearance and disappearance during the cell cycle,46 function as positive regulatory subunits while the active site of the kinase complex is on the CDK component. CDKs were genetically defined in yeasts as cell division cycle (cdc) mutants, cdc2 for S. pombe and CDC28 for S. cerevisiae.47–50 Although the prototypical CDK1/A has a primary function as a central cell cycle regulator, the other groups have been implicated in additional diverse cellular processes like regulation of transcription, translation and mRNA processing.51,52
Higher eukaryotes possess a number of cdc2/CDC28-related genes,53 now known as CDKs, with the different classes designated by numbers (animals) or letters and numbers (plants). Thus, in Arabidopsis, seven different classes of CDKs named CDKA through CDKF have been described based on their cyclin binding motif. In addition, there is a class of CDK-like proteins designated as CKLs.54 The CDKB family is unique to plants and contains 2 sub-classes, B1 and B2. Members of the B1 group are expressed from the onset of S phage until mitosis while those belonging to the B2 sub-class are expressed during G2 to M transition. Additionally, the members of this CDK family seem to control the balance between mitotically dividing and endoreduplicating cells.55–59 The C-class contains two genes, CDKC1 and CDKC2, that are expressed constantly during the cell cycle.54 The plant CDKC-CycT complex was reported to phosphorylate the C-terminal domain (CTD) of RNA Polymerase II (RNAP II), suggesting a role similar to the human CDK9-CycT complex of the P-TEFb transcription factor.60,61 Recently, CDKC function has been implicated in the regulation of mRNA processing in the nucleus.62 CDKD genes belong to the family of CDK-activating kinases (CAKs). One role is to regulate activation of CDKs through phosphorylation of a conserved residue within the T-loop (Thr161 in CDKA1). There are four potential CAK orthologs in Arabidopsis. The three members of the D-class are closely related to the human CDK-activating kinase CDK7 and the fourth potential CDKD gene has been proposed to define a distinct class, designated as CDKF1.63,64 The G-class of Arabidopsis CDKs is closely related to the human galactosyltransferase-associated protein kinase p58/GTA, an isoform of human CDK11 with a characteristic PITSLRE motif,54,65 suggesting a role in regulation of transcription and mRNA processing.52 Finally, the CDK-like proteins (CKLs) encompass 15 members (CKL1-15) that are more closely related to each other than to any of the other cdc2-related classes and their function is still unknown.54 Different members of the CDK have, therefore, diverse roles in cell cycle progression and in regulating gene expression. As regulators of these essential processes, they are good candidates as targets of stress responsive signalling cascades. In the next section we discuss the evidence for this.
Abiotic Stress Negatively Affects CDK Function
Stress negatively affects plant growth—plants grown under stressful conditions are almost always smaller than they would be under favorable conditions. Often the reduction in plant size can be attributed to a reduction in cell number, although in many cases, cell growth is also affected. Post proliferative cell enlargement is modulated in response to stress by the plant growth hormone gibberellin.66 Reduction in cell number has been attributed to inhibition of CDK activity, resulting in cell cycle arrest at the G1/S and G2/M checkpoints, prolonged S-phase progression or delayed entry into mitosis.67 In parsley cell suspensions, the expression of mitotic CDK and cyclin genes was suppressed following treatment with both UV irradiation and fungal elicitors.68 Although the perception mechanisms differ, the end result is very similar.
In the following section, we discuss three types of abiotic stress with agronomic importance to the farmers; heat stress, drought stress and salt stress and the effect of such stresses on CDK activity.
Heat stress.
Transient exposure to elevated temperatures can disturb reproductive development in many crop species leading to reduced yields. Prolonged exposure can lead to irreversible damage, mainly due to increased tissue temperature and excessive evaporation demands.69 At the molecular level, heat stress was reported to affect the organization of microtubules, cell division and eventually results to the production of reactive oxygen species.69
Under heat stress, proliferating cells generally accumulate at G1/S and G2/M, as it was originally shown in studies with the model protozoa species of Tetrahymena.70 It was also suggested that heat shock could be applied for the synchronization of cell populations.71,72 However, heat stress leads to increased permeability of mitochondrial membranes, resulting in ATP depletion and disruption of phosphorylation-dependent signaling cascades.73 Thus, heat-stressed animal cells arrest at G1/S transition due to inhibition of CDK-mediated phosphorylation and inactivation of the retinoblastoma protein (Rb), which is required for S-phase progression.74 Additionally, heat stress was reported to enhance inhibitory phosphorylation of CDKs by inhibitors and reduce the expression of cyclins, suggesting an alternative regulatory mechanism of CDK function.75–78
In plants, the molecular mechanisms the regulate CDK function in response to heat stress are beginning to emerge. Mild heat stress (30°C) induced a transient cell cycle arrest at G1/S or G2/M in BY2 cell suspensions, depending on the stage at which the stress was applied.79 The expression of CycA and CycB and, as a consequence, the activity of their corresponding CDKs was transiently inhibited. On the other hand, application of very high temperature (55°C) activated a signaling pathway that resulted in programmed cell death.80,81 The EL2 protein from rice (Orysa; E2L) may couple cell cycle progression to cold stress response pathway. Orysa; E2L belongs to a plant specific family of CDK inhibitors that inhibit the activity of CDKA1 during G1/S transition, both directly, and indirectly through binding to CycD.82 This gene was transcriptionally induced by low temperatures but remained constant under heat stress. Similarly, cold stress was reported to induce the rice transcription factor OsMYB3R-2 that resulted to higher transcript levels of several G2/M phase specific genes, including OsCycB1;1. Additionally, plants overexpressing OsCycB1;1 displayed enhanced resistance to cold stress, suggesting that this gene is involved in the regulation of cell cycle progression under chilling conditions.83 Such findings indicate that response to different temperature-related stresses might involve different molecular components.
Drought stress.
Increasing temperatures reduces plant water content by increasing the rate of respiration, and can have a negative impact on plant development and productivity. Under water deficit, plants experience drought stress that stimulates a series of physiological and biochemical responses including stomatal closure, inhibition of cell growth, repression of photosynthesis and activation of respiration.12 At the cellular level, plants deploy a mechanism of water stress response and adaptation that involves the expression and/or repression of genes with various functions including different types of transcription factors, protein kinases, phosphatases, metabolic enzymes and cis-acting elements.84–86
Drought-stressed pea leaves displayed reduced rates of cell division and cell expansion, depending on the developmental stage of the leaves.87 Drought-stressed maize roots displayed longer (larger) meristematic cells suggesting that entry into mitosis is delayed, while the rate and zone of cell division were reduced at the root meristem.88 The decrease of mitotic activity following imposition of drought stress has been reported in root meristems.89,90 The link with reduced CDK activity was provided by studies on wheat leaves where a reduction in mesophyll cell division was correlated with inhibition of CDKA1 activity. CDKA was phosphorylated on Tyr-15, a post-translational modification known to reduce kinase activity. Additionally, cyclin expression was also reduced and cells accumulated in G1 or G2.91 Similarly, mild water deficit reduced both the cell division rate and CDKA1 activity almost by half in maize leaves. However, the amount of CDKA1 protein remained unaffected implying again that post-translational regulation of CDKA1 was responsible.92 On the other hand, water stress affected maize CDKA1 activity at the transcriptional and post transcriptional level and resulted to inhibition of endosperm development and, to a less extend, of the endoreduplication cycle.93 It was suggested that early endosperm development, during which mitosis predominates, is more sensitive to stress compared to later stages where cells are committed to repeated rounds of S-phase, in the absence of mitosis that results to DNA replication.
Salt stress.
Salt contamination of the soil is another major agricultural problem that takes much land out of cultivation and marginalizes even more. This is a particular problem in hot areas with insufficient rain or poor drainage and currently affects almost 20% of the cultivated and nearly half of the irrigated land worldwide.94 Excess soluble salts disturb the natural process of water uptake by plants and elevated salt concentrations in the soil can draw out water from plant cells resulting to wilting or even premature death. Some plants known as halophytes can flourish in salt concentrations higher than 400 mM.95,96
In general, high salinity causes ion toxicity, nutritional disorders, water deficit and oxidative stress to plants.94,97 Additionally, it can adversely affects all aspects of plant development and limits productivity of crop species, disturbing cell cycle progression and differentiation.98–101 More specifically, salt stress decreased the transcript levels of CDKA/CDKB and CycA/CycB resulting in transient downregulation of mitotic activity in the shoot and root apex of Arabidopsis plants and cell cycle arrest. As a result, fewer cells of smaller size were produced resulting to a smaller meristem and limited growth.102,103 However, CDKA and CDKB proteins levels were minimally affected, suggesting post-translational level regulation. The period of reduced proliferative activity was considered adaptive, since the promoter activities and transcripts of the cell cycle genes eventually returned to their original levels.
Participation of Non Cell Cycle CDKs in the Plant Stress Response
CDKs with non-cell cycle functions are also involved in the stress response. Orysa;CDKC1 was proposed to be involved in the plant salt stress response mechanism through an ABA-signaling pathway. The transcript level of the rice gene Orysa;CDKC1 was increased by ABA treatment, unlike the levels of mitotic CDKB2;1 transcript that were considerably decreased.104 Furthermore, elevated NaCl also triggered increased expression of Orysa; CDKC1 in cell suspensions, while the activity of Orysa;CDKB2;1 was transiently reduced but recovered. The authors concluded that Orysa;CDKC1 expression might be a primary response to salt stress and could participate in the transcriptional regulation of salt-induced genes for the induction of defense and/or tolerance in a saline environment.104 Such a regulatory role is certainly possible as the Arabidopsis CDKC gene has been implicated in the regulation of mRNA processing. CDKC co-localizes with splicing factor SRp34 and their spatial organization was dependent on CDKC kinase activity.62 Reversible phosphorylation is important for the functional distribution and dynamics of plant SR-splicing factors105,106 so it is likely that CDKC is an important regulator. Abiotic stress modulates the splicing profiles of SR-splicing factors resulting in the production of different isoforms with (possibly) distinct functions. Stress may affect their phosphorylation status and result to altered ratios of their splicing variants and other pre-mRNAs that participate in the acquisition of stress tolerance.107,108 Indeed, non-functional forms of stress-inducible genes were accumulated in plant cells under non-stress conditions, while alternatively spliced functional forms of the same genes were found under abiotic stress.109,110 All the above suggest that CDKC contributes to plant stress tolerance by regulating the phosphorylation of SR-splicing factors, which in turn triggers the alternative splicing of their pre-mRNAs and of stress-related genes, resulting to the induction of stress response.
Conclusion
Plants have developed highly sophisticated stress tolerance mechanisms to better adapt to a variable environment, from which they cannot escape. Such mechanisms involve complex signaling cascades that facilitate the perception of stress signals and the activation of hundreds of stress-related genes and transcription factors.111–113 A common cellular response upon stress application is transient inhibition of cell cycle progression or even cell cycle exit. The signaling pathways that integrate abiotic stress with cell division control remains to be fully elucidated, but evidence suggests that post-transcriptional modulation of CDK kinase activity is important for transient inhibition. Other kinases including CDKs with functions other than cell cycle control also are crucial in the stress response. Such findings suggest novel intrinsic functions for CDKs in the regulation of plant response upon biotic and abiotic stress.
References
- 1.Vranova E, Inze D, Van Breusegem F. Signal transduction during oxidative stress. J Exp Bot. 2002;53:1227–1236. [PubMed] [Google Scholar]
- 2.Vinebrooke Rolf D, Cottingham Kathryn L, Norberg Jon, et al. Impacts of multiple stressors on biodiversity and ecosystem functioning: the role of species co-tolerance. Oxford, ROYAUME-UNI: Blackwell; 2004. [Google Scholar]
- 3.Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Arch Biochem Biophys. 2005;444:139–158. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 4.Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr Opin Biotechnol. 2006;17:113–122. doi: 10.1016/j.copbio.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, et al. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- 6.Ma S, Bohnert HJ. Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures and cell-specific expression. Genome Biology. 2007;8:49. doi: 10.1186/gb-2007-8-4-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weston DJ, Gunter LE, Rogers A, Wullschleger SD. Connecting genes, coexpression modules and molecular signatures to environmental stress phenotypes in plants. BMC Systems Biology. 2008;2:16. doi: 10.1186/1752-0509-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yata K, Esashi F. Dual role of CDKs in DNA repair: to be, or not to be. DNA repair. 2009;8:6–18. doi: 10.1016/j.dnarep.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Huang D, Moffat J, Andrews B. Dissection of a complex phenotype by functional genomics reveals roles for the yeast cyclin-dependent protein kinase Pho85 in stress adaptation and cell integrity. Mol Cell Biol. 2002;22:5076–5088. doi: 10.1128/MCB.22.14.5076-5088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishizawa M, Katou Y, Shirahige K, Toh-e A. Yeast Pho85 kinase is required for proper gene expression during the diauxic shift. Yeast (Chichester, England) 2004;21:903–918. doi: 10.1002/yea.1138. [DOI] [PubMed] [Google Scholar]
- 11.Nishizawa M, Tanigawa M, Hayashi M, Maeda T, Yazaki Y, Saeki Y, et al. Pho85 kinase, a cyclin-dependent kinase, regulates nuclear accumulation of the Rim101 transcription factor in the stress response of Saccharomyces cerevisiae. Eukaryotic Cell. 2010;9:943–951. doi: 10.1128/EC.00247-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- 13.Wise MJ, Tunnacliffe A. POPP the question: what do LEA proteins do? Trends Plant Sci. 2004;9:13–17. doi: 10.1016/j.tplants.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K. Overexpression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000;23:319–327. doi: 10.1046/j.1365-313x.2000.00787.x. [DOI] [PubMed] [Google Scholar]
- 15.Boudsocq M, Barbier-Brygoo H, Lauriere C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J Biol Chem. 2004;279:41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell. 2004;16:1163–1177. doi: 10.1105/tpc.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umezawa T, Yoshida R, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2004;101:17306–17311. doi: 10.1073/pnas.0407758101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi HI, Park HJ, Park JH, Kim S, Im MY, Seo HH, et al. Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression and modulates its activity. Plant Physiol. 2005;139:1750–1761. doi: 10.1104/pp.105.069757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, et al. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005;44:939–949. doi: 10.1111/j.1365-313X.2005.02583.x. [DOI] [PubMed] [Google Scholar]
- 20.Boudsocq M, Droillard MJ, Barbier-Brygoo H, Lauriere C. Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol Biol. 2007;63:491–503. doi: 10.1007/s11103-006-9103-1. [DOI] [PubMed] [Google Scholar]
- 21.Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, et al. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell. 2007;19:3019–3036. doi: 10.1105/tpc.107.050666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diedhiou CJ, Popova OV, Dietz KJ, Golldack D. The SNF1-type serine-threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice. BMC plant biology. 2008;8:49. doi: 10.1186/1471-2229-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao X, Zhang H, Tian S, Chang X, Jing R. TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis. J Exp Bot. 2010;61:683–696. doi: 10.1093/jxb/erp331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Mao X, Jing R, Chang X, Xie H. Characterization of a common wheat (Triticum aestivum L.) TaSnRK2.7 gene involved in abiotic stress responses. J Exp Bot. 2010 doi: 10.1093/jxb/erq328. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ichimura K, Mizoguchi T, Irie K, Morris P, Giraudat J, Matsumoto K, et al. Isolation of ATMEKK1 (a MAP kinase kinase kinase)-interacting proteins and analysis of a MAP kinase cascade in Arabidopsis. Biochem Biophys Res Commun. 1998;253:532–543. doi: 10.1006/bbrc.1998.9796. [DOI] [PubMed] [Google Scholar]
- 26.Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K. Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 2000;24:655–665. doi: 10.1046/j.1365-313x.2000.00913.x. [DOI] [PubMed] [Google Scholar]
- 27.Teige M, Scheikl E, Eulgem T, Doczi R, Ichimura K, Shinozaki K, et al. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Molecular cell. 2004;15:141–152. doi: 10.1016/j.molcel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 28.Kovtun Y, Chiu WL, Tena G, Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA. 2000;97:2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong L, Yang Y. Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell. 2003;15:745–759. doi: 10.1105/tpc.008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehlmer N, Wurzinger B, Stael S, Hofmann-Rodrigues D, Csaszar E, Pfister B, et al. The Ca(2+)-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J. 2010 doi: 10.1111/j.1365-313X.2010.04257.x. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osmani SA, Ye XS. Cell cycle regulation in Aspergillus by two protein kinases. Biochem J. 1996;317:633–641. doi: 10.1042/bj3170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayward DG, Fry AM. Nek2 kinase in chromosome instability and cancer. Cancer Letters. 2006;237:155–166. doi: 10.1016/j.canlet.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 33.Pnueli L, Gutfinger T, Hareven D, Ben-Naim O, Ron N, Adir N, et al. Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell. 2001;13:2687–2702. doi: 10.1105/tpc.010293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cloutier M, Vigneault F, Lachance D, Seguin A. Characterization of a poplar NIMA-related kinase PNek1 and its potential role in meristematic activity. FEBS Lett. 2005;579:4659–4665. doi: 10.1016/j.febslet.2005.07.036. [DOI] [PubMed] [Google Scholar]
- 35.Vigneault F, Lachance D, Cloutier M, Pelletier G, Levasseur C, Seguin A. Members of the plant NIMA-related kinases are involved in organ development and vascularization in poplar, Arabidopsis and rice. Plant J. 2007;51:575–588. doi: 10.1111/j.1365-313x.2007.03161.x. [DOI] [PubMed] [Google Scholar]
- 36.Motose H, Tominaga R, Wada T, Sugiyama M, Watanabe Y. A NIMA-related protein kinase suppresses ectopic outgrowth of epidermal cells through its kinase activity and the association with microtubules. Plant J. 2008;54:829–844. doi: 10.1111/j.1365-313X.2008.03445.x. [DOI] [PubMed] [Google Scholar]
- 37.Sakai T, Honing H, Nishioka M, Uehara Y, Takahashi M, Fujisawa N, et al. Armadillo repeat-containing kinesins and a NIMA-related kinase are required for epidermal-cell morphogenesis in Arabidopsis. Plant J. 2008;53:157–171. doi: 10.1111/j.1365-313X.2007.03327.x. [DOI] [PubMed] [Google Scholar]
- 38.Kim S, Choi HI, Ryu HJ, Park JH, Kim MD, Kim SY. ARIA, an Arabidopsis arm repeat protein interacting with a transcriptional regulator of abscisic acid-responsive gene expression, is a novel abscisic acid signaling component. Plant Physiol. 2004;136:3639–3648. doi: 10.1104/pp.104.049189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim SY. The role of ABF family bZIP class transcription factors in stress response. Physiologia Plantarum. 2006;126:519–527. [Google Scholar]
- 40.Lee SJ, Cho DI, Kang JY, Kim MD, Kim SY. AtNEK6 interacts with ARIA and is involved in ABA response during seed germination. Mol Cells. 2010;29:559–566. doi: 10.1007/s10059-010-0070-7. [DOI] [PubMed] [Google Scholar]
- 41.Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, et al. An update on abscisic acid signaling in plants and more. Mol Plant. 2008;1:198–217. doi: 10.1093/mp/ssm022. [DOI] [PubMed] [Google Scholar]
- 42.Fujii H, Zhu JK. Arabidopsis mutant deficient in 3-abscisic acid-activated protein kinases reveals critical roles in growth, reproduction and stress. Proc Natl Acad Sci USA. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50:1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- 44.John PC, Mews M, Moore R. Cyclin/Cdk complexes: their involvement in cell cycle progression and mitotic division. Protoplasma. 2001;216:119–142. doi: 10.1007/BF02673865. [DOI] [PubMed] [Google Scholar]
- 45.Francis D. The plant cell cycle—15 years on. New Phytol. 2007;174:261–278. doi: 10.1111/j.1469-8137.2007.02038.x. [DOI] [PubMed] [Google Scholar]
- 46.Evans T, Rosenthal ET, Youngblom J, Distel D, Hunt T. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983;33:389–396. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- 47.Hartwell LH, Culotti J, Pringle JR, Reid BJ. Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- 48.Nasmyth KA, Reed SI. Isolation of genes by complementation in yeast: molecular cloning of a cell cycle gene. Proc Natl Acad Sci USA. 1980;77:2119–2123. doi: 10.1073/pnas.77.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nurse P, Thuriaux P. Regulatory genes controlling mitosis in the fission yeast Schizosaccharomyces pombe. Genetics. 1980;96:627–637. doi: 10.1093/genetics/96.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hindley J, Phear GA. Sequence of the cell division gene CDC2 from Schizosaccharomyces pombe; patterns of splicing and homology to protein kinases. Gene. 1984;31:129–134. doi: 10.1016/0378-1119(84)90203-8. [DOI] [PubMed] [Google Scholar]
- 51.Joubes J, Chevalier C, Dudits D, Heberle-Bors E, Inze D, Umeda M, et al. CDK-related protein kinases in plants. Plant Mol Biol. 2000;43:607–620. doi: 10.1023/a:1006470301554. [DOI] [PubMed] [Google Scholar]
- 52.Doonan JH, Kitsios G. Functional evolution of cyclin-dependent kinases. Mol Biotechnol. 2009;42:14–29. doi: 10.1007/s12033-008-9126-8. [DOI] [PubMed] [Google Scholar]
- 53.Morgan DO. Cyclin-dependent kinases: engines, clocks and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 54.Menges M, de Jager SM, Gruissem W, Murray JA. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 2005;41:546–566. doi: 10.1111/j.1365-313X.2004.02319.x. [DOI] [PubMed] [Google Scholar]
- 55.Fobert PR, Gaudin V, Lunness P, Coen ES, Doonan JH. Distinct classes of cdc2-related genes are differentially expressed during the cell division cycle in plants. Plant Cell. 1996;8:1465–1476. doi: 10.1105/tpc.8.9.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Segers G, Gadisseur I, Bergounioux C, de Almeida Engler J, Jacqmard A, Van Montagu M, et al. The Arabidopsis cyclin-dependent kinase gene cdc2bAt is preferentially expressed during S and G2 phases of the cell cycle. Plant J. 1996;10:601–612. doi: 10.1046/j.1365-313x.1996.10040601.x. [DOI] [PubMed] [Google Scholar]
- 57.Porceddu A, Stals H, Reichheld JP, Segers G, De Veylder L, Barroco RP, et al. A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. J Biol Chem. 2001;276:36354–36360. doi: 10.1074/jbc.M011060200. [DOI] [PubMed] [Google Scholar]
- 58.Boudolf V, Vlieghe K, Beemster GT, Magyar Z, Torres Acosta JA, Maes S, et al. The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell. 2004;16:2683–2692. doi: 10.1105/tpc.104.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inze D, De Veylder L. Cell cycle regulation in plant development. Annu Rev Genet. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- 60.Barroco RM, De Veylder L, Magyar Z, Engler G, Inze D, Mironov V. Novel complexes of cyclin-dependent kinases and a cyclin-like protein from Arabidopsis thaliana with a function unrelated to cell division. Cell Mol Life Sci. 2003;60:401–412. doi: 10.1007/s000180300033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fulop K, Pettko-Szandtner A, Magyar Z, Miskolczi P, Kondorosi E, Dudits D, et al. The Medicago CDKC;1-CYCLINT;1 kinase complex phosphorylates the carboxy-terminal domain of RNA polymerase II and promotes transcription. Plant J. 2005;42:810–820. doi: 10.1111/j.1365-313X.2005.02421.x. [DOI] [PubMed] [Google Scholar]
- 62.Kitsios G, Alexiou KG, Bush M, Shaw P, Doonan JH. A cyclin-dependent protein kinase, CDKC2, colocalizes with and modulates the distribution of spliceosomal components in Arabidopsis. Plant J. 2008;54:220–235. doi: 10.1111/j.1365-313X.2008.03414.x. [DOI] [PubMed] [Google Scholar]
- 63.Shimotohno A, Matsubayashi S, Yamaguchi M, Uchimiya H, Umeda M. Differential phosphorylation activities of CDK-activating kinases in Arabidopsis thaliana. FEBS Lett. 2003;534:69–74. doi: 10.1016/s0014-5793(02)03780-8. [DOI] [PubMed] [Google Scholar]
- 64.Shimotohno A, Umeda-Hara C, Bisova K, Uchimiya H, Umeda M. The plant-specific kinase CDKF;1 is involved in activating phosphorylation of cyclin-dependent kinase-activating kinases in Arabidopsis. Plant Cell. 2004;16:2954–2966. doi: 10.1105/tpc.104.025601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi J, Feng Y, Goulet AC, Vaillancourt RR, Sachs NA, Hershey JW, et al. The p34cdc2-related cyclin-dependent kinase 11 interacts with the p47 subunit of eukaryotic initiation factor 3 during apoptosis. J Biol Chem. 2003;278:5062–5071. doi: 10.1074/jbc.M206427200. [DOI] [PubMed] [Google Scholar]
- 66.Razem FA, Baron K, Hill RD. Turning on gibberellin and abscisic acid signaling. Curr Opin Plant Biol. 2006;9:454–459. doi: 10.1016/j.pbi.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 67.De Veylder L, Beeckman T, Inze D. The ins and outs of the plant cell cycle. Nat Rev Mol Cell Biol. 2007;8:655–665. doi: 10.1038/nrm2227. [DOI] [PubMed] [Google Scholar]
- 68.Logemann E, Wu SC, Schroder J, Schmelzer E, Somssich IE, Hahlbrock K. Gene activation by UV light, fungal elicitor or fungal infection in Petroselinum crispum is correlated with repression of cell cycle-related genes. Plant J. 1995;8:865–876. doi: 10.1046/j.1365-313x.1995.8060865.x. [DOI] [PubMed] [Google Scholar]
- 69.Wahid A, Gelani S, Ashraf M, Foolad MR. Heat tolerance in plants: An overview. Environ Exp Bot. 2007;61:199–223. [Google Scholar]
- 70.Kramhoft B, Zeuthen E. Synchronization of cell divisions in the fission yeast, Schizosaccharomyces pombe, using heat shocks. Comptes-rendus des travaux du Laboratoire Carlsberg. 1971;38:351–368. (Fre). [PubMed] [Google Scholar]
- 71.Zeuthen E. Synchronization of the Tetrahymena cell cycle. Adv Cell Biol. 1971;2:111–152. doi: 10.1007/978-1-4615-9588-5_3. [DOI] [PubMed] [Google Scholar]
- 72.Read RA, Fox MH, Bedford JS. The cell cycle dependence of thermotolerance. II. CHO cells heated at 45.0 degrees C. Radiation Research. 1984;98:491–505. [PubMed] [Google Scholar]
- 73.Kuhl NM, Rensing L. Heat shock effects on cell cycle progression. Cell Mol Life Sci. 2000;57:450–463. doi: 10.1007/PL00000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khandjian EW. Heat treatment induces dephosphorylation of pRb and dissociation of T-antigen/pRb complex during transforming infection with SV40. Oncogene. 1995;10:359–367. [PubMed] [Google Scholar]
- 75.Rowley A, Johnston GC, Butler B, Werner-Washburne M, Singer RA. Heat shock-mediated cell cycle blockage and G1 cyclin expression in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:1034–1041. doi: 10.1128/mcb.13.2.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fuse T, Yamada K, Asai K, Kato T, Nakanishi M. Heat shock-mediated cell cycle arrest is accompanied by induction of p21 CKI. Biochem Biophys Res Commun. 1996;225:759–763. doi: 10.1006/bbrc.1996.1247. [DOI] [PubMed] [Google Scholar]
- 77.Schnier JB, Nishi K, Goodrich DW, Bradbury EM. G1 arrest and downregulation of cyclin E/cyclin-dependent kinase 2 by the protein kinase inhibitor staurosporine are dependent on the retinoblastoma protein in the bladder carcinoma cell line 5637. Proc Natl Acad Sci USA. 1996;93:5941–5946. doi: 10.1073/pnas.93.12.5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nitta M, Okamura H, Aizawa S, Yamaizumi M. Heat shock induces transient p53-dependent cell cycle arrest at G1/S. Oncogene. 1997;15:561–568. doi: 10.1038/sj.onc.1201210. [DOI] [PubMed] [Google Scholar]
- 79.Jang SJ, Shin SH, Yee ST, Hwang B, Im KH, Park KY. Effects of abiotic stresses on cell cycle progression in tobacco BY-2 cells. Mol Cells. 2005;20:136–141. [PubMed] [Google Scholar]
- 80.Larkindale J, Knight MR. Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene and salicylic acid. Plant Physiol. 2002;128:682–695. doi: 10.1104/pp.010320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vacca RA, de Pinto MC, Valenti D, Passarella S, Marra E, De Gara L. Production of reactive oxygen species, alteration of cytosolic ascorbate peroxidase and impairment of mitochondrial metabolism are early events in heat shock-induced programmed cell death in tobacco Bright-Yellow 2 cells. Plant Physiol. 2004;134:1100–1112. doi: 10.1104/pp.103.035956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peres A, Churchman ML, Hariharan S, Himanen K, Verkest A, Vandepoele K, et al. Novel plant-specific cyclin-dependent kinase inhibitors induced by biotic and abiotic stresses. J Biol Chem. 2007;282:25588–25596. doi: 10.1074/jbc.M703326200. [DOI] [PubMed] [Google Scholar]
- 83.Ma Q, Dai X, Xu Y, Guo J, Liu Y, Chen N, et al. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 2009;150:244–256. doi: 10.1104/pp.108.133454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- 85.Bartels D, Sunkar R. Drought and Salt Tolerance in Plants. Crit Rev Plant Sci. 2005;24:23–58. [Google Scholar]
- 86.Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 87.Lecoeur Jrm, Wery J, Turc O, Tardieu Fo. Expansion of pea leaves subjected to short water deficit: cell number and cell size are sensitive to stress at different periods of leaf development. J Exp Bot. 1995;46:1093–1101. [Google Scholar]
- 88.Sacks MM, Silk WK, Burman P. Effect of Water Stress on Cortical Cell Division Rates within the Apical Meristem of Primary Roots of Maize. Plant Physiol. 1997;114:519–527. doi: 10.1104/pp.114.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Robertson JM, Yeung EC, Reid DM, Hubick KT. Developmental Responses to Drought and Abscisic Acid in Sunflower Roots. J Exp Bot. 1990;41:339–350. [Google Scholar]
- 90.Bracale M, Levi M, Savini C, Dicorato W, Galli MG. Water Deficit in Pea Root Tips: Effects on the Cell Cycle and on the Production of Dehydrin-like Proteins. Ann Bot. 1997;79:593–600. [Google Scholar]
- 91.Schuppler U, He PH, John PC, Munns R. Effect of water stress on cell division and cell-division-cycle 2-like cell cycle kinase activity in wheat leaves. Plant Physiol. 1998;117:667–678. doi: 10.1104/pp.117.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Granier C, Inze D, Tardieu F. Spatial distribution of cell division rate can be deduced from that of p34(cdc2) kinase activity in maize leaves grown at contrasting temperatures and soil water conditions. Plant Physiol. 2000;124:1393–1402. doi: 10.1104/pp.124.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Setter TL, Flannigan BA. Water deficit inhibits cell division and expression of transcripts involved in cell proliferation and endoreduplication in maize endosperm. J Exp Bot. 2001;52:1401–1408. doi: 10.1093/jexbot/52.360.1401. [DOI] [PubMed] [Google Scholar]
- 94.Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought and salt stress. Plant Cell. 2002;14:165–183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hammel HT, Schlegel WM. Osmosis and solute-solvent drag: fluid transport and fluid exchange in animals and plants. Cell Biochem Biophys. 2005;42:277–345. doi: 10.1385/CBB:42:3:277. [DOI] [PubMed] [Google Scholar]
- 96.Flowers TJ, Colmer TD. Salinity tolerance in halophytes. New Phytol. 2008;179:945–963. doi: 10.1111/j.1469-8137.2008.02531.x. [DOI] [PubMed] [Google Scholar]
- 97.Sun J, Chen SL, Dai SX, Wang RG, Li NY, Shen X, et al. Ion flux profiles and plant ion homeostasis control under salt stress. Plant Signal Behav. 2009;4:261–264. doi: 10.4161/psb.4.4.7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Munns R, Rawson HM. Effect of salinity on salt accumulation and reproductive development in the apical meristem of wheat and barley. Functional Plant Biology. 1999;26:459–464. [Google Scholar]
- 99.Zeng L, Shannon MC. Salinity Effects on Seedling Growth and Yield Components of Rice. Crop Sci. 2000;40:996–1003. [Google Scholar]
- 100.Netondo GW, Onyango JC, Beck E. Sorghum and Salinity. Crop Sci. 2004;44:806–811. [Google Scholar]
- 101.Sun K, Hunt K, Hauser BA. Ovule abortion in Arabidopsis triggered by stress. Plant Physiol. 2004;135:2358–2367. doi: 10.1104/pp.104.043091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Burssens S, Himanen K, van de Cotte B, Beeckman T, Van Montagu M, Inze D, et al. Expression of cell cycle regulatory genes and morphological alterations in response to salt stress in Arabidopsis thaliana. Planta. 2000;211:632–640. doi: 10.1007/s004250000334. [DOI] [PubMed] [Google Scholar]
- 103.West G, Inze D, Beemster GT. Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiol. 2004;135:1050–1058. doi: 10.1104/pp.104.040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huang YW, Tsay WS, Chen CC, Lin CW, Huang HJ. Increased expression of the rice C-type cyclin-dependent protein kinase gene, Orysa;CDKC;1, in response to salt stress. Plant Physiol Biochem. 2008;46:71–81. doi: 10.1016/j.plaphy.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 105.Tillemans V, Dispa L, Remacle C, Collinge M, Motte P. Functional distribution and dynamics of Arabidopsis SR splicing factors in living plant cells. Plant J. 2005;41:567–582. doi: 10.1111/j.1365-313X.2004.02321.x. [DOI] [PubMed] [Google Scholar]
- 106.Tillemans V, Leponce I, Rausin G, Dispa L, Motte P. Insights into nuclear organization in plants as revealed by the dynamic distribution of Arabidopsis SR splicing factors. Plant Cell. 2006;18:3218–3234. doi: 10.1105/tpc.106.044529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, et al. Genome-wide analysis of alternative pre-mRNA splicing in. Nucleic Acids Res. 2004;32:5096–5103. doi: 10.1093/nar/gkh845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Palusa SG, Ali GS, Reddy AS. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J. 2007;49:1091–1107. doi: 10.1111/j.1365-313X.2006.03020.x. [DOI] [PubMed] [Google Scholar]
- 109.Qin F, Kakimoto M, Sakuma Y, Maruyama K, Osakabe Y, Tran LS, et al. Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 2007;50:54–69. doi: 10.1111/j.1365-313X.2007.03034.x. [DOI] [PubMed] [Google Scholar]
- 110.Matsukura S, Mizoi J, Yoshida T, Todaka D, Ito Y, Maruyama K, et al. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol Genet Genomics. 2010;283:185–196. doi: 10.1007/s00438-009-0506-y. [DOI] [PubMed] [Google Scholar]
- 111.Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic and cold stress. Plant Physiol. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, et al. Monitoring the expression profiles of 7,000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- 113.Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, et al. Monitoring expression profiles of rice genes under cold, drought and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gelblot analyses. Plant Physiol. 2003;133:1755–1767. doi: 10.1104/pp.103.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]


