Abstract
X-linked sideroblastic anemia with ataxia (XLSA/A) is a rare inherited disorder characterized by mild anemia and ataxia. XLSA/A is caused by mutations in the ABCB7 gene, which encodes a member of the ATP-binding cassette transporter family. Studies in yeast, mammalian cells, and mice have shown that ABCB7 functions in the transport of iron-sulfur (Fe-S) clusters into the cytoplasm. To further investigate the mechanism of this disease, we have identified and characterized the Caenorhabditis elegans homologue of the ABCB7 gene, abtm-1. We have studied the function of abtm-1 using mutants and RNAi. abtm-1-depleted animals produce arrested embryos that have morphogenetic defects and unusual premature, putative apoptotic events. abtm-1(RNAi) animals also show accumulation of ferric iron and increased oxidative stress. Despite the increased level of oxidative stress in abtm-1(RNAi) animals, they have an increased life span. We observed accumulation of DAF-16/FOXO in the nuclei of affected animals and elevation of the expression of SOD-3, a well established target of DAF-16, which may explain the increased life span extension of these animals. abtm-1 is strongly expressed in tissues with a high energy demand, and abtm-1(RNAi) animals have phenotypes that reflect the need for abtm-1 in these tissues. Finally, we show that reducing the function of other genes involved in Fe-S cluster production produces similar phenotypic consequences to abtm-1 loss of function. Therefore, ablation of abtm-1 in C. elegans provides a model in which to investigate the mechanism underlying XLSA/A.
Keywords: ABC Transporter, Apoptosis, Ataxia, C. elegans, Iron Metabolism, Oxidative Stress, RNA Interference (RNAi), Frataxin, abtm-1
Introduction
X-linked sideroblastic anemia with ataxia (XLSA/A2; OMIM 301310) is a rare inherited disorder in which male patients suffer from mild anemia together with a nonprogressive and early onset ataxia characterized by dysmetria and dysdiadochokinesis (1). Other reported symptoms include dysarthria, intention tremor, mild learning disability, and depression. XLSA/A-associated anemia is mild and asymptomatic in males. Female carriers usually do not show anemia or neurological conditions. XLSA/A is caused by mutations in ABCB7, a gene located at position Xql3 (2). ABCB7 encodes a highly conserved protein belonging to the ABCB family of ATP-binding cassette (ABC) transporters (3). Members of the ABC superfamily are transmembrane proteins that use the hydrolysis of ATP to facilitate transport of a range of substrates across membranes.
Analyses of three different families with XLSA/A have identified three associated mutations in the ABCB7 gene. Two of these are missense mutations, which cause the substitution of residues within the ABCB7 transmembrane domains: V411L (4) and I400M (2). The third mutation produces a more substantial amino acid change E433K (5). A human cDNA containing the E433K change is able to partially rescue yeast carrying a mutation in ATM1, the homologue of ABCB7, suggesting that this change does not cause a complete loss of function (5). Thus, no complete loss of function mutations in this gene have been described, suggesting that ABCB7 is an essential molecule. In support of this suggestion, ABCB7 knock-out stem cells, hemizygous mice, and mice with conditional knock-outs in the central nervous system or bone marrow are all inviable (6, 7).
Studies in yeast have shown that Atm1p localizes to the mitochondrial inner membrane, and it has been suggested that this molecule is involved in the transport of iron-sulfur (Fe-S) clusters from the lumen of mitochondria to the cytosol (8, 9). Moreover, work performed with conditional knock-out ABCB7 mice suggests that mammalian ABCB7 transporters are also involved in Fe-S cluster assembly (6). Thus, mammalian (10) and yeast (8) ABCB7/ATM1-deficient cells show mitochondrial iron accumulation presumably because Fe-S clusters cannot be transferred to the cytosol. It is believed that this accumulation of mitochondrial iron causes oxidative stress, probably by the catalytic production of hydroxyl radicals, which then react with other biologically important molecules such as proteins, lipids, or DNA. This mitochondrial stress may then cause cellular dysfunction in the nervous system of patients. In addition, the impairment of iron homeostasis due to a lack of ABCB7 activity, directly or indirectly, disrupts the heme synthesis pathway, because Fe-S cluster assembly is essential for heme production (11), and it is this change that is responsible for the anemia of XLSA/A patients (7). Therefore, XLSA/A is a mitochondrial disease caused by a mutation of a nuclear gene involved in Fe-S cluster biosynthesis.
In this study, we used Caenorhabditis elegans to establish a model for the investigation of the molecular basis of XLSA/A. To do this, we have investigated the effect of depletion of abtm-1/ABCB7 using mutant worms and by RNAi. We report that reduction of abtm-1 function causes embryonic lethality during morphogenesis. We show that there is premature, putative apoptosis in abtm-1(RNAi) embryos that compromises the development of some cell lineages and may thus account for the increased embryonic lethality. abtm-1(RNAi) animals that reach adulthood show evidence of increased oxidative stress and accumulate ferric iron (Fe3+), which may produce free radicals. We have also found that abtm-1(RNAi) worms have a pattern of alterations in life span, defecation, motility, and other behaviors indicative of mitochondrial impairment. We show increased expression of SOD-3 (superoxide dismutase), a well known DAF-16/FOXO target, and nuclear localization of DAF-16/FOXO in abtm-1(RNAi) animals. These adaptations to stress may partially account for the observed life span extension. Analysis of the expression pattern of abtm-1 shows that the gene is strongly expressed in tissues that are likely to have a high demand for ATP. We also show that ablating other components of the Fe-S cluster synthesis pathway recapitulates some of the phenotypes observed in abtm-1-deficient animals.
EXPERIMENTAL PROCEDURES
Isolation of the abtm-1 cDNA
To identify the 5′ end of the cDNA, we used the sequence of the spliced leader, SL1, as a forward primer, and a gene-specific oligonucleotide as the reverse primer. The 3′ end was determined using rapid amplification of 3′-complementary DNA ends. Both products were cloned into pGEM-T (Promega, Madison, WI) and sequenced. We used primers designed using the information obtained from these clones to amplify the full-length cDNA. All products were cloned and sequenced to obtain the whole structure of abtm-1. The coding sequence of the gene does not differ significantly from the predicted spliced gene in the WormBase (release WS181) (NCBI accession number AF490975) (supplemental Fig. S1A).
Worm Culture and Strains
Worms were cultured using standard techniques and media (12). Strains used in this work are listed in supplemental Table S2. We out-crossed abtm-1(tm2721), a gift from S. Mitani, four times before any phenotypic analysis was performed. After outcrossing, abtm-1(tm2721) males were crossed to KR344, which carries the free duplication sDp2. F2 hermaphrodites carrying sDp2 and tm2721 in homozygosis were isolated. All strains were maintained at 20 °C, unless otherwise stated.
Construction of Transgenic Worms
We used a PCR-based fusion approach to make abtm-1::GFP constructs. Both constructs also contain the 3′-UTR from unc-54. To produce the mitochondrial construct, abtm-1::GFP1, we fused a region containing the putative promoter and the first three exons of the abtm-1 gene to GFP (Fig. 1A). The primers used were as follows: forward primer FP1175, 5′ CTC ACG ATT GAA ACG GAC CCG, and reverse primer FP784, 5′ AGT CGA CCT GCA GGC ATG CAA GCT TAC TTG ACT ACT GGC TCC CGG. The peptide produced by this construct contains the putative mitochondrial signal peptide. To obtain the cytoplasmic construct, abtm-1::GFP2 (Fig. 1A), we amplified the region contained between the upstream gene, eif-3, and the first codon of abtm-1 (forward primer FP1178, 5′ CCT ATT TTT GAA GGT TTC TGC G; reverse primer FP814, 5′ AGT CGA CCT GCA GGC ATG CAA GCT CAT TAT CGA TCT CTG AAA ACT GGA TTC GG). Both reverse primers used to make the constructs contain a sequence that overlaps the GFP sequence to allow fusion by PCR. To obtain transgenic animals, we injected 1–2 ng/μl of the test DNA together with 60 ng/μl of pRF4 as a marker, and 60 ng/μl of genomic DNA from wild type worms, digested with XhoI, as a DNA carrier, using methods described before (13).
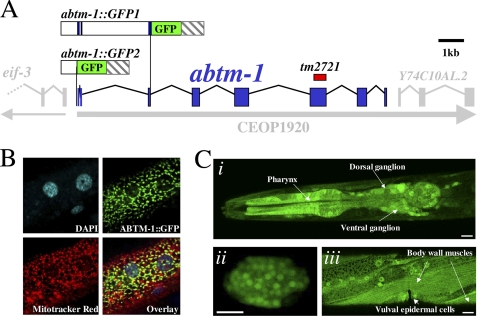
FIGURE 1.
abtm-1 gene is the homologue of human ABCB7. A, genomic structure of abtm-1. The gray arrow below the gene indicates that abtm-1 is contained within an operon, OP1920, which contains another gene, Y74C10AL.2. The red box above the gene shows the position of the deletion in abtm-1(tm2721). This diagram also shows the two GFP translational fusions used in this work. Blue boxes indicate exons of abtm-1. B, confocal microscope images showing co-localization of ABTM-1::GFP with Mitotracker Red, in muscle cells from adult hermaphrodites. C, abtm-1::GFP2 is highly expressed in the pharynx and neurons of the ventral and dorsal ganglions (panel i), developing embryos (panel ii), and body wall muscles and epidermis (panel iii).
Cellular Localization of ABTM-1
Hermaphrodites carrying extrachromosomal arrays containing abtm-1::GFP1 were incubated at 25 °C for an hour in 50 μm MitoTracker® Red CMXRos (Invitrogen) and 1.8 mm 4′,6-diamidino-2-phenylindole (DAPI), diluted in M9 buffer (12). These animals were allowed to recover on fresh NGM plates for an hour at 20 °C. Then worms were anesthetized using 20 mm sodium azide in M9 buffer and mounted for microscopy on 2% agarose pads. Images were acquired using a Leica SP5 confocal microscope (Leica Microsystems GmbH, Wetzlar, Germany).
RNA Interference
RNAi was carried out by injection of dsRNA. dsRNA was synthesized in vitro, using template fragments between 400 and 700 bp, from the appropriate ORF, amplified by PCR using primers described in supplemental Table S3, and cloned into pGEM-T (Promega). dsRNA was made using T7 and SP6 RNA polymerases (Invitrogen) and combining the two single-stranded molecules, as described before (14). As a control, we used dsRNA from the Escherichia coli chloramphenicol acetyltransferase (cat) gene. Several young adult hermaphrodites were injected in the gonads (13). Injected worms were transferred after 18 h and phenotypes analyzed in the post-18-h offspring. Phenotypes were scored as described below.
Analysis of Embryonic Lethality and Larval Arrest
To investigate embryonic lethality in RNAi experiments, we collected embryos from animals after 24 and 38 h following the injection of dsRNA. Then we removed the parents and allowed the embryos to develop fully for 24 h before counting the number of arrested embryos and larvae. To investigate lethality from mutant strains, we did the same, but for the entire fertile period of the adults. We used 10–12 parental animals per strain per experiment. We considered embryos as arrested when they failed to hatch within 24 h. We considered larvae as arrested when they did not progress to adulthood within 48 h of hatching.
To further analyze embryonic development, embryos were isolated by dissection and then mounted in embryo culture medium (15). To determine the terminal phenotype, embryos were left for 18 h at 20 °C. Confocal microscopy was performed using a Leica SP5 confocal microscope. Embryos for four-dimensional microscopy analysis were prepared as described (16). Eight abtm-1(RNAi) and three wild type embryos were mounted, at a very early stage (usually the two-cell stage), and recording of their development was carried out at 25 °C for 10 h at intervals of 60 s, with 25 different focal planes (1 μm separation), using a Zeiss Axioplan microscope (Carl Zeiss STM GmbH, Germany) equipped with Nomarski optics adapted as described (16). Cell lineage of the recorded embryos was subsequently traced using SIMI Biocell (SIMI GmbH, Germany).
Protein Extraction
Worms were collected from plates by washing with M9 buffer and transferred to screw cap tubes. The tubes were shaken gently for 20 min at room temperature to allow digestion of bacteria. Worms were collected by sedimentation and washed three times with M9 buffer. Five volumes of ice-cold lysis buffer (150 mm NaCl, 50 mm Tris-HCl, pH 8, 1% Nonidet P-40, and protease inhibitor mixture) were added to the worm pellets, and the mixture was homogenized using glass homogenizer during a 30-min period to allow cuticle breakdown. Lysates were centrifuged at 10,000 × g for 30 min, and the supernatant was collected for use in further analysis. Protein concentrations were determined by the Bradford method.
Iron Measurement
To determine the iron content of the worms, we used a BioVision iron assay kit (BioVision, Mountain View, CA). Protein samples from ∼3000 atm-1(RNAi), or cat(RNAi), or frh-1(ok610), or N2 worms were tested for ferrous (Fe2+), ferric (Fe3+), and total (Fe2+ + Fe3+) iron following the manufacturer's instructions.
Oxidative Stress Assays
To measure sensitivity to oxidative stress, 80 abtm-1(RNAi) or cat(RNAi) L4 larvae were incubated in the presence of 0, 0.5, 5, and 10 mm paraquat (methyl viologen, Sigma). The experiments were carried out for 3 days at 25 °C, as described previously (17). We scored worm survival each day, using the same protocol as for life span assays.
To analyze protein carbonylation we used an OxyblotTM protein oxidation detection kit (Millipore, Billerica, MA). Briefly, worm lysate, prepared as above, containing 15 μg of protein was incubated in 12% SDS supplemented with 2,4-dinitrophenylhydrazine for 10 min at room temperature. Samples were resolved on a 12% SDS-PAGE, and 2,4-dinitrophenylhydrazine-derivatized proteins were detected by immunoblot using an anti-2,4-dinitrophenylhydrazine antibody.
In vivo analysis of sod-3 and daf-16 reporters-sod-3 expression was analyzed using the KN259 (huIs33[sod-3p::sod-3::GFP+pRF4(rol-6(su1006)]) reporter strain. To quantify sod-3::GFP expression, we collected fluorescence images of more than 30 animals for each condition (abtm-1(RNAi) or cat(RNAi)). We then measured the pixels produced by fluorescence in the whole body of animals and determined the fluorescence level relative to worm area, using ImageJ. To investigate the cellular localization of DAF-16, we used the strain TJ365 (zIs356[daf-16p::daf-16::GFP; rol-6(su1006)]). We analyzed control and abtm-1(RNAi) young adults produced by independently injected hermaphrodites (number of observed animals per strain ≥300). We observed the worms under a dissecting microscope, equipped with fluorescence. Animals were kept on plates with food, at 20 °C, and positives were scored when the animals presented fluorescent nuclei in one or more tissues. As a positive control we used animals under starving conditions.
Life Span Assays
To measure life span, L4 larvae were cultured at 20 °C on NGM with 0.1 g/ml of 5-fluorodeoxyuridine (Sigma) until death. Death was assessed by the response of the animal to gentle nose touch. Worm survival was counted every 24 h. Missing worms were scored as censored data. We scored more than 200 worms per sample.
Statistical Analyzes
All data are presented as means ± S.E. or as % of population. To assess statistical significance, we compared different populations using Student's two-tailed t test using GraphPad on line. To compare survival curves from life span and oxidative stress assays, we used the log-rank (Mantel-Cox) test, contained within the GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA).
RESULTS
C. elegans ABCB7 Homologue, ABTM-1, Is a Mitochondrial Protein That Is Highly Expressed in Mitochondrion-rich Tissues
To identify homologues of ABCB7 in C. elegans, we used the human ABCB7 (hABCB7) sequence to search the C. elegans genome. We found that abtm-1 (Y74C10AR.3) (Fig. 1A) shows high similarity to hABCB7. We cloned the abtm-1 cDNA using RT-PCR. The abtm-1 cDNA is 2504 bp long (supplemental Fig. S1A) and encodes a predicted protein of 703 residues. The C-terminal region of the predicted peptide, ABTM-1, is 51% identical to hABCB7 (supplemental Fig. S1B). ABTM-1 has the typical ABC superfamily structure, with six transmembrane domains and an ATP binding domain (supplemental Fig. S1B). Alignment of the protein sequence with sequences from the human and yeast ABCB7 family shows that ABTM-1 clusters within this family (supplemental Fig. S1C), strongly suggesting that this protein belongs to the ABCB7 transporter group. Analysis of ABTM-1 using the Mitoprot program (18) gave a probability of 99.4% that ABTM-1 is mitochondrial. To demonstrate that ABTM-1 is indeed a mitochondrial protein, we produced transgenic animals carrying an abtm-1::GFP fusion (abtm-1::GFP1, Fig. 1A) in which GFP is fused to the end of the third exon of abtm-1. The predicted peptide should contain the first 67 amino acids of the protein, including the predicted mitochondrial localization signal. We incubated transgenic worms carrying abtm-1::GFP1 with a mitochondrial marker, MitoTracker® Red CMXRos (Molecular Probes). Confocal microscopy of these worms shows that ABTM-1::GFP1 clearly co-localizes with the mitochondrial marker (Fig. 1B) confirming that ABTM-1 is a mitochondrial protein and the C. elegans homologue of hABCB7.
We sought to investigate the expression pattern of abmt-1 using the abtm-1::GFP1 fusion; however, this construct resulted in some toxicity (data not shown). We therefore produced a cytoplasmic GFP construct, abtm-1::GFP2, in which the first codon of abtm-1 was fused to GFP (Fig. 1A). Transgenic animals containing abtm-1::GFP2 show widespread expression in the tissues of adult hermaphrodites, including the intestine, spermatheca, epidermis, and coelomocytes, among others (data not shown). Interestingly, abtm-1::GFP2 is highly expressed in tissues that are predicted to require high levels of energy production such as pharyngeal muscles (Fig. 1C, panel i), neurons (Fig. 1C, panel i), developing embryos (Fig. 1C, panel ii), and body wall muscles (Fig. 1C, panel iii), among others.
Worms Carrying a Deletion in abtm-1 Are Not Viable
We obtained a strain carrying a deletion and, probably, null allele of abtm-1, abtm-1(tm2721) (Fig. 1A). The tm2721 allele was out-crossed and balanced with sDp2, a duplication of the region of chromosome I carrying the abtm-1 gene thus, allowing us to maintain tm2721. sDp2 is a free duplication that undergoes random, non-Mendelian segregation with either copy of chromosome I during meiosis (19). Hermaphrodites carrying mutations that map in the region balanced with sDp2 segregate, about 38% unbalanced mutant homozygotes (19). We isolated and analyzed three abtm-1(tm2721)I; sDp2(I;f) strains from independent crosses. We were unable to isolate viable homozygous unbalanced animals from these strains. We observed that the balanced animals produced 28.1 ± 1.1% arrested embryos and 7.1 ± 0.6% arrested L1-L2 larvae (wild type 1.2 ± 0.2% and 0% arrested embryos and larvae). The total lethality is therefore 35.2%, which is close to the predicted value of 38% for homozygous animals. This experiment strongly suggests that abmt-1(tm2721) homozygous animals exhibit early lethality and are unable to develop to adulthood.
RNAi of abtm-1 Induces Embryonic Arrest during Late Embryogenesis
Because complete ablation of abtm-1 is lethal, we sought to emulate the condition of XLSA/A patients by reducing, rather than completely removing, the function of the abtm-1 gene. To do this we used RNAi (abtm-1(RNAi)). We injected double-stranded abtm-1 RNA into young wild type hermaphrodites to induce RNAi and then examined the resulting offspring. In agreement with results obtained using abmt-1(tm2721) embryos, we observed that reducing ABTM-1 in wild type hermaphrodites induces embryonic arrest in their offspring, 28 ± 1.24% (Fig. 2A). This lethality is further increased to 56 ± 2.7% in daf-16/FOXO mutants (Fig. 2A). DAF-16 is widely involved in insulin signaling and stress responses in C. elegans (20). Thus daf-16 can protect against abtm-1(RNAi)-induced lethality, suggesting that the lethality may result from increased stress of some kind.
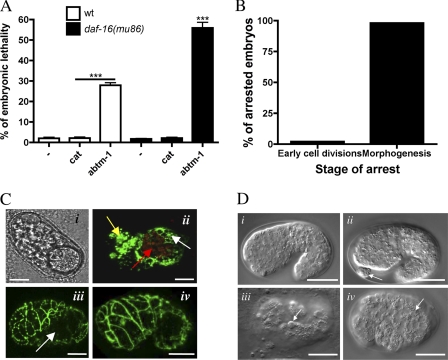
FIGURE 2.
Disruption of abtm-1 produces embryonic lethality, which is further increased by depletion of daf-16. A, embryonic lethality in the offspring of abtm-1(RNAi) worms in wild type and daf-16(mu86) backgrounds. abtm-1(RNAi) hermaphrodites produce 28% dead embryos, whereas depletion of abtm-1 in daf-16 mutants produce 56%. ***, Student's t test, p <0.0001. B, abtm-1(RNAi) produces arrest of embryos mostly during morphogenesis (96%). C, representative examples of abtm-1(RNAi)-arrested embryos. C, panel i, transmitted light image of an embryo arrested during the proliferation stage. C, panel ii, confocal image of an embryo arrested during morphogenesis, showing signs of cellular differentiation. The yellow and white arrows point to AJM-1::GFP expression, a cell-specific marker of epithelia, in the pharyngeal and epidermal cells, respectively. The red arrow indicates autofluorescent granules, characteristic of the intestinal cells. C, panel iii, confocal image of an embryo in which the epidermis has failed to migrate during ventral enclosure leaving the embryo incompletely enclosed as indicated by the arrow. C, panel iv, an embryo arrested during the last step of morphogenesis, elongation. The epidermis has enclosed most of the embryo, but it is highly disorganized. D, transmitted light images, from four-dimensional microscopic analysis. D, panel i, wild type embryo. D, panel ii, embryo arrested during morphogenesis showing the presence of internal tissues external to the embryo. D, panels iii and iv, embryos showing premature apoptotic events in the 9th generation. Apoptotic cells are indicated by arrows. All scale bars represent 10 μm.
To further analyze the nature of the defects in abtm-1(RNAi), we observed them under a differential interference contrast microscope. Some abtm-1(RNAi) embryos, 7.7%, arrested during the early proliferative stages (Fig. 2B); however, most of the arrested embryos showed clear signs of cell differentiation (Fig. 2C). For example, abtm-1(RNAi) embryos had gut granules indicating that the intestinal cells had differentiated (Fig. 2C, panel ii) and also showed vigorous twitching, an indicator of muscle cell differentiation. As the embryos arrested after gastrulation, we analyzed the behavior of epidermis, which plays a major role during embryonic morphogenesis (21, 22). To do this we used an epithelial cell-specific marker, ajm-1::GFP (22). 92.3% of arrested abtm-1(RNAi) embryos showed correctly (i.e. apically) localized AJM-1::GFP (n = 104) (Fig. 2C, panels ii–iv), suggesting that the epidermal cells are also differentiated. Nevertheless, analysis of the pattern of AJM-1::GFP revealed that most embryos show a highly disorganized structure, including a retracted epidermis (Fig. 2C, panels ii and iii), and the presence of internal tissues on the exterior of the embryo (Fig. 2C, panel ii). Therefore, ABTM-1 is required for appropriate embryonic morphogenesis in C. elegans.
abtm-1(RNAi) Embryos Have Premature, Putative Apoptotic Events
To investigate possible causes of the morphogenetic defects in abtm-1(RNAi) embryos, we used four-dimensional video microscopy and lineage analysis. The cell lineage of C. elegans is well defined so that every somatic cell can be traced (16, 57). Moreover, four-dimensional imaging is a well established tool for identifying and studying apoptosis (23, 24). In a wild type embryo the progeny of the “founder cell” AB (the anterior cell in the two-cell stage embryo) divide, in near synchrony, after each cell generation and can be readily tracked by four-dimensional video microscopy imaging. We recorded abtm-1(RNAi) and wild type embryos to identify differences in cell cycle length, fate specification, morphogenesis, and cell death. Six of the abtm-1(RNAi) embryos were analyzed in detail with respect to cell lineage and timing. We observed four phenotypic classes of abtm-1(RNAi) embryos (Table 1). abtm-1(RNAi) embryos have delayed cell divisions, cells that are excluded from the embryo and in extreme cases burst. In addition, two of the embryos showed a striking defect. Some cells in the 9th cell generation (ABalaaapp, ABprappap, and ABprppapa in one embryo, and most probably ABplpppapp in the other) underwent cell death, most probably by apoptosis (Fig. 2D). In normal embryos apoptosis occurs in specific cells in the 10th and the 11th cell generation, as a consequence of specific cell fate decisions. The presence of these putative premature apoptotic events may indicate that cells are activating the apoptotic pathway as consequence of cellular stress. abtm-1(RNAi) embryos also show increases in the cell cycle length (Table 1). The severity of this defect increases as development proceeds with late cells dividing much more slowly than the early blastomeres. On average the division of ABxxxxxx, the last cell division recorded, was nearly twice as long as the wild type division, and in extreme cases was nearly three times as long (Table 1). Again, this suggests that cellular metabolism is disrupted. In addition, in three embryos some cells were excluded from the body during morphogenesis (Fig. 2D, panel ii). Four-dimensional analysis, in one embryo, clearly identified 3 of 10 of these cells as neuronal cells of the ring ganglion (the specific cells are ABalapappap, ABalappaapa, and ABalapppaap).
TABLE 1.
Delayed cell divisions in abtm-1(RNAi) embryos
abtm-1(RNAi) embryos were analyzed by four-dimensional video microscopy and subsequent lineage analysis. Embryos were divided into four types based on the nature of the developmental defects observed. Delays during the development of all the embryos were measured, although we only show data of the four representative types. Numbers indicate the time (in minutes) to complete the cell cycle for the cell in the left column. Although some variability in the timing of cell cleavages occurs from embryo to embryo in the wild type (57), the difference compared with the reference published by Sulston in 1983 (57) is always smaller than 10% of the cell cycle length. We consider that a cell is dividing significantly slower than wild type when the length of its cell cycle is at least 20% longer than the wild type reference.
| Cycle cell | Time for cell division (mins) (% of increment in cell cycle)a |
|||||
|---|---|---|---|---|---|---|
| Wild type average mean time | abtm-1(RNAi) type 1 | abtm-1(RNAi) type 2 | abtm-1(RNAi) type 3 | abtm-1(RNAi) type 4 | abtm-1(RNAi) average mean time | |
| min | min | |||||
| AB | ||||||
| ABx | 12.5 | 17 (36%) | 19 (52%) | 20 (60%) | 16.4 (31%) | |
| ABxx | 14.6 | 20 (37%) | 21 (44%) | 24 (65%) | 18.8 (29%) | |
| ABxxx | 16.4 | 27 (64%) | 24 (45%) | 24 (46%) | 32 (94%) | 23.0 (39%) |
| ABxxxx | 23.9 | 41 (71%) | 33 (37%) | 36 (51%) | 42 (76%) | 33.0 (38%) |
| ABxxxxx | 25.3 | 57 (125%) | 45 (78%) | 47 (86%) | 63 (149%) | 44.3 (75%) |
| ABxxxxxx | 33.2 | 93 (179%) | 55 (65%) | - | 82 (146%) | 61.0 (67%) |
| Phenotypeb | (3) | Delayed development alone (2) | Delayed development and excluded cells (3) | Delayed development and explosion (1) | Delayed development and premature apoptosis (2) | |
a The increment of the time in delayed cycles is shown in parentheses. To obtain the average of the increment, we included the cycles of all available embryos (n = 8).
b The lower row shows the phenotype for each type of abtm-1(RNAi) embryo. All types showed delayed development. Type 1 did not have any other apparent defects. Type 2 embryos had internal tissues on the exterior (Gex phenotype). Type 3 embryos burst, probably due to a defect in morphogenesis. Type 4 showed some premature apoptosis as well as morphogenetic defects. The number of embryos of each type that we observed is shown in parentheses.
abtm-1(RNAi) Animals Show Accumulation of Fe3+
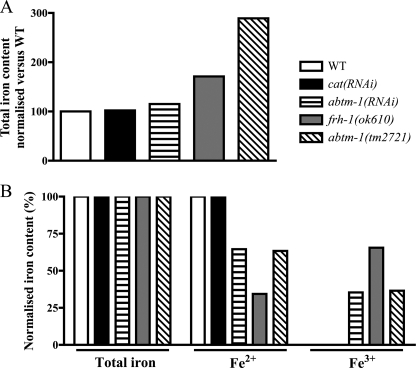
Our results above suggest that abtm-1 animals have severe cellular defects. We sought to determine whether those abtm-1(RNAi) animals that reached adulthood have changes in their physiology reflecting the function of ABTM-1. Deletion of the yeast ABCB7 homologue, ATM1 (8), results in ferric iron (Fe3+) overload. We therefore hypothesized that if abtm-1 function is conserved in C. elegans, abtm-1 knockdown should lead to a similar phenotype. To test this hypothesis, we sought to measure iron levels in abtm-depleted animals. We also tested frh-1(ok610) worms that carry a loss of function allele in the frataxin (frh-1) gene (17) responsible for Friedreich ataxia (25). This molecule has been suggested to be involved in Fe-S cluster synthesis (reviewed by Stemmler et al. (26)), among other hypotheses (reviewed by Gonzalez-Cabo et al. (27)). Disruption of frataxin induces iron accumulation in many organisms, for example in yeast (28). Although total iron levels (Fe2+ + Fe3+) in abtm-1(RNAi) animals did not show any significant difference to control animals, abtm-1(tm2721)/+ mutants showed a sensible increase (n ≥3000 for each strain tested) (Fig. 3A). In both cases Fe3+ was present at much increased levels in abtm-1(RNAi) (10-fold), abtm-1(tm2721)/+ (16-fold), and also in frh-1(ok610)/+ (18-fold) animals compared with wild type and cat(RNAi) controls (Fig. 3B).
FIGURE 3.
Depletion of abtm-1 induces accumulation of ferric iron. A, graph shows total iron content, normalized to the wild type level in wild type, cat(RNAi), abtm-1(RNAi), frh-1(ok610) animals, and abtm-1(tm2721). B, graph shows the percentage of ferrous and ferric iron in the same strains normalized to the total iron content in that strain. Fe3+ is increased in animals with reduced ABTM-1 or frataxin (FRH-1).
Depletion of abtm-1 Causes Increased Oxidative Stress
As abtm-1(RNAi) animals show accumulation of Fe3+, we sought to investigate if this caused increased oxidative stress. First, we tested the ability of abtm-1(RNAi) animals to cope with an externally induced increase in free radicals by measuring survival on exposure to paraquat (29, 30). Worm survival in the presence of a range of paraquat concentrations was measured. On 0 and 0.5 mm paraquat, the survival of abtm-1(RNAi) animals is unaltered (data not shown). However, at 5 and 10 mm paraquat, abtm-1(RNAi) worms show substantial reductions in survival (log-rank (Mantel-Cox) test p < 0.0001 for both paraquat concentrations) (Fig. 4A), suggesting that reductions in ABTM-1 cause increased sensitivity to oxidative stress.
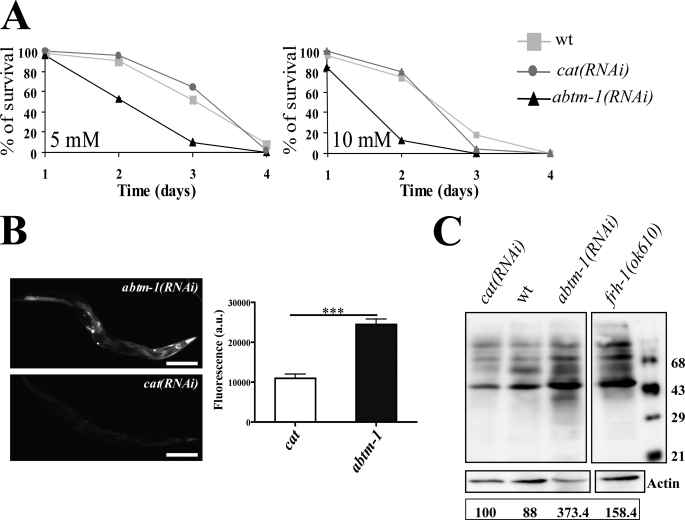
FIGURE 4.
abtm-1(RNAi) worms are under oxidative stress. A, survival graphs of wild type, abtm-1(RNAi), and control animals cat(RNAi) grown in different concentrations of paraquat. At 5 and 10 mm, the survival of abtm-1 depleted animals is significantly lower (p < 0.0001 in both cases) than control and wild type animals. B, transmission and fluorescent images of representative offspring from RNAi-treated worms containing the sod-3::GFP construct. abtm-1(RNAi) animals show an obvious increase in fluorescence (i.e. sod-3 expression) compared with controls. Scale bars represent 50 μm. Quantification of this increase is shown below where the average fluorescence intensity (represented by arbitrary units), relative to the body area of each worm, in sod-3::GFP worms is plotted. abtm-1-depleted animals show a significant increase of expression compared with control worms (p < 0.001) (n ≥30). C, OxyblotTM assay on cat(RNAi), wild type abtm-1(RNAi), and frh-1(ok610) animals. Carbonylated proteins were quantified for each lane using MultiGauge software (FujiFilm). To allow for loading variation, values were normalized to the actin control. Final values are expressed as a percentage of the wild type value and are shown below each lane. It can be seen that frh-1(ok610) and the abtm-1(RNAi) worms show a marked increase in carbonylated proteins.
We next examined whether abtm-1(RNAi) animals had increased levels of endogenous free radicals. We examined the expression of a sod-3::GFP reporter gene. SOD-3 is a mitochondrial manganese-dependent superoxide dismutase homologue and a known free radical scavenger. Expression of sod-3::GFP transgenes is known to be increased in response to increases in free radical production (31). abtm-1 RNAi on animals carrying the sod-3::GFP transgene resulted in a greater than 2-fold increase in fluorescence compared with controls (p < 0.001; n = 30) (Fig. 4B).
To confirm that ablation of abtm-1 induces oxidative stress, we sought to investigate the effects of increased free radical levels in vivo. Thus, we measured protein carbonylation as amino acid carbonylation is caused by free radicals. To do this, we used an OxyblotTM assay on abtm-1(RNAi), wild type, and cat(RNAi) worms, together with frataxin mutant animals. cat(RNAi) and wild type worms show similar amounts of carbonylated proteins (Fig. 4C, 1st and 2nd lanes), although the frataxin worms show an increase of 1.5-fold (Fig. 4C, 4th lane). Interestingly, the abtm-1(RNAi) worms show a 3.5-fold increase in carbonylated proteins (Fig. 4C, 3rd lane). Thus, the above evidence suggests that abtm-1-depleted animals are under significantly increased levels of oxidative stress, presumably resulting from increased Fe3+ levels.
abtm-1(RNAi) Animals Have Increased Life Span, which Is Partially Dependent on daf-16/FOXO
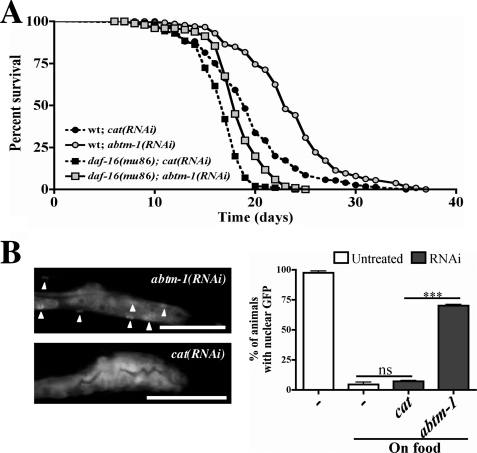
Increased oxidative stress might be expected to reduce life span. However, the life span of abtm-1 deficient worms is significantly increased compared with control animals (abtm-1(RNAi)) (median life span (ML) = 23 days versus cat(RNAi) ML = 19 days, p < 0.0001; supplemental Table S1) (Fig. 5A). Changes in the activity of the transcription factor daf-16/FOXO alter longevity in many organisms, including humans (32). Furthermore, daf-16 is known to mediate both insulin and stress responses (20). Therefore, we tested for interactions between the effects on life span of abtm-1 and daf-16. Interestingly, depletion of abtm-1 in a daf-16 loss of function background shows a partial increase in life span (Fig. 5A) (daf-16(mu86); abtm-1(RNAi) ML = 18 daf-16(mu86); cat(RNAi) ML = 17; supplemental Table S1), suggesting that the life span increase in abtm-1(RNAi) animals is partially dependent on DAF-16 function.
FIGURE 5.
Life span of abtm-1(RNAi) adults is increased in a partially daf-16–dependent manner. A, life span curves of abtm-1(RNAi) worms in wild type and daf-16(mu86) mutant backgrounds. The life span of abtm-1-depleted animals is significantly longer than control worms (p < 0.0001). Induction of abtm-1 RNAi also increases the life span of daf-16(mu86) mutants (p < 0.0001), but to a lesser extent (also see supplemental Table S1). B, abtm-1 RNAi induces localization of DAF-16 in the nucleus in some tissues in 70% of worms. Starved animals (first bar) were used as positive control, as they show strong nuclear DAF-16 localization.
DAF-16 is known to relocate from the cytoplasm to the nucleus when activated (33). Therefore, we investigated the cellular localization of DAF-16 in abtm-1 depleted worms by inducing abtm-1 RNAi in animals that carry a functional DAF-16::GFP fusion (33). We observed that most abtm-1(RNAi) animals showed nuclear localization of DAF-16 in one or more tissues, suggesting that ablation of abtm-1 induces signals that activate DAF-16.
abtm-1-depleted Adult Animals Have a Pleiotropic Phenotype, Including Defects in Locomotion and Rhythmic Behavior
Phenotypic analysis of adults (and embryos) is a prerequisite to our aim of establishing a model for XLSA/A that can be used for both mechanistic studies and screening for compounds that may be developed for clinical use. In C. elegans, mitochondrial mutants usually show a range of phenotypes, including developmental, physiological, and behavioral defects (reviewed by Tsang and Lemire (34)). This is indeed the case of abtm-1(RNAi) worms, which show a pleiotropic phenotype that includes developmental defects, growth, egg laying, ultradian rhythms (35), and locomotion defects (see above and supplemental material). Therefore abtm-1(RNAi) show similar traits to other mitochondrial mutants.
Disruption of Other Components of the Fe-S Cluster Biosynthesis Pathway Induces Similar Phenotypes to Depletion of abtm-1
In yeast, cytosolic Fe-S clusters are produced exclusively in the lumen of mitochondria. Fe-S clusters are produced using sulfur from cysteine in a reaction catalyzed by cysteine desulfurase (Nfs1p). Production also requires chaperones and scaffold proteins (Isa1p, Isa2p, Isu1p, and Isu2p) and redox proteins (Arh1p, Yah1p, and glutaredoxin-5), which supply Fe2+ (reviewed in Refs. 36, 37). Once produced, Fe-S clusters are either used in mitochondrial Fe-S-containing proteins or they are transported to the cytosol, by means of Atm1p/ABCB7, where they are used to produce cytosolic Fe-S-containing proteins (Fig. 6) (36). We investigated whether disrupting other steps in Fe-S synthesis would phenocopy the traits described above for abtm-1(RNAi) animals. We performed RNAi on the C. elegans orthologues of six Fe-S cluster biosynthesis genes (Wormbase release WS185) (Table 2 and Fig. 6). Knocking down these genes produces a range of phenotypes (e.g. disrupted defecation, extended life span, and larval arrest) all compatible with a mitochondrial phenotype (Table 2). Moreover, in all six cases, depletion produces significant embryonic arrest showing that disrupting the Fe-S cluster pathway compromises embryonic development in C. elegans. Both the mitochondrial phenotype and embryonic lethality recapitulate the phenotypes produced by ablation of abtm-1.
FIGURE 6.
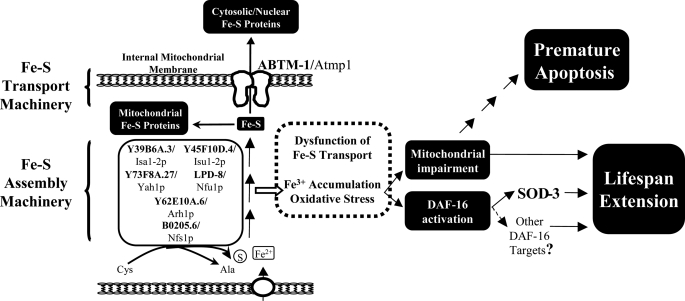
Fe-S cluster synthesis pathway and our working model for the life span extension and apoptotic events caused by depletion of abtm-1 in C. elegans. The diagram shows the ABC transporter ABTM-1/Atm1p and some of the enzymes (ferredoxin reductase, Y62E10A.6/Arh1p; ferredoxin, Y73F8A.27/Yahp) and scaffolding proteins (LPD-8/Nfu1p, B0205.6/Nfs1p, Y39B6A.3/Isa1p, and Y45F10D.4/Isu1p) involved in the synthesis of Fe-S clusters. These molecules are represented by the name of the protein in C. elegans (boldface), and the corresponding S. cerevisiae homologue. Defects in Fe-S cluster synthesis caused by abtm-1(RNAi), as indicated by the box surrounded by a dotted line, cause accumulation of iron, which in turn induces free radicals. The oxidative stress promoted by the free radicals is responsible for mitochondrial damage, which may lead to premature apoptosis and activation of DAF-16. Both the mitochondrial damage and DAF-16 activation may induce life span extension in individuals that survive embryogenesis. The life span extension induced by DAF-16 may be due to the expression of genes such as sod-3.
TABLE 2.
Disruption of genes involved in Fe-S cluster synthesis produces a range of phenotypes
The coefficient of variation (CV) is reported as a percentage and is calculated from the average and S.D. as follows: 100 × S.D./average.
| Strain | Yeast putative homologue gene | % of Embryonic arrest (n) | Larval arrest | Defecation cycle (CV) | Life span |
|---|---|---|---|---|---|
| % | |||||
| Wild type | 2.1 (1597) | 0 | 48 ± 3 (6 ± 1) | Normal | |
| cat(RNAi) | NAa | 1.9 (1641) | 0 | 48 ± 1 (5 ± 1) | Normal |
| abtm-1(RNAi) | ATM1 | 27.9 (945) | <5 | 60 ± 3 (17 ± 3) | Long lived (p < 0.001)b |
| Y62E10A.6(RNAi) | ARH1 | 11.8 (543) | <5 | 77 ± 5 (14 ± 2) | Long lived (p < 0.001)b |
| lpd-8(RNAi) | NFU1 | 18.0 (389) | <5 | 50 ± 2 (8 ± 1) | Long-lived (p < 0.001)b |
| B0205.6(RNAi) | NFS1 | 39.5 (440) | 100 | NA | NA |
| Y73F8A.27(RNAi) | YAH1 | 48.8 (540) | 100 | NA | NA |
| Y39B6A.3(RNAi) | ISA1 | 63.1 (367) | 100 | NA | NA |
| Y45F10D.4(RNAi) | ISU1 | 84.1 (414) | 100 | NA | NA |
a NA means not applicable.
b Log rank (Mantel-Cox) test was used.
DISCUSSION
XLSA/A is an untreatable disease caused by mutations in the ABCB7 gene. ABCB7 is believed to function in the mitochondrial transport of Fe-S clusters into the cytoplasm (8, 9). It is thought that defective Fe-S cluster transport leads to mitochondrial iron overload with subsequent free radical production and to reduced heme synthesis, which in turn results in reduced Fe-S-containing enzyme activity. These deficits may then lead to mitochondrial dysfunction and to the pathology of XLSA/A (11). Thus, XLSA/A is one of a growing number of mitochondrial diseases (38, 39). Using mutants and RNAi, we analyzed the effects of total and partial loss of function in abtm-1, the C. elegans homologue of ABCB7. Complete loss of abtm-1 function is lethal, as is the loss of ABCB7 in mice (6), and partial loss of function also leads to a substantial level of embryonic and larval arrest. Thus, as in other systems, ABCB7/abtm-1 is an essential gene. The use of RNAi enabled us to produce a model that recapitulates the partial loss of function found in XLSA/A patients. Partial loss of function results in arrested embryos and adults with increased Fe3+ levels, higher oxidative stress, increased life span, and a range of phenotypes characteristic of mutations that cause mitochondrial dysfunction. Expression analysis showed that ABTM-1 is a widely expressed mitochondrial protein that it is produced at particularly high levels in tissues that are expected to have high energy requirements, as is the case in humans and mice (3, 6). Thus our data show that, as in other models, abtm-1 is an essential mitochondrial gene.
Depletion of abtm-1 caused substantial embryonic arrest. Analysis of abtm-1(RNAi) embryos showed that arrest mostly occurs after the initial proliferative phase and usually during morphogenesis. Detailed analysis of early embryonic development, using four-dimensional imaging, showed that the embryos have slower cell divisions and that, strikingly, they exhibit putative premature apoptosis. These early defects may underlie later defects in morphogenesis. For example, the loss of cells due to putative apoptosis could result in the loss of cells or cell types that are required for morphogenesis. In particular the putative apoptotic cells observed in the abtm-1(RNAi) embryos belong to lineages that will give rise to neuroblasts, a cell type that is essential for proper epidermal migration during morphogenesis (21). In addition, disorganized cell division may result in the failure of important inductive developmental signaling events in the early embryo, again leading to the loss of particular cells and cell types. The early defects in these embryos could all result from an abnormal mitochondrial function. For example, apoptosis may result from defective mitochondrial function (40). Similarly the cell cycle has a high energy demand (41) and may be retarded due to reduced ATP levels caused by poor mitochondrial function. Interestingly, a delay in cell cycle progression has been described in Drosophila as a result of a mutation in tenured, a gene encoding mitochondrial cytochrome oxidase subunit Va (42). Thus defects in abtm-1 may cause lethality in embryos due to a pyramid of effects, which emanate from mitochondrial dysfunction.
Although depletion of abtm-1 by RNAi produced substantial lethality, a significant proportion of worms were able to survive to adulthood. This presumably reflects different degrees of abtm-1 depletion in different animals. Animals that survived to adulthood demonstrated a range of phenotypes, including disrupted defecation, egg laying, and locomotion suggesting a widespread perturbation of physiological function in several tissues, including neurons. This range of phenotypes is reminiscent of the pleiotropy observed in other mitochondrial mutants, such as frh-1, the C. elegans homologue of frataxin (17, 43). Interestingly, the role of frataxin, although still controversial (see Refs. 27, 44, 45), has also been linked to Fe-S cluster production (for a review see Rouault and Tong (46)). Therefore, phenotypes caused by depletion of both molecules may be due to similar cellular stresses. We propose that this pleiotropic phenotype results from defects in Fe-S cluster metabolism. In support of this, depleting other genes involved in Fe-S biogenesis and transport results in phenotypes that share some of the features of abtm-1(RNAi) worms. It is interesting to note that RNAi on four of these genes (B0205.6/NFS1, Y73F8A.27/YAH1, Y39B6A.3/ISA1, and Y45F10D.4/ISU1) produced 100% embryonic and larval arrest in the offspring. This may reflect a more severe mitochondrial defect in these worms, perhaps because they affect key steps in the synthesis of all Fe-S clusters in contrast to abtm-1, which only mediates transport of cytosolic Fe-S clusters. Differences in the strength of the RNAi may also explain differences in the severity of phenotypes.
We observed a substantial increase in Fe3+ in abtm-1 animals. abtm-1 mutant animals also showed an increase in total iron content. Disruption of iron homeostasis underlies many human disorders. This is probably also the case in XLSA/A where it has been suggested that the pathological condition may be a consequence of mitochondrial iron accumulation. In this regard, the yeast mutant atm1Δ shows accumulation of ferric phosphate (47). It is suggested that ferrous iron (Fe2+) enters mitochondria where it is oxidized to Fe3+. Excess ferric iron, due to inactivity of ABTM-1, has been shown in Atm1/ABCB7-depleted yeast (28, 47, 48). Thus our results are compatible with previous studies.
abtm-1 animals have increased oxidative stress as measured by a number of approaches. One implication of the increase in oxidative stress might be expected to be a decrease in life span; however, these worms actually have increased life span. This paradox may be explained by hormesis. In hormesis, a beneficial biological response in a cell or organism is initiated by a nonlethal detrimental stress. The response is primarily aimed at counteracting the damage produced by that stress, but it also results in an increased life expectancy. This phenomenon has been documented in yeast (49) and C. elegans (50, 51) and indeed many mitochondrial mutants in C. elegans that do not compromise survival also show life span extension (10, 52). Thus, in abtm-1(RNAi) animals, the activation of scavenging molecules that promote cell survival, in response to the production of free radicals in mitochondria, may in turn increase life span. This may be reflected in the nuclear localization of DAF-16 and the increased expression of sod-3, both of which are markers of increased oxidative stress but are also, presumably, protective.
daf-16 is a master regulator of life and health span (53), and it is widely implicated in stress and related responses. DAF-16 appears to play an important role in the effects of abtm-1 depletion suggesting that increased stress is a key determinant of the effects of abtm-1 ablation, probably in response to free radical production. Increased nuclear localization of DAF-16 in abtm-1(RNAi) worms strongly suggests that it is being activated. DAF-16 may in turn induce the expression of sod-3, among other protective genes. The embryonic arrest caused by depletion of abtm-1 is enhanced in a daf-16 null background, suggesting that the presence of daf-16 is able to protect against abtm-1(RNAi)-mediated damage caused by Fe3+ accumulation and free radical production. In addition, the increase in life span observed in abtm-1 ablated animals is, at least, partially dependent on DAF-16. This may be explained if the putative hormesis discussed above is daf-16-dependent. Indeed, DAF-16 may act as a mediator of hormesis in C. elegans, upon activation by cellular stress (54). The increase in life span cannot be completely attributed to the action of DAF-16 and its downstream targets, as daf-16(mu86);abtm-1(RNAi) animals still have an increase in life span, suggesting that other forms of life span extension such as mitochondrial malfunction (Fig. 5C) or calorific restriction could be involved.
Model systems are critical to our ability to understand these processes. We have established a C. elegans model of the human disease XLSA/A. The ability to use a genetically tractable metazoan in the study of XLSA/A, and other mitochondrial diseases, should improve our understanding of the mechanisms that underlie these conditions. C. elegans enables the use of high throughput automated RNAi screens, using for example embryonic lethality, to identify genetic interactions (55). C. elegans can also be used in high throughput compound screens (56) or in the testing of drug candidates such as free radical scavenging molecules or activators of cell survival pathways that diminish cellular stress.
Supplementary Material
Acknowledgments
We are grateful to S. Mitani and the Caenorhabditis Genetic Centre for the provision of strains. We also thank A. Fire for vectors. We thank M. Artal-Sanz for helpful discussions.
This work was supported by The Spanish Ministry of Education and Science Grants SAF2006-01147, BFU2008-01808, BFU2010-21794, and Consolider CSD2007-00015, Generalitat Valenciana, Junta de Castilla y León Grants CSI03A08 and Grupo de Excelencia GR 265, The Welcome Trust, and Medical Research Council. The CIBERER is an initiative of the Instituto de Salud Carlos III.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” “Results,” Figs. S1–S3, and Tables S1–S3.
- XLSA/A
- X-linked sideroblastic anemia with ataxia
- ABC
- ATP-binding cassette
- cat
- chloramphenicol acetyltransferase
- ML
- median life span.
REFERENCES
- 1. Pagon R. A., Bird T. D., Detter J. C., Pierce I. (1985) J. Med. Genet. 22, 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allikmets R., Raskind W. H., Hutchinson A., Schueck N. D., Dean M., Koeller D. M. (1999) Hum. Mol. Genet. 8, 743–749 [DOI] [PubMed] [Google Scholar]
- 3. Savary S., Allikmets R., Denizot F., Luciani M. F., Mattei M. G., Dean M., Chimini G. (1997) Genomics 41, 275–278 [DOI] [PubMed] [Google Scholar]
- 4. Maguire A., Hellier K., Hammans S., May A. (2001) Br. J. Haematol. 115, 910–917 [DOI] [PubMed] [Google Scholar]
- 5. Bekri S., Kispal G., Lange H., Fitzsimons E., Tolmie J., Lill R., Bishop D. F. (2000) Blood 96, 3256–3264 [PubMed] [Google Scholar]
- 6. Pondarré C., Antiochos B. B., Campagna D. R., Clarke S. L., Greer E. L., Deck K. M., McDonald A., Han A. P., Medlock A., Kutok J. L., Anderson S. A., Eisenstein R. S., Fleming M. D. (2006) Hum. Mol. Genet. 15, 953–964 [DOI] [PubMed] [Google Scholar]
- 7. Pondarre C., Campagna D. R., Antiochos B., Sikorski L., Mulhern H., Fleming M. D. (2007) Blood 109, 3567–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kispal G., Csere P., Guiard B., Lill R. (1997) FEBS Lett. 418, 346–350 [DOI] [PubMed] [Google Scholar]
- 9. Kispal G., Csere P., Prohl C., Lill R. (1999) EMBO J. 18, 3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cavadini P., Biasiotto G., Poli M., Levi S., Verardi R., Zanella I., Derosas M., Ingrassia R., Corrado M., Arosio P. (2007) Blood 109, 3552–3559 [DOI] [PubMed] [Google Scholar]
- 11. Wingert R. A., Galloway J. L., Barut B., Foott H., Fraenkel P., Axe J. L., Weber G. J., Dooley K., Davidson A. J., Schmid B., Schmidt B., Paw B. H., Shaw G. C., Kingsley P., Palis J., Schubert H., Chen O., Kaplan J., Zon L. I. (2005) Nature 436, 1035–1039 [DOI] [PubMed] [Google Scholar]
- 12. Lewis J. A., Fleming J. T. (1995) Methods Cell Biol. 48, 3–29 [PubMed] [Google Scholar]
- 13. Mello C., Fire A. (1995) Methods Cell Biol. 48, 451–482 [PubMed] [Google Scholar]
- 14. Miguel-Aliaga I., Culetto E., Walker D. S., Baylis H. A., Sattelle D. B., Davies K. E. (1999) Hum. Mol. Genet. 8, 2133–2143 [DOI] [PubMed] [Google Scholar]
- 15. Zwaal R. R., Ahringer J., van Luenen H. G., Rushforth A., Anderson P., Plasterk R. H. (1996) Cell 86, 619–629 [DOI] [PubMed] [Google Scholar]
- 16. Schnabel R., Hutter H., Moerman D., Schnabel H. (1997) Dev. Biol. 184, 234–265 [DOI] [PubMed] [Google Scholar]
- 17. Vázquez-Manrique R. P., González-Cabo P., Ros S., Aziz H., Baylis H. A., Palau F. (2006) FASEB J. 20, 172–174 [DOI] [PubMed] [Google Scholar]
- 18. Claros M. G., Vincens P. (1996) Eur. J. Biochem. 241, 779–786 [DOI] [PubMed] [Google Scholar]
- 19. Rose A. M., Baillie D. L., Curran J. (1984) Mol. Gen. Genet. 195, 52–56 [DOI] [PubMed] [Google Scholar]
- 20. Baumeister R., Schaffitzel E., Hertweck M. (2006) J. Endocrinol. 190, 191–202 [DOI] [PubMed] [Google Scholar]
- 21. Chisholm A. D., Hardin J. (2005) in WormBook (Priess J. R., Seydoux G. eds) pp. 1–22, doi/ 10.1895/wormbook.1.35.1 [DOI] [Google Scholar]
- 22. Köppen M., Simske J. S., Sims P. A., Firestein B. L., Hall D. H., Radice A. D., Rongo C., Hardin J. D. (2001) Nat. Cell Biol. 3, 983–991 [DOI] [PubMed] [Google Scholar]
- 23. Ellis H. M., Horvitz H. R. (1986) Cell 44, 817–829 [DOI] [PubMed] [Google Scholar]
- 24. Kinchen J. M., Cabello J., Klingele D., Wong K., Feichtinger R., Schnabel H., Schnabel R., Hengartner M. O. (2005) Nature 434, 93–99 [DOI] [PubMed] [Google Scholar]
- 25. Campuzano V., Montermini L., Moltò M. D., Pianese L., Cossée M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., Zara F., Cañizares J., Koutnikova H., Bidichandani S. I., Gellera C., Brice A., Trouillas P., De Michele G., Filla A., De Frutos R., Palau F., Patel P. I., Di Donato S., Mandel J. L., Cocozza S., Koenig M., Pandolfo M. (1996) Science 271, 1423–1427 [DOI] [PubMed] [Google Scholar]
- 26. Stemmler T. L., Lesuisse E., Pain D., Dancis A. (2010) J. Biol. Chem. 285, 26737–26743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. González-Cabo P., Llorens J. V., Palau F., Moltó M. D. (2009) Adv. Exp. Med. Biol. 652, 247–261 [DOI] [PubMed] [Google Scholar]
- 28. Lesuisse E., Santos R., Matzanke B. F., Knight S. A., Camadro J. M., Dancis A. (2003) Hum. Mol. Genet. 12, 879–889 [DOI] [PubMed] [Google Scholar]
- 29. Ishii N., Takahashi K., Tomita S., Keino T., Honda S., Yoshino K., Suzuki K. (1990) Mutat. Res. 237, 165–171 [DOI] [PubMed] [Google Scholar]
- 30. Yamamoto K., Honda S., Ishii N. (1996) Mutat. Res. 358, 1–6 [DOI] [PubMed] [Google Scholar]
- 31. Essers M. A., de Vries-Smits L. M., Barker N., Polderman P. E., Burgering B. M., Korswagen H. C. (2005) Science 308, 1181–1184 [DOI] [PubMed] [Google Scholar]
- 32. Willcox B. J., Donlon T. A., He Q., Chen R., Grove J. S., Yano K., Masaki K. H., Willcox D. C., Rodriguez B., Curb J. D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13987–13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Henderson S. T., Johnson T. E. (2001) Curr. Biol. 11, 1975–1980 [DOI] [PubMed] [Google Scholar]
- 34. Tsang W. Y., Lemire B. D. (2003) Biochim. Biophys. Acta 1638, 91–105 [DOI] [PubMed] [Google Scholar]
- 35. Baylis H. A. (2005) Cell 123, 5–7 [DOI] [PubMed] [Google Scholar]
- 36. Lill R., Mühlenhoff U. (2006) Annu. Rev. Cell Dev. Biol. 22, 457–486 [DOI] [PubMed] [Google Scholar]
- 37. Rouault T. A., Tong W. H. (2005) Nat. Rev. Mol. Cell Biol. 6, 345–351 [DOI] [PubMed] [Google Scholar]
- 38. Zeviani M., Carelli V. (2003) Curr. Opin. Neurol. 16, 585–594 [DOI] [PubMed] [Google Scholar]
- 39. Zeviani M., Carelli V. (2007) Curr. Opin. Neurol. 20, 564–571 [DOI] [PubMed] [Google Scholar]
- 40. Liu X., Kim C. N., Yang J., Jemmerson R., Wang X. (1996) Cell 86, 147–157 [DOI] [PubMed] [Google Scholar]
- 41. Murray A., Hunt T. (1993) The Cell Cycle, pp. 42–65, Oxford University Press, Oxford [Google Scholar]
- 42. Mandal S., Guptan P., Owusu-Ansah E., Banerjee U. (2005) Dev. Cell 9, 843–854 [DOI] [PubMed] [Google Scholar]
- 43. Ventura N., Rea S., Henderson S. T., Condo I., Johnson T. E., Testi R. (2005) Aging Cell 4, 109–112 [DOI] [PubMed] [Google Scholar]
- 44. Palau F. (2001) Int. J. Mol. Med. 7, 581–589 [DOI] [PubMed] [Google Scholar]
- 45. Wilson R. B. (2006) Semin. Pediatr. Neurol. 13, 166–175 [DOI] [PubMed] [Google Scholar]
- 46. Rouault T. A., Tong W. H. (2008) Trends Genet. 24, 398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miao R., Kim H., Koppolu U. M., Ellis E. A., Scott R. A., Lindahl P. A. (2009) Biochemistry 48, 9556–9568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miao R., Martinho M., Morales J. G., Kim H., Ellis E. A., Lill R., Hendrich M. P., Münck E., Lindahl P. A. (2008) Biochemistry 47, 9888–9899 [DOI] [PubMed] [Google Scholar]
- 49. Kharade S. V., Mittal N., Das S. P., Sinha P., Roy N. (2005) FEBS Lett. 579, 6809–6813 [DOI] [PubMed] [Google Scholar]
- 50. Cypser J. R., Johnson T. E. (2002) J. Gerontol. A Biol. Sci. Med. Sci. 57, B109–114 [DOI] [PubMed] [Google Scholar]
- 51. Schulz T. J., Zarse K., Voigt A., Urban N., Birringer M., Ristow M. (2007) Cell Metab. 6, 280–293 [DOI] [PubMed] [Google Scholar]
- 52. Feng J., Bussière F., Hekimi S. (2001) Dev. Cell 1, 633–644 [DOI] [PubMed] [Google Scholar]
- 53. Hsu A. L., Murphy C. T., Kenyon C. (2003) Science 300, 1142–1145 [DOI] [PubMed] [Google Scholar]
- 54. Kenyon C. (2005) Cell 120, 449–460 [DOI] [PubMed] [Google Scholar]
- 55. Lehner B., Crombie C., Tischler J., Fortunato A., Fraser A. G. (2006) Nat. Genet. 38, 896–903 [DOI] [PubMed] [Google Scholar]
- 56. Ségalat L. (2006) ACS Chem. Biol. 1, 277–278 [DOI] [PubMed] [Google Scholar]
- 57. Sulston J. E., Schierenberg E., White J. G., Thomson J. N. (1983) Dev. Biol. 100, 64–119 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.